The Concept of Cancer Stem Cells: Elaborating on ALDH1B1 as an Emerging Marker of Cancer Progression
Abstract
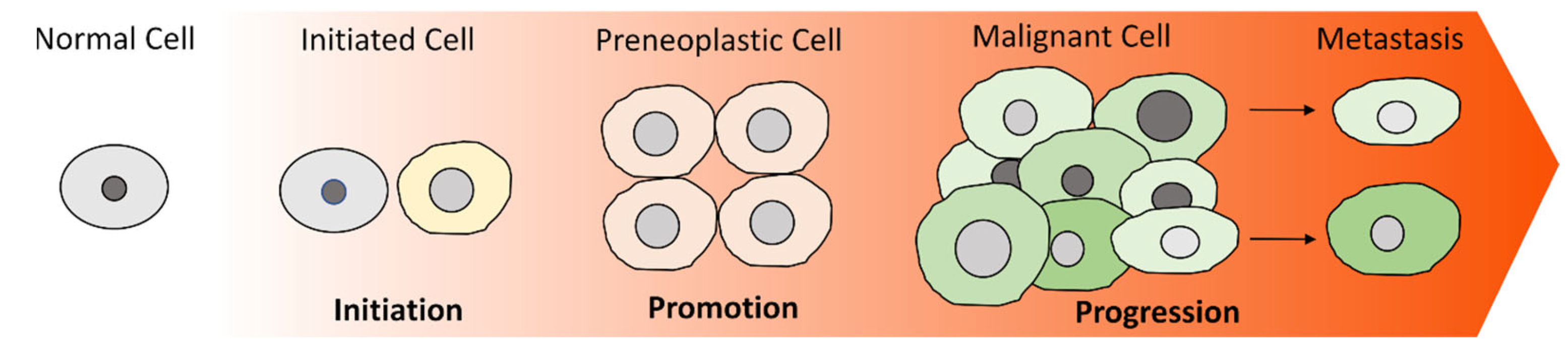
1. Introduction
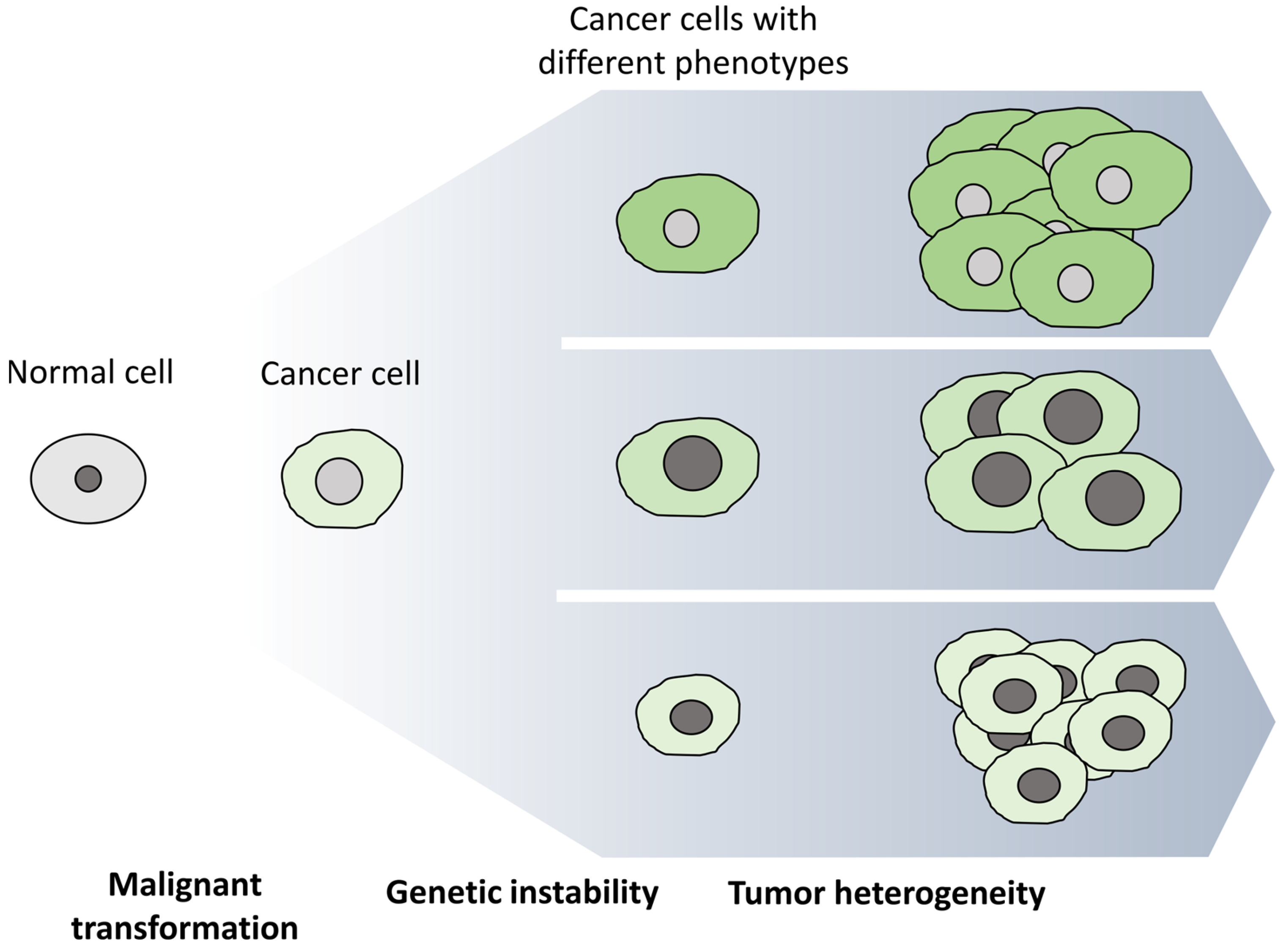
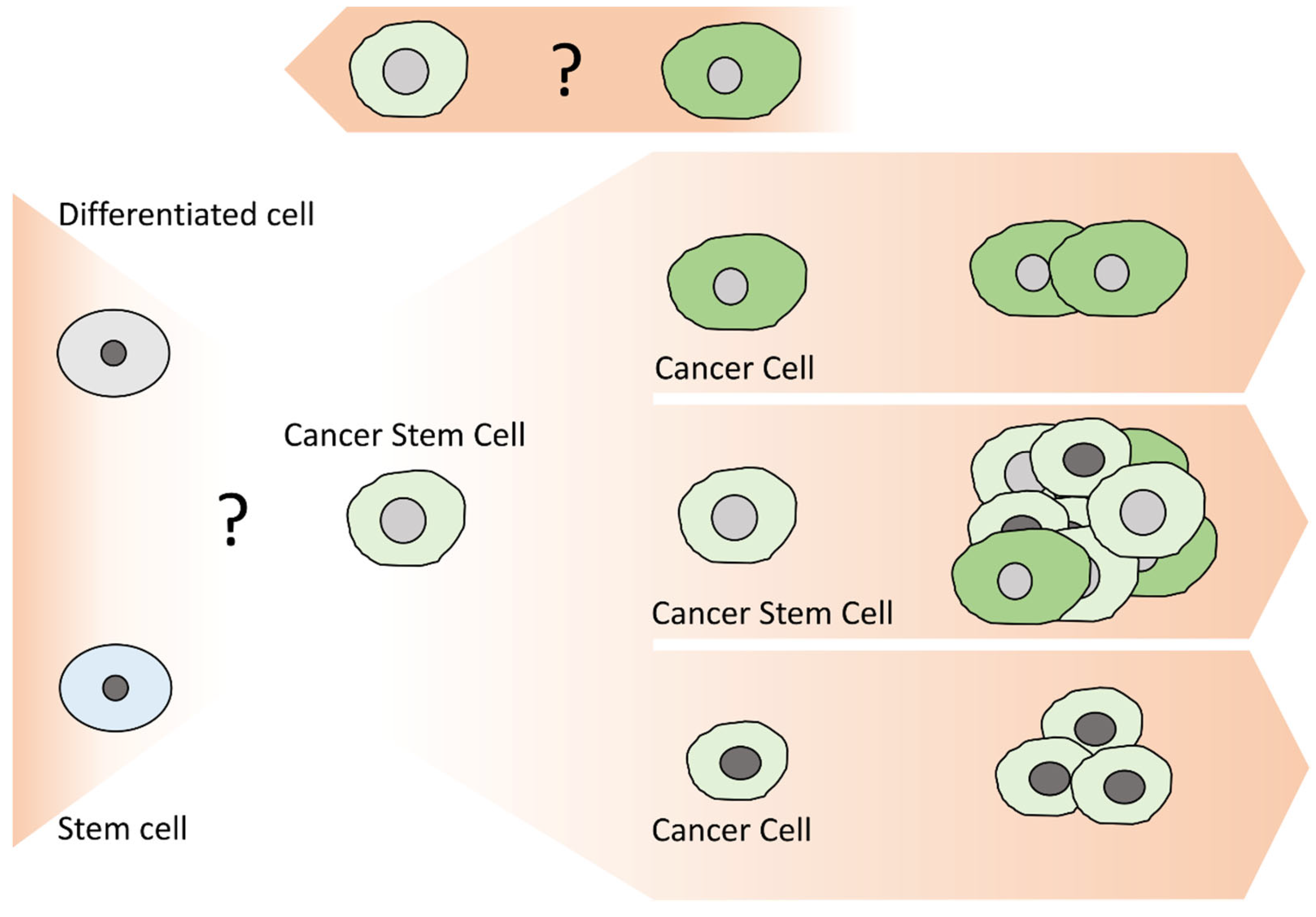
2. Cancer Stem Cells (CSCs)
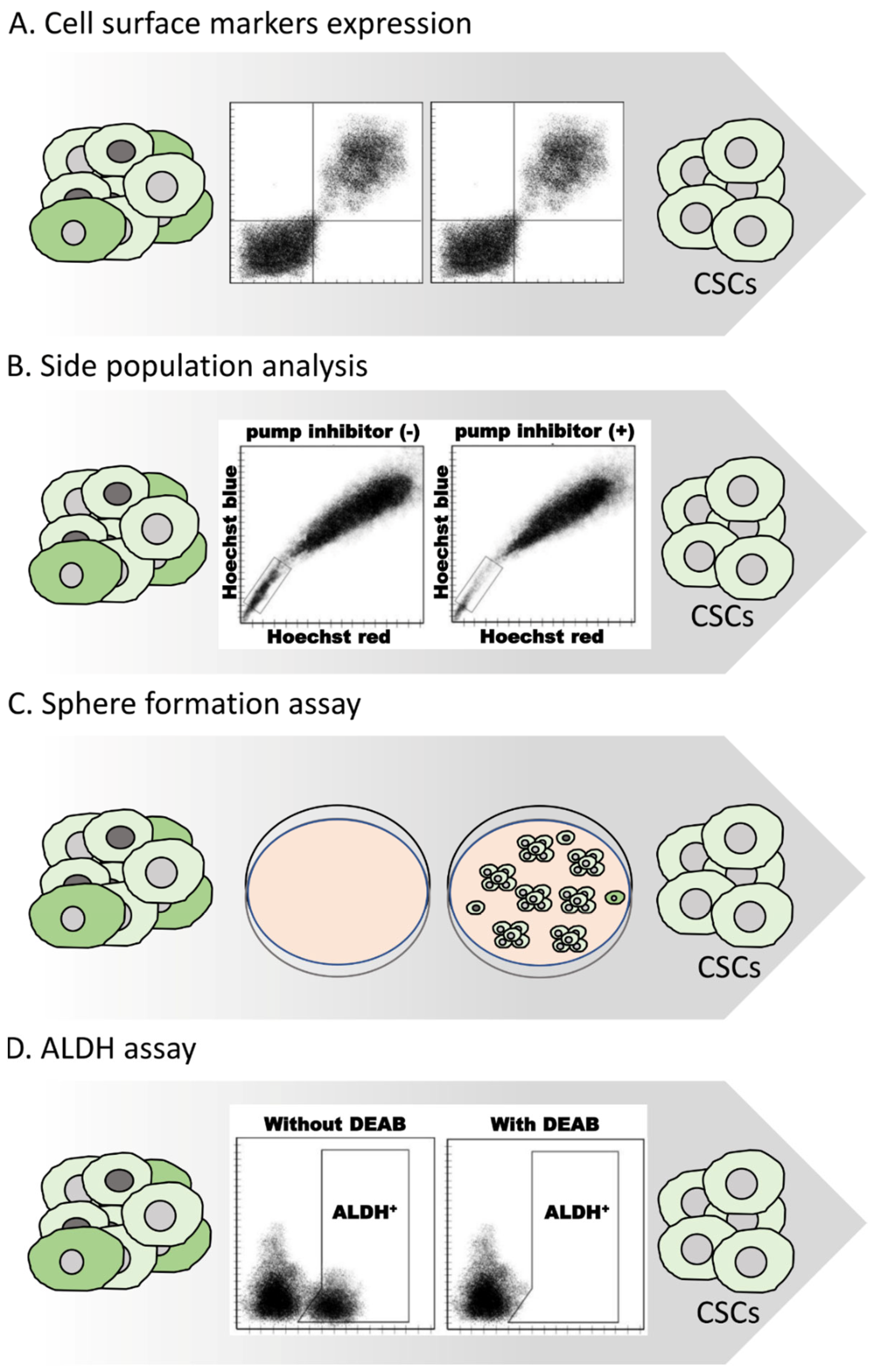
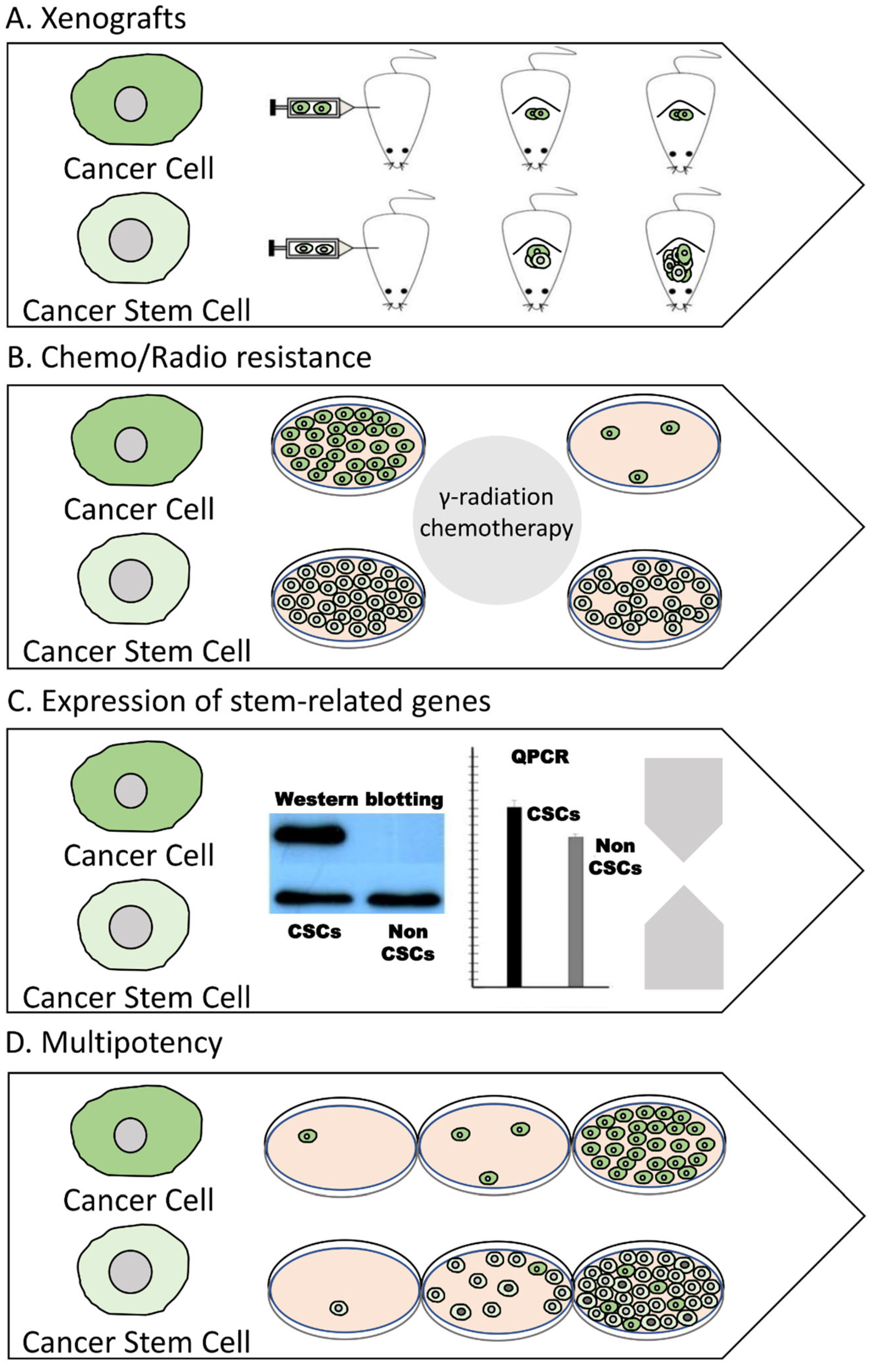
2.1. Methods for Isolating CSCs
2.2. Methods for Characterizing CSCs
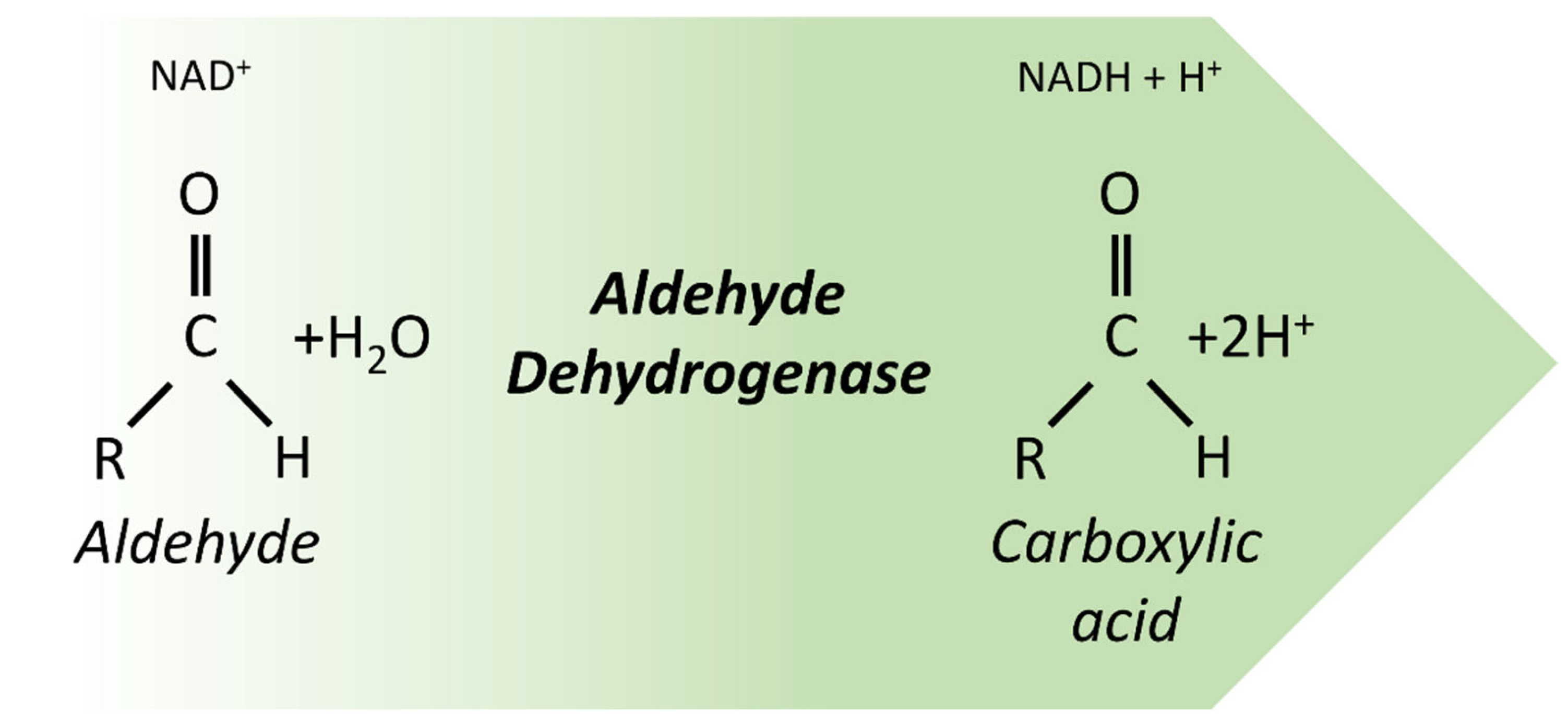
3. Aldehyde Dehydrogenase 1B1
3.1. Aldehyde Dehydrogenases
3.2. Historical Overview of ALDH1B1 Discovery and Research
4. Current Knowledge on the Association of ALDH1B1 with Cancer Progression and CSC Phenotype
4.1. Colorectal Cancer
4.2. Pancreatic Cancer
4.3. Other Types of Cancer
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhu, W.; Thompson, P.; Hannun, Y.A. Evaluating intrinsic and non-intrinsic cancer risk factors. Nat. Commun. 2018, 9, 3490. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Horn, G.; Moulton, K.; Oza, A.; Byler, S.; Kokolus, S.; Longacre, M. Cancer development, progression, and therapy: An epigenetic overview. Int. J. Mol. Sci. 2013, 14, 21087–21113. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Burgio, E.; Migliore, L. Towards a systemic paradigm in carcinogenesis: Linking epigenetics and genetics. Mol. Biol. Rep. 2015, 42, 777–790. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Morris, L.G.T.; Chan, T.A. Therapeutic targeting of tumor suppressor genes. Cancer 2015, 121, 1357–1368. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Wang, Z. Oxidative stress in carcinogenesis. Curr. Opin. Toxicol. 2018, 7, 116–121. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T. Cancer Epigenetics: From Disruption of Differentiation Programs to the Emergence of Cancer Stem Cells. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 251. [Google Scholar] [CrossRef]
- López-Lázaro, M. The stem cell division theory of cancer. Crit. Rev. Oncol. Hematol. 2018, 123, 95–113. [Google Scholar] [CrossRef]
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 2018, 18, 669–680. [Google Scholar] [CrossRef]
- Shlush, L.I.; Zandi, S.; Mitchell, A.; Chen, W.C.; Brandwein, J.M.; Gupta, V.; Kennedy, J.A.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Identification of pre-leukemic hematopoietic stem cells in acute leukemia. Nature 2014, 506, 328. [Google Scholar] [CrossRef]
- Blaas, L.; Pucci, F.; Messal, H.A.; Andersson, A.B.; Ruiz, E.J.; Gerling, M.; Douagi, I.; Spencer-Dene, B.; Musch, A.; Mitter, R.; et al. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat. Cell Biol. 2016, 18, 1346. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Figueroa, M.E.; Sinha, A.U.; Stubbs, M.C.; Feng, Z.; Valk, P.J.M.; Delwel, R.; Döhner, K.; Bullinger, L.; Kung, A.L.; et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia 2013, 27, 852–860. [Google Scholar] [CrossRef]
- Lenzi, M.; Fimognari, C.; Hrelia, P.; Fimognari, C.; Lenzi, M.; Hrelia, Á.P. Sulforaphane as a Promising Molecule for Fighting Cancer. Cancer Treat. Res. 2014, 159, 207–223. [Google Scholar] [CrossRef]
- Baxter, E.; Windloch, K.; Gannon, F.; Lee, J.S. Epigenetic regulation in cancer progression. Cell Biosci. 2014, 4, 45. [Google Scholar] [CrossRef]
- Gilson, P.; Merlin, J.L.; Harlé, A. Deciphering Tumour Heterogeneity: From Tissue to Liquid Biopsy. Cancers 2022, 14, 1384. [Google Scholar] [CrossRef]
- Wang, T.; Narayanaswamy, R.; Ren, H.; Torchilin, V.P. Combination therapy targeting both cancer stem-like cells and bulk tumor cells for improved efficacy of breast cancer treatment. Cancer Biol. Ther. 2016, 17, 698. [Google Scholar] [CrossRef]
- Stanta, G.; Bonin, S. Overview on clinical relevance of intra-tumor heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef]
- Schulte, L.A.; López-Gil, J.C.; Sainz, B.; Hermann, P.C. The Cancer Stem Cell in Hepatocellular Carcinoma. Cancers 2020, 12, 684. [Google Scholar] [CrossRef] [PubMed]
- Van Niekerk, G.; Davids, L.M.; Hattingh, S.M.; Engelbrecht, A.M. Cancer stem cells: A product of clonal evolution? Int. J. Cancer 2017, 140, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Hu, Z.; Curtis, C. Big Bang Tumor Growth and Clonal Evolution. Cold Spring Harb. Perspect. Med. 2018, 8, a028381. [Google Scholar] [CrossRef] [PubMed]
- Bielecka, Z.F.; Maliszewska-Olejniczak, K.; Safir, I.J.; Szczylik, C.; Czarnecka, A.M. Three-dimensional cell culture model utilization in cancer stem cell research. Biol. Rev. Camb. Philos. Soc. 2017, 92, 1505–1520. [Google Scholar] [CrossRef] [PubMed]
- Afify, S.M.; Seno, M. Conversion of stem cells to cancer stem cells: Undercurrent of cancer initiation. Cancers 2019, 11, 345. [Google Scholar] [CrossRef]
- Shackleton, M.; Quintana, E.; Fearon, E.R.; Morrison, S.J. Heterogeneity in Cancer: Cancer Stem Cells versus Clonal Evolution. Cell 2009, 138, 822–829. [Google Scholar] [CrossRef]
- Dick, J.E. Stem cell concepts renew cancer research. Blood 2008, 112, 4793–4807. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Alison, M.R.; Lim, S.M.; Nicholson, L.J. Cancer stem cells: Problems for therapy? J. Pathol. 2011, 223, 148–162. [Google Scholar] [CrossRef]
- Prabavathy, D.; Swarnalatha, Y.; Ramadoss, N. Lung cancer stem cells-origin, characteristics and therapy. Stem Cell Investig. 2018, 5, 6. [Google Scholar] [CrossRef]
- Desai, A.; Yan, Y.; Gerson, S.L. Concise Reviews: Cancer Stem Cell Targeted Therapies: Toward Clinical Success. Stem Cells Transl. Med. 2019, 8, 75–81. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer stem cells (CSCs) in cancer progression and therapy. J. Cell. Physiol. 2019, 234, 8381–8395. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef]
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells—Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef]
- Gimple, R.C.; Yang, K.; Halbert, M.E.; Agnihotri, S.; Rich, J.N. Brain cancer stem cells: Resilience through adaptive plasticity and hierarchical heterogeneity. Nat. Rev. Cancer 2022, 22, 497–514. [Google Scholar] [CrossRef]
- Thankamony, A.P.; Saxena, K.; Murali, R.; Jolly, M.K.; Nair, R. Cancer Stem Cell Plasticity—A Deadly Deal. Front. Mol. Biosci. 2020, 7, 79. [Google Scholar] [CrossRef]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef]
- Biddle, A.; Gammon, L.; Liang, X.; Costea, D.E.; Mackenzie, I.C. Phenotypic Plasticity Determines Cancer Stem Cell Therapeutic Resistance in Oral Squamous Cell Carcinoma. eBioMedicine 2016, 4, 138–145. [Google Scholar] [CrossRef]
- Palomeras, S.; Ruiz-Martínez, S.; Puig, T. Targeting breast cancer stem cells to overcome treatment resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kuşoğlu, A.; Biray Avcı, Ç. Cancer stem cells: A brief review of the current status. Gene 2019, 681, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W.K. Aldehyde dehydrogenase its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011, 10, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.T.; Ryu, C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017, 50, 285–298. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, H.Y.; Wang, D.X.; Zeng, Q.; Fan, Z.; Xi, J.F.; Nan, X.; He, L.J.; Zhou, J.N.; Pei, X.T.; et al. A new protocol for long-term culture of a specific subpopulation of liver cancer stem cells enriched by cell surface markers. FEBS Open Bio 2020, 10, 1737–1747. [Google Scholar] [CrossRef]
- Tirino, V.; Desiderio, V.; Paino, F.; De Rosa, A.; Papaccio, F.; La Noce, M.; Laino, L.; De Francesco, F.; Papaccio, G. Cancer stem cells in solid tumors: An overview and new approaches for their isolation and characterization. FASEB J. 2013, 27, 13–24. [Google Scholar] [CrossRef]
- Liu, L.; Borlak, J. Advances in Liver Cancer Stem Cell Isolation and their Characterization. Stem Cell Rev. Rep. 2021, 17, 1215–1238. [Google Scholar] [CrossRef]
- Salinas-Jazmín, N.; Rosas-Cruz, A.; Velasco-Velazquez, M. Reporter gene systems for the identification and characterization of cancer stem cells. World J. Stem Cells 2021, 13, 861. [Google Scholar] [CrossRef]
- Lv, D.; Ma, Q.H.; Duan, J.J.; Wu, H.B.; Zhao, X.L.; Yu, S.C.; Bian, X.W. Optimized dissociation protocol for isolating human glioma stem cells from tumorspheres via fluorescence-activated cell sorting. Cancer Lett. 2016, 377, 105–115. [Google Scholar] [CrossRef]
- Hashimoto, O.; Shimizu, K.; Semba, S.; Chiba, S.; Ku, Y.; Yokozaki, H.; Hori, Y. Hypoxia induces tumor aggressiveness and the expansion of CD133-positive cells in a hypoxia-inducible factor-1α-dependent manner in pancreatic cancer cells. Pathobiology 2011, 78, 181–192. [Google Scholar] [CrossRef]
- Wang, N.; Wang, S.; Li, M.Y.; Hu, B.G.; Liu, L.P.; Yang, S.L.; Yang, S.; Gong, Z.; Lai, P.B.S.; Chen, G.G. Cancer stem cells in hepatocellular carcinoma: An overview and promising therapeutic strategies. Ther. Adv. Med. Oncol. 2018, 10, 1–25. [Google Scholar] [CrossRef]
- Satar, N.A.; Fakiruddin, K.S.; Lim, M.N.; Mok, P.L.; Zakaria, N.; Fakharuzi, N.A.; Abd Rahman, A.Z.; Zakaria, Z.; Yahaya, B.H.; Baharuddin, P. Novel triple-positive markers identified in human non-small cell lung cancer cell line with chemotherapy-resistant and putative cancer stem cell characteristics. Oncol. Rep. 2018, 40, 669–681. [Google Scholar] [CrossRef]
- Elkashty, O.A.; Elghanam, G.A.; Su, X.; Liu, Y.; Chauvin, P.J.; Tran, S.D. Cancer stem cells enrichment with surface markers CD271 and CD44 in human head and neck squamous cell carcinomas. Carcinogenesis 2020, 41, 458–466. [Google Scholar] [CrossRef]
- Golebiewska, A.; Brons, N.H.C.; Bjerkvig, R.; Niclou, S.P. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell 2011, 8, 136–147. [Google Scholar] [CrossRef]
- Unno, K.; Jain, M.; Liao, R. Cardiac Side Population Cells: Moving toward the Center Stage in Cardiac Regeneration. Circ. Res. 2012, 110, 1355. [Google Scholar] [CrossRef]
- Roy, L.; Dahl, K.D.C. Can Stemness and Chemoresistance Be Therapeutically Targeted via Signaling Pathways in Ovarian Cancer? Cancers 2018, 10, 241. [Google Scholar] [CrossRef]
- Yin, W.; Xiang, D.; Wang, T.; Zhang, Y.; Pham, C.V.; Zhou, S.; Jiang, G.; Hou, Y.; Zhu, Y.; Han, Y.; et al. The inhibition of ABCB1/MDR1 or ABCG2/BCRP enables doxorubicin to eliminate liver cancer stem cells. Sci. Rep. 2021, 11, 10791. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, X.; Chen, P.; Wu, X.; Duan, A.; Qin, Y. Combined effects of octreotide and cisplatin on the proliferation of side population cells from anaplastic thyroid cancer cell lines. Oncol. Lett. 2018, 16, 4033–4042. [Google Scholar] [CrossRef]
- Fultang, N.; Illendula, A.; Lin, J.; Pandey, M.K.; Klase, Z.; Peethambaran, B. ROR1 regulates chemoresistance in Breast Cancer via modulation of drug efflux pump ABCB1. Sci. Rep. 2020, 10, 1821. [Google Scholar] [CrossRef]
- Xie, T.; Mo, L.; Li, L.; Mao, N.; Li, D.; Liu, D.; Zuo, C.; Huang, D.; Pan, Q.; Yang, L.; et al. Identification of side population cells in human lung adenocarcinoma A549 cell line and elucidation of the underlying roles in lung cancer. Oncol. Lett. 2018, 15, 4900–4906. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wu, Z.; Bao, C.; Li, W.; He, H.; Sun, Y.; Chen, Z.; Zhang, H.; Ning, Z. Cancer stem cells in esophageal squamous cell cancer (review). Oncol. Lett. 2019, 18, 5022–5032. [Google Scholar] [CrossRef] [PubMed]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes Wide Open: A Critical Review of Sphere-Formation as an Assay For Stem Cells. Cell Stem Cell 2011, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, M.; Maroufi, N.F.; Tazehkand, A.P.; Akbarzadeh, M.; Bastani, S.; Safdari, R.; Farzane, A.; Fattahi, A.; Nejabati, H.R.; Nouri, M.; et al. Current approaches in identification and isolation of cancer stem cells. J. Cell. Physiol. 2019, 234, 14759–14772. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.; Chen, H.; Lo, P.-K. In vitro Tumorsphere Formation Assays. Bio-protocol 2013, 3, e325. [Google Scholar] [CrossRef]
- Ma, X.L.; Sun, Y.F.; Wang, B.L.; Shen, M.N.; Zhou, Y.; Chen, J.W.; Hu, B.; Gong, Z.J.; Zhang, X.; Cao, Y.; et al. Sphere-forming culture enriches liver cancer stem cells and reveals Stearoyl-CoA desaturase 1 as a potential therapeutic target. BMC Cancer 2019, 19, 760. [Google Scholar] [CrossRef]
- Su, P.; Yang, Y.; Wang, G.; Chen, X.; Ju, Y. Curcumin attenuates resistance to irinotecan via induction of apoptosis of cancer stem cells in chemoresistant colon cancer cells. Int. J. Oncol. 2018, 53, 1343–1353. [Google Scholar] [CrossRef]
- Zhang, X.; Powell, K.; Li, L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers 2020, 12, 3765. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Cheaito, K.; Chalhoub, R.M.; Hadadeh, O.; Monzer, A.; Ballout, F.; El-Hajj, A.; Mukherji, D.; Liu, Y.N.; Daoud, G.; et al. Sphere-Formation Assay: Three-Dimensional in vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front. Oncol. 2018, 8, 347. [Google Scholar] [CrossRef]
- Opdenaker, L.M.; Modara, S.R.; Boman, B.M. The Proportion of ALDEFLUOR-Positive Cancer Stem Cells Changes with Cell Culture Density Due to the Expression of Different ALDH Isoforms. Cancer Stud. Mol. Med. Open J. 2015, 2, 87–95. [Google Scholar] [CrossRef]
- Mele, L.; Liccardo, D.; Tirino, V. Evaluation and Isolation of Cancer Stem Cells Using ALDH Activity Assay. Methods Mol. Biol. 2018, 1692, 43–48. [Google Scholar] [CrossRef]
- Pors, K.; Moreb, J.S. Aldehyde dehydrogenases in cancer: An opportunity for biomarker and drug development? Drug Discov. Today 2014, 19, 1953–1963. [Google Scholar] [CrossRef]
- Morgan, C.A.; Parajuli, B.; Buchman, C.D.; Dria, K.; Hurley, T.D. N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem. Biol. Interact. 2015, 234, 18. [Google Scholar] [CrossRef]
- Zhou, L.; Sheng, D.; Wang, D.; Ma, W.; Deng, Q.; Deng, L.; Liu, S. Identification of cancer-type specific expression patterns for active aldehyde dehydrogenase (ALDH) isoforms in ALDEFLUOR assay. Cell Biol. Toxicol. 2019, 35, 161–177. [Google Scholar] [CrossRef]
- Garaycoechea, J.I.; Crossan, G.P.; Langevin, F.; Daly, M.; Arends, M.J.; Patel, K.J. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 2012, 489, 571–575. [Google Scholar] [CrossRef]
- Moreb, J.S.; Ucar, D.; Han, S.; Amory, J.K.; Goldstein, A.S.; Ostmark, B.; Chang, L.J. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem. Biol. Interact. 2012, 195, 52–60. [Google Scholar] [CrossRef]
- Storms, R.W.; Trujillo, A.P.; Springer, J.B.; Shah, L.; Colvin, O.M.; Ludeman, S.M.; Smith, C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9118–9123. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Fulawka, L.; Donizy, P.; Halon, A. Cancer stem cells—The current status of an old concept: Literature review and clinical approaches. Biol. Res. 2014, 47, 66. [Google Scholar] [CrossRef]
- Kuwata, T.; Yanagihara, K.; Iino, Y.; Komatsu, T.; Ochiai, A.; Sekine, S.; Taniguchi, H.; Katai, H.; Kinoshita, T.; Ohtsu, A. Establishment of Novel Gastric Cancer Patient-Derived Xenografts and Cell Lines: Pathological Comparison between Primary Tumor, Patient-Derived, and Cell-Line Derived Xenografts. Cells 2019, 8, 585. [Google Scholar] [CrossRef]
- Lai, Y.; Wei, X.; Lin, S.; Qin, L.; Cheng, L.; Li, P. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 2017, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Keysar, S.B.; Eagles, J.R.; Miller, B.; Jackson, B.C.; Chowdhury, F.N.; Reisinger, J.; Chimed, T.S.; Le, P.N.; Morton, J.J.; Somerset, H.L.; et al. Salivary Gland Cancer Patient-Derived Xenografts Enable Characterization of Cancer Stem Cells and New Gene Events Associated with Tumor Progression. Clin. Cancer Res. 2018, 24, 2935–2943. [Google Scholar] [CrossRef] [PubMed]
- Giraud, J.; Bouriez, D.; Seeneevassen, L.; Rousseau, B.; Sifré, E.; Giese, A.; Mégraud, F.; Lehours, P.; Dubus, P.; Gronnier, C.; et al. Orthotopic Patient-Derived Xenografts of Gastric Cancer to Decipher Drugs Effects on Cancer Stem Cells and Metastatic Dissemination. Cancers 2019, 11, 560. [Google Scholar] [CrossRef] [PubMed]
- Sargent, J.K.; Warner, M.A.; Low, B.E.; Schott, W.H.; Hoffert, T.; Coleman, D.; Woo, X.Y.; Sheridan, T.; Erattupuzha, S.; Henrich, P.P.; et al. Genetically diverse mouse platform to xenograft cancer cells. Dis. Model. Mech. 2022, 15, dmm049457. [Google Scholar] [CrossRef] [PubMed]
- Rycaj, K.; Tang, D.G. Cell-of-Origin of Cancer versus Cancer Stem Cells: Assays and Interpretations. Cancer Res. 2015, 75, 4003–4011. [Google Scholar] [CrossRef]
- Schulenburg, A.; Blatt, K.; Cerny-Reiterer, S.; Sadovnik, I.; Herrmann, H.; Marian, B.; Grunt, T.W.; Zielinski, C.C.; Valent, P. Cancer stem cells in basic science and in translational oncology: Can we translate into clinical application? J. Hematol. Oncol. 2015, 8, 16. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497. [Google Scholar] [CrossRef]
- Nunes, T.; Hamdan, D.; Leboeuf, C.; El Bouchtaoui, M.; Gapihan, G.; Nguyen, T.T.; Meles, S.; Angeli, E.; Ratajczak, P.; Lu, H.; et al. Targeting Cancer Stem Cells to Overcome Chemoresistance. Int. J. Mol. Sci. 2018, 19, 4036. [Google Scholar] [CrossRef]
- Bousquet, G.; El Bouchtaoui, M.; Sophie, T.; Leboeuf, C.; De Bazelaire, C.; Ratajczak, P.; Giacchetti, S.; De Roquancourt, A.; Bertheau, P.; Verneuil, L.; et al. Targeting autophagic cancer stem-cells to reverse chemoresistance in human triple negative breast cancer. Oncotarget 2017, 8, 35205. [Google Scholar] [CrossRef]
- Kathawala, R.J.; Gupta, P.; Ashby, C.R.; Chen, Z.S. The modulation of ABC transporter-mediated multidrug resistance in cancer: A review of the past decade. Drug Resist. Updates 2015, 18, 1–17. [Google Scholar] [CrossRef]
- Yuan, M.; Eberhart, C.G.; Kai, M. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget 2014, 5, 2820–2826. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Tuy, K.; Rickenbacker, L.; Hjelmeland, A.B. Reactive oxygen species produced by altered tumor metabolism impacts cancer stem cell maintenance. Redox Biol. 2021, 44, 101953. [Google Scholar] [CrossRef]
- Arnold, C.R.; Mangesius, J.; Skvortsova, I.I.; Ganswindt, U. The Role of Cancer Stem Cells in Radiation Resistance. Front. Oncol. 2020, 10, 164. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, M.H.; Kim, K.S.; Park, M.J.; Jeong, J.H.; Park, S.W.; Ji, Y.H.; Kim, K.I.; Lee, T.S.; Ryu, P.Y.; et al. In vivo monitoring of CD44+ cancer stem-like cells by γ-irradiation in breast cancer. Int. J. Oncol. 2016, 48, 2277–2286. [Google Scholar] [CrossRef]
- Ghisolfi, L.; Keates, A.C.; Hu, X.; Lee, D.K.; Li, C.J. Ionizing radiation induces stemness in cancer cells. PLoS ONE 2012, 7, e43628. [Google Scholar] [CrossRef]
- Reynolds, D.S.; Tevis, K.M.; Blessing, W.A.; Colson, Y.L.; Zaman, M.H.; Grinstaff, M.W. Breast Cancer Spheroids Reveal a Differential Cancer Stem Cell Response to Chemotherapeutic Treatment. Sci. Rep. 2017, 7, 10382. [Google Scholar] [CrossRef]
- Zhao, Z.L.; Zhang, L.; Huang, C.F.; Ma, S.R.; Bu, L.L.; Liu, J.F.; Yu, G.T.; Liu, B.; Gutkind, J.S.; Kulkarni, A.B.; et al. NOTCH1 inhibition enhances the efficacy of conventional chemotherapeutic agents by targeting head neck cancer stem cell. Sci. Rep. 2016, 6, 24704. [Google Scholar] [CrossRef]
- Kida, K.; Ishikawa, T.; Yamada, A.; Shimada, K.; Narui, K.; Sugae, S.; Shimizu, D.; Tanabe, M.; Sasaki, T.; Ichikawa, Y.; et al. Effect of ALDH1 on prognosis and chemoresistance by breast cancer subtype. Breast Cancer Res. Treat. 2016, 156, 261–269. [Google Scholar] [CrossRef]
- Li, J.; Zhang, B.; Yang, Y.F.; Jin, J.; Liu, Y.H. Aldehyde dehydrogenase 1 as a predictor of the neoadjuvant chemotherapy response in breast cancer: A meta-analysis. Medicine (Baltimore) 2018, 97, e12056. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, M.; Si, J.; Xiong, Y.; Lu, F.; Zhang, J.; Zhang, L.; Zhang, P.; Yang, Y. Blockade of Notch3 inhibits the stem-like property and is associated with ALDH1A1 and CD44 via autophagy in non-small lung cancer. Int. J. Oncol. 2016, 48, 2349–2358. [Google Scholar] [CrossRef] [PubMed]
- Awad, O.; Yustein, J.T.; Shah, P.; Gul, N.; Katuri, V.; O’Neill, A.; Kong, Y.; Brown, M.L.; Toretsky, J.A.; Loeb, D.M. High ALDH activity identifies chemotherapy-resistant Ewing’s sarcoma stem cells that retain sensitivity to EWS-Fli1 inhibition. PLoS ONE 2010, 5, e13943. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.; Ray, P. Cisplatin triggers cancer stem cell enrichment in platinum-resistant cells through NF-κB-TNFα-PIK3CA loop. J. Exp. Clin. Cancer Res. 2017, 36, 164. [Google Scholar] [CrossRef]
- Wang, Y.C.; Yo, Y.T.; Lee, H.Y.; Liao, Y.P.; Chao, T.K.; Su, P.H.; Lai, H.C. ALDH1-bright epithelial ovarian cancer cells are associated with CD44 expression, drug resistance, and poor clinical outcome. Am. J. Pathol. 2012, 180, 1159–1169. [Google Scholar] [CrossRef]
- Kozovska, Z.; Patsalias, A.; Bajzik, V.; Durinikova, E.; Demkova, L.; Jargasova, S.; Smolkova, B.; Plava, J.; Kucerova, L.; Matuskova, M. ALDH1A inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer 2018, 18, 656. [Google Scholar] [CrossRef]
- Liu, K.; Ou, J.H.J. Regulators of liver cancer stem cells. World J. Stem Cells 2021, 13, 1127. [Google Scholar] [CrossRef]
- Mohan, A.; Raj Rajan, R.; Mohan, G.; Kollenchery Puthenveettil, P.; Maliekal, T.T. Markers and Reporters to Reveal the Hierarchy in Heterogeneous Cancer Stem Cells. Front. Cell Dev. Biol. 2021, 9, 668851. [Google Scholar] [CrossRef]
- Bley, N.; Hmedat, A.; Müller, S.; Rolnik, R.; Rausch, A.; Lederer, M.; Hüttelmaier, S. Musashi-1—A stemness RBP for cancer therapy? Biology 2021, 10, 407. [Google Scholar] [CrossRef]
- Che, N.; Yang, Z.; Liu, X.; Li, M.; Feng, Y.; Zhang, C.; Li, C.; Cui, Y.; Xuan, Y. Suppression of LETM1 inhibits the proliferation and stemness of colorectal cancer cells through reactive oxygen species-induced autophagy. J. Cell. Mol. Med. 2021, 25, 2110–2120. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem Cells 2015, 7, 1150–1184. [Google Scholar] [CrossRef]
- Vasefifar, P.; Motafakkerazad, R.; Maleki, L.A.; Najafi, S.; Ghrobaninezhad, F.; Najafzadeh, B.; Alemohammad, H.; Amini, M.; Baghbanzadeh, A.; Baradaran, B. Nanog, as a key cancer stem cell marker in tumor progression. Gene 2022, 827, 146448. [Google Scholar] [CrossRef]
- Wang, R.; Fan, H.; Sun, M.; Lv, Z.; Yi, W. Roles of BMI1 in the Initiation, Progression, and Treatment of Hepatocellular Carcinoma. Technol. Cancer Res. Treat. 2022, 21, 1–9. [Google Scholar] [CrossRef]
- Badarinath, K.; Dam, B.; Kataria, S.; Zirmire, R.K.; Dey, R.; Kansagara, G.; Ajnabi, J.; Hegde, A.; Singh, R.; Masudi, T.; et al. Snail maintains the stem/progenitor state of skin epithelial cells and carcinomas through the autocrine effect of matricellular protein Mindin. Cell Rep. 2022, 40, 111390. [Google Scholar] [CrossRef]
- Ihemelandu, C.; Naeem, A.; Parasido, E.; Berry, D.; Chaldekas, K.; Harris, B.T.; Rodriguez, O.; Albanese, C. Clinicopathologic and prognostic significance of LGR5, a cancer stem cell marker in patients with colorectal cancer. Color. Cancer 2019, 8, CRC11. [Google Scholar] [CrossRef]
- Li, X.; Bu, W.; Meng, L.; Liu, X.; Wang, S.; Jiang, L.; Ren, M.; Fan, Y.; Sun, H. CXCL12/CXCR4 pathway orchestrates CSC-like properties by CAF recruited tumor associated macrophage in OSCC. Exp. Cell Res. 2019, 378, 131–138. [Google Scholar] [CrossRef]
- Zeng, Y.T.; Liu, X.F.; Yang, W.T.; Zheng, P.S. REX1 promotes EMT-induced cell metastasis by activating the JAK2/STAT3-signaling pathway by targeting SOCS1 in cervical cancer. Oncogene 2019, 38, 6940–6957. [Google Scholar] [CrossRef]
- Lemma, S.; Avnet, S.; Salerno, M.; Chano, T.; Baldini, N. Identification and Validation of Housekeeping Genes for Gene Expression Analysis of Cancer Stem Cells. PLoS ONE 2016, 11, e0149481. [Google Scholar] [CrossRef]
- Boumahdi, S.; Driessens, G.; Lapouge, G.; Rorive, S.; Nassar, D.; Le Mercier, M.; Delatte, B.; Caauwe, A.; Lenglez, S.; Nkusi, E.; et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014, 511, 246–250. [Google Scholar] [CrossRef]
- Wang, M.C.; Jiao, M.; Wu, T.; Jing, L.; Cui, J.; Guo, H.; Tian, T.; Ruan, Z.; Wei, Y.C.; Jiang, L.L.; et al. Polycomb complex protein BMI-1 promotes invasion and metastasis of pancreatic cancer stem cells by activating PI3K/AKT signaling, an ex vivo, in vitro, and in vivo study. Oncotarget 2016, 7, 9586–9599. [Google Scholar] [CrossRef]
- Es-haghi, M.; Soltanian, S.; Dehghani, H. Perspective: Cooperation of Nanog, NF-κΒ, and CXCR4 in a regulatory network for directed migration of cancer stem cells. Tumour Biol. 2016, 37, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Ota, I.; Masui, T.; Kurihara, M.; Yook, J.I.; Mikami, S.; Kimura, T.; Shimada, K.; Konishi, N.; Yane, K.; Yamanaka, T.; et al. Snail-induced EMT promotes cancer stem cell-like properties in head and neck cancer cells. Oncol. Rep. 2016, 35, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, H.; Sueoka, E.; Rawangkan, A.; Suganuma, M. Human cancer stem cells are a target for cancer prevention using (-)-epigallocatechin gallate. J. Cancer Res. Clin. Oncol. 2017, 143, 2401–2412. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, M.; Katsuda, T.; Yamada, Y.; Ochiya, T. In vitro reconstitution of breast cancer heterogeneity with multipotent cancer stem cells using small molecules. Biochem. Biophys. Res. Commun. 2017, 482, 750–757. [Google Scholar] [CrossRef] [PubMed]
- O’Brien-Ball, C.; Biddle, A. Reprogramming to developmental plasticity in cancer stem cells. Dev. Biol. 2017, 430, 266–274. [Google Scholar] [CrossRef]
- Jia, X.; Shen, G.; Jia, J.; Zhang, Y.; Zhang, D.; Li, W.; Zhang, J.; Huang, X.; Tian, J. Lineage Tracing and Molecular Real-Time Imaging of Cancer Stem Cells. Biosensors 2022, 12, 703. [Google Scholar] [CrossRef]
- Ueno, H. Identification of normal and neoplastic stem cells by the multicolor lineage tracing methods. Pathol. Int. 2016, 66, 423–430. [Google Scholar] [CrossRef]
- Qin, J.; Liu, X.; Laffin, B.; Chen, X.; Choy, G.; Jeter, C.R.; Calhoun-Davis, T.; Li, H.; Palapattu, G.S.; Pang, S.; et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell 2012, 10, 556–569. [Google Scholar] [CrossRef]
- Marchitti, S.A.; Brocker, C.; Stagos, D.; Vasiliou, V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 2008, 4, 697–720. [Google Scholar] [CrossRef]
- Jackson, B.; Brocker, C.; Thompson, D.C.; Black, W.; Vasiliou, K.; Nebert, D.W.; Vasiliou, V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genom. 2011, 5, 283–303. [Google Scholar] [CrossRef]
- Vasiliou, V.; Thompson, D.C.; Smith, C.; Fujita, M.; Chen, Y. Aldehyde dehydrogenases: From eye crystallins to metabolic disease and cancer stem cells. Chem.-Biol. Interact. 2013, 202, 2–10. [Google Scholar] [CrossRef]
- Charkoftaki, G.; Chen, Y.; Han, M.; Sandoval, M.; Yu, X.; Zhao, H.; Orlicky, D.J.; Thompson, D.C.; Vasiliou, V. Transcriptomic analysis and plasma metabolomics in Aldh16a1-null mice reveals a potential role of ALDH16A1 in renal function. Chem. Biol. Interact. 2017, 276, 15–22. [Google Scholar] [CrossRef]
- Chute, J.P.; Muramoto, G.G.; Whitesides, J.; Colvin, M.; Safi, R.; Chao, N.J.; McDonnell, D.P. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 11707. [Google Scholar] [CrossRef]
- Tsochantaridis, I.; Kontopoulos, A.; Voulgaridou, G.P.; Tsifintaris, M.; Triantafyllou, C.; Pappa, A. Aldehyde Dehydrogenase 1B1 Is Implicated in DNA Damage Response in Human Colorectal Adenocarcinoma. Cells 2022, 11, 2017. [Google Scholar] [CrossRef]
- Vassalli, G. Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef]
- Toledo-Guzmán, M.E.; Hernández, M.I.; Gómez-Gallegos, Á.A.; Ortiz-Sánchez, E. ALDH as a Stem Cell Marker in Solid Tumors. Curr. Stem Cell Res. Ther. 2018, 14, 375–388. [Google Scholar] [CrossRef]
- Rodriguez-Torres, M.; Allan, A.L. Aldehyde dehydrogenase as a marker and functional mediator of metastasis in solid tumors. Clin. Exp. Metastasis 2016, 33, 97–113. [Google Scholar] [CrossRef]
- Xu, X.; Chai, S.; Wang, P.; Zhang, C.; Yang, Y.; Yang, Y.; Wang, K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015, 369, 50–57. [Google Scholar] [CrossRef]
- Marcato, P.; Dean, C.A.; Liu, R.Z.; Coyle, K.M.; Bydoun, M.; Wallace, M.; Clements, D.; Turner, C.; Mathenge, E.G.; Gujar, S.A.; et al. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol. Oncol. 2015, 9, 17–31. [Google Scholar] [CrossRef]
- Marcato, P.; Dean, C.A.; Pan, D.; Araslanova, R.; Gillis, M.; Joshi, M.; Helyer, L.; Pan, L.; Leidal, A.; Gujar, S.; et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 2011, 29, 32–45. [Google Scholar] [CrossRef]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Seung, J.K.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef]
- Liu, C.; Qiang, J.; Deng, Q.; Xia, J.; Deng, L.; Zhou, L.; Wang, D.; He, X.; Liu, Y.; Zhao, B.; et al. ALDH1A1 Activity in Tumor-Initiating Cells Remodels Myeloid-Derived Suppressor Cells to Promote Breast Cancer Progression. Cancer Res. 2021, 81, 5919–5934. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Kwok, W.C.; Lee, T.K.W.; Kwan, H.T.; Wo, J.Y.H.; Zheng, B.J.; Guan, X.Y. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol. Cancer Res. 2008, 6, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, C.; Xu, H.; Gao, Y. Aldehyde Dehydrogenase, Liver Disease and Cancer. Int. J. Biol. Sci. 2020, 16, 921. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Arcaroli, J.; Chen, Y.; Thompson, D.C.; Messersmith, W.; Jimeno, A.; Vasiliou, V. ALDH1B1 Is Crucial for Colon Tumorigenesis by Modulating Wnt/β-Catenin, Notch and PI3K/Akt Signaling Pathways. PLoS ONE 2015, 10, e0121648. [Google Scholar] [CrossRef]
- Tsochantaridis, I.; Roupas, A.; Voulgaridou, G.-P.; Giatromanolaki, A.; Koukourakis, M.I.; Panayiotidis, M.I.; Pappa, A. Aldehyde Dehydrogenase 1B1 Is Associated with Altered Cell Morphology, Proliferation, Migration and Chemosensitivity in Human Colorectal Adenocarcinoma Cells. Biomedicines 2021, 9, 44. [Google Scholar] [CrossRef]
- Feng, H.; Liu, Y.; Bian, X.; Zhou, F.; Liu, Y. ALDH1A3 affects colon cancer in vitro proliferation and invasion depending on CXCR4 status. Br. J. Cancer 2018, 118, 224–232. [Google Scholar] [CrossRef]
- Durinikova, E.; Kozovska, Z.; Poturnajova, M.; Plava, J.; Cierna, Z.; Babelova, A.; Bohovic, R.; Schmidtova, S.; Tomas, M.; Kucerova, L.; et al. ALDH1A3 upregulation and spontaneous metastasis formation is associated with acquired chemoresistance in colorectal cancer cells. BMC Cancer 2018, 18, 848. [Google Scholar] [CrossRef]
- Moreb, J.S.; Baker, H.V.; Chang, L.J.; Amaya, M.; Lopez, M.C.; Ostmark, B.; Chou, W. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol. Cancer 2008, 7, 87. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Z.; Wong, S.K.-M.; Tin, V.P.-C.; Ho, K.-Y.; Wang, J.; Sham, M.-H.; Wong, M.P. Lung cancer tumorigenicity and drug resistance are maintained through ALDH(hi)CD44(hi) tumor initiating cells. Oncotarget 2013, 4, 1698–1711. [Google Scholar] [CrossRef]
- Yan, J.; De Melo, J.; Cutz, J.C.; Aziz, T.; Tang, D. Aldehyde dehydrogenase 3A1 associates with prostate tumorigenesis. Br. J. Cancer 2014, 110, 2593–2603. [Google Scholar] [CrossRef]
- Le Magnen, C.; Bubendorf, L.; Rentsch, C.A.; Mengus, C.; Gsponer, J.; Zellweger, T.; Rieken, M.; Thalmann, G.N.; Cecchini, M.G.; Germann, M.; et al. Characterization and clinical relevance of ALDHbright populations in prostate cancer. Clin. Cancer Res. 2013, 19, 5361–5371. [Google Scholar] [CrossRef]
- Singh, S.; Arcaroli, J.J.; Orlicky, D.J.; Chen, Y.; Messersmith, W.A.; Bagby, S.; Purkey, A.; Quackenbush, K.S.; Thompson, D.C.; Vasiliou, V. Aldehyde Dehydrogenase 1B1 as a Modulator of Pancreatic Adenocarcinoma. Pancreas 2016, 45, 117–122. [Google Scholar] [CrossRef]
- Nie, S.; Qian, X.; Shi, M.; Li, H.; Peng, C.; Ding, X.; Zhang, S.; Zhang, B.; Xu, G.; Lv, Y.; et al. ALDH1A3 Accelerates Pancreatic Cancer Metastasis by Promoting Glucose Metabolism. Front. Oncol. 2020, 10, 915. [Google Scholar] [CrossRef]
- Kuroda, T.; Hirohashi, Y.; Torigoe, T.; Yasuda, K.; Takahashi, A.; Asanuma, H.; Morita, R.; Mariya, T.; Asano, T.; Mizuuchi, M.; et al. ALDH1-high ovarian cancer stem-like cells can be isolated from serous and clear cell adenocarcinoma cells, and ALDH1 high expression is associated with poor prognosis. PLoS ONE 2013, 8, e65158. [Google Scholar] [CrossRef]
- Ajani, J.A.; Wang, X.; Song, S.; Suzuki, A.; Taketa, T.; Sudo, K.; Wadhwa, R.; Hofstetter, W.L.; Komaki, R.; Maru, D.M.; et al. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol. Oncol. 2014, 8, 142–149. [Google Scholar] [CrossRef]
- Wang, Y.; Zhe, H.; Gao, P.; Zhang, N.; Li, G.; Qin, J. Cancer stem cell marker ALDH1 expression is associated with lymph node metastasis and poor survival in esophageal squamous cell carcinoma: A study from high incidence area of northern China. Dis. Esophagus Off. J. Int. Soc. Dis. Esophagus 2012, 25, 560–565. [Google Scholar] [CrossRef]
- Zhu, T.; He, J.Y.; Liu, Y.P.; Deng, K.; Zuo, J.H.; Ai, X.H. ALDH1B1 predicts poor survival for locally advanced nasopharyngeal carcinoma patients. Transl. Cancer Res. 2022, 11, 382–391. [Google Scholar] [CrossRef]
- Li, X.; Xu, Q.; Fu, X.; Luo, W. ALDH1A1 overexpression is associated with the progression and prognosis in gastric cancer. BMC Cancer 2014, 14, 705. [Google Scholar] [CrossRef]
- Wu, D.; Mou, Y.P.; Chen, K.; Cai, J.Q.; Zhou, Y.C.; Pan, Y.; Xu, X.W.; Zhou, W.; Gao, J.Q.; Chen, D.W.; et al. Aldehyde dehydrogenase 3A1 is robustly upregulated in gastric cancer stem-like cells and associated with tumorigenesis. Int. J. Oncol. 2016, 49, 611–622. [Google Scholar] [CrossRef]
- Greco, N.; Schott, T.; Mu, X.; Rothenberg, A.; Voigt, C.; McGough, R.L., III; Goodman, M.; Huard, J.; Weiss, K.R. ALDH Activity Correlates with Metastatic Potential in Primary Sarcomas of Bone. J. Cancer Ther. 2014, 5, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, Y.; He, Y.; Cai, Q.; Gao, S.; Yao, W.; Liu, Z.; Tian, Z.; Han, Q.; Wang, W.; et al. Upregulation of ALDH1B1 promotes tumor progression in osteosarcoma. Oncotarget 2018, 9, 2502–2514. [Google Scholar] [CrossRef] [PubMed]
- Flahaut, M.; Jauquier, N.; Chevalier, N.; Nardou, K.; Balmas Bourloud, K.; Joseph, J.M.; Barras, D.; Widmann, C.; Gross, N.; Renella, R.; et al. Aldehyde dehydrogenase activity plays a Key role in the aggressive phenotype of neuroblastoma. BMC Cancer 2016, 16, 781. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Dallaglio, K.; Chen, Y.; Robinson, W.A.; Robinson, S.E.; McCarter, M.D.; Wang, J.; Gonzalez, R.; Thompson, D.C.; Norris, D.A.; et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells 2012, 30, 2100–2113. [Google Scholar] [CrossRef] [PubMed]
- Ran, D.; Schubert, M.; Pietsch, L.; Taubert, I.; Wuchter, P.; Eckstein, V.; Bruckner, T.; Zoeller, M.; Ho, A.D. Aldehyde dehydrogenase activity among primary leukemia cells is associated with stem cell features and correlates with adverse clinical outcomes. Exp. Hematol. 2009, 37, 1423–1434. [Google Scholar] [CrossRef]
- Cheung, A.M.S.; Wan, T.S.K.; Leung, J.C.K.; Chan, L.Y.Y.; Huang, H.; Kwong, Y.L.; Liang, R.; Leung, A.Y.H. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia 2007, 21, 1423–1430. [Google Scholar] [CrossRef]
- Voulgaridou, G.P.; Kiziridou, M.; Mantso, T.; Chlichlia, K.; Galanis, A.; Koukourakis, M.I.; Franco, R.; Panayiotidis, M.I.; Pappa, A. Aldehyde dehydrogenase 3A1 promotes multi-modality resistance and alters gene expression profile in human breast adenocarcinoma MCF-7 cells. Int. J. Biochem. Cell Biol. 2016, 77, 120–128. [Google Scholar] [CrossRef]
- Cojoc, M.; Peitzsch, C.; Kurth, I.; Trautmann, F.; Kunz-Schughart, L.A.; Telegeev, G.D.; Stakhovsky, E.A.; Walker, J.R.; Simin, K.; Lyle, S.; et al. Aldehyde dehydrogenase is regulated by β-Catenin/TCF and promotes radioresistance in prostate cancer progenitor cells. Cancer Res. 2015, 75, 1482–1494. [Google Scholar] [CrossRef]
- Yao, T.; Weng, X.; Yao, Y.; Huang, C.; Li, J.; Peng, Y.; Lin, R.; Lin, Z. ALDH-1-positive cells exhibited a radioresistant phenotype that was enhanced with hypoxia in cervical cancer. BMC Cancer 2020, 20, 891. [Google Scholar] [CrossRef]
- Kim, R.J.; Park, J.R.; Roh, K.J.; Choi, A.R.; Kim, S.R.; Kim, P.H.; Yu, J.H.; Lee, J.W.; Ahn, S.H.; Gong, G.; et al. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2α. Cancer Lett. 2013, 333, 18–31. [Google Scholar] [CrossRef]
- Hsu, L.C.; Chang, W.-C. Cloning and Characterization of a New Functional Human Aldehyde Dehydrogenase Gene. J. Biol. Chem. 1991, 266, 12257–12265. [Google Scholar] [CrossRef]
- Hiraoka, L.R.; Hsu, L.; Hsieh, C.L. Assignment of ALDH3 to human chromosome 17p11.2 and ALDH5 to human chromosome 9p13. Genomics 1995, 25, 323–325. [Google Scholar] [CrossRef]
- Jackson, B.C.; Holmes, R.S.; Backos, D.S.; Reigan, P.; Thompson, D.C.; Vasiliou, V. Comparative genomics, molecular evolution and computational modeling of ALDH1B1 and ALDH2. Chem.-Biol. Interact. 2013, 202, 11–21. [Google Scholar] [CrossRef]
- Steinmetz, C.G.; Xie, P.; Weiner, H.; Hurley, T.D. Structure of mitochondrial aldehyde dehydrogenase: The genetic component of ethanol aversion. Structure 1997, 5, 701–711. [Google Scholar] [CrossRef]
- Larson, H.N.; Zhou, J.; Chen, Z.; Stamler, J.S.; Weiner, H.; Hurley, T.D. Structural and functional consequences of coenzyme binding to the inactive Asian variant of mitochondrial aldehyde dehydrogenase: Roles of residues 475 and 487. J. Biol. Chem. 2007, 282, 12940–12950. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Yu, Z.; Morais, D.; Ivanga, M.; Harrison, P.M. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinform. 2007, 8, 308. [Google Scholar] [CrossRef]
- Lutfullah, G.; Qaisar Khan, N.; Amin, F.; Kakakhel, L.; Azhar, N. Structural Modeling Studies of Aldehyde Dehydrogenase X: Insights into Key Interactions in the Tetrameric Assembly of the Isoenzyme. Protein Pept. Lett. 2011, 18, 41–57. [Google Scholar] [CrossRef]
- Stewart, M.J.; Malek, K.; Xiao, Q.; Dipple, K.M.; Crabb, D.W. The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem. Biophys. Res. Commun. 1995, 211, 144–151. [Google Scholar] [CrossRef]
- Stagos, D.; Chen, Y.; Brocker, C.; Donald, E.; Jackson, B.C.; Orlicky, D.J.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenase 1B1: Molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab. Dispos. 2010, 38, 1679–1687. [Google Scholar] [CrossRef]
- Sheikh, S.; Weiner, H. Allosteric inhibition of human liver aldehyde dehydrogenase by the isoflavone prunetin. Biochem. Pharmacol. 1997, 53, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.C.; Reigan, P.; Miller, B.; Thompson, D.C.; Vasiliou, V. Human ALDH1B1 Polymorphisms may Affect the Metabolism of Acetaldehyde and All-trans retinaldehyde–In Vitro Studies and Computational Modeling. Pharm. Res. 2015, 32, 1648–1662. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Hom, M.E.; Bearrood, T.E.; Rosenthal, Z.C.; Fernández, D.; Ondrus, A.E.; Gu, Y.; McCormick, A.K.; Tomaske, M.G.; Marshall, C.R.; et al. Targeting colorectal cancer with small-molecule inhibitors of ALDH1B1. Nat. Chem. Biol. 2022, 18, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Benzaldehyde|C6H5CHO—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/240 (accessed on 5 December 2022).
- Acetaldehyde|CH3CHO—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/177 (accessed on 5 December 2022).
- Hexanal|C6H12O—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/6184 (accessed on 5 December 2022).
- Nonanal|C9H18O—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/31289 (accessed on 5 December 2022).
- Propionaldehyde|CH3CH2CHO—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/527 (accessed on 5 December 2022).
- Nitroglycerin|C3H5(NO3)3—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/4510 (accessed on 5 December 2022).
- Retinal|C20H28O—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/638015 (accessed on 5 December 2022).
- Chen, Y.; Orlicky, D.J.; Matsumoto, A.; Singh, S.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem. Biophys. Res. Commun. 2011, 405, 173–179. [Google Scholar] [CrossRef]
- Ioannou, M.; Serafimidis, I.; Arnes, L.; Sussel, L.; Singh, S.; Vasiliou, V.; Gavalas, A. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev. Biol. 2013, 374, 153. [Google Scholar] [CrossRef]
- Anastasiou, V.; Ninou, E.; Alexopoulou, D.; Stertmann, J.; Müller, A.; Dahl, A.; Solimena, M.; Speier, S.; Serafimidis, I.; Gavalas, A. Aldehyde dehydrogenase activity is necessary for beta cell development and functionality in mice. Diabetologia 2016, 59, 139–150. [Google Scholar] [CrossRef]
- Wang, Y.; Du, F.; Zhao, H.; Yu, X.; Liu, J.; Xiao, Y.; Lu, C.; Li, X.; Wang, Y.; Wang, B.; et al. Synergistic association between two alcohol metabolism relevant genes and coronary artery disease among Chinese hypertensive patients. PLoS ONE 2014, 9, e103161. [Google Scholar] [CrossRef]
- Mastrokolias, A.; Pool, R.; Mina, E.; Hettne, K.M.; van Duijn, E.; van der Mast, R.C.; van Ommen, G.J.; t Hoen, P.A.C.; Prehn, C.; Adamski, J.; et al. Integration of targeted metabolomics and transcriptomics identifies deregulation of phosphatidylcholine metabolism in Huntington’s disease peripheral blood samples. Metabolomics 2016, 12, 137. [Google Scholar] [CrossRef]
- Song, Y.N.; Dong, S.; Wei, B.; Liu, P.; Zhang, Y.Y.; Su, S.B. Metabolomic mechanisms of gypenoside against liver fibrosis in rats: An integrative analysis of proteomics and metabolomics data. PLoS ONE 2017, 12, e0173598. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Wang, Q.; Tesfaye, D.; Schellander, K.; Hoelker, M.; Hossain, M.M.; Tsang, B.K. Polycystic ovarian syndrome is accompanied by repression of gene signatures associated with biosynthesis and metabolism of steroids, cholesterol and lipids. J. Ovarian Res. 2015, 8, 24. [Google Scholar] [CrossRef]
- Singh, S.; Chen, Y.; Matsumoto, A.; Orlicky, D.J.; Dong, H.; Thompson, D.C.; Vasiliou, V. ALDH1B1 links alcohol consumption and diabetes. Biochem. Biophys. Res. Commun. 2015, 463, 768–773. [Google Scholar] [CrossRef]
- Li, H.; Toth, E.; Cherrington, N.J. Alcohol Metabolism in the Progression of Human Nonalcoholic Steatohepatitis. Toxicol. Sci. 2018, 164, 428–438. [Google Scholar] [CrossRef]
- Husemoen, L.L.N.; Fenger, M.; Friedrich, N.; Tolstrup, J.S.; Beenfeldt Fredriksen, S.; Linneberg, A. The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol. Clin. Exp. Res. 2008, 32, 1984–1991. [Google Scholar] [CrossRef]
- Cui, Y.Q.; Xiang, Y.; Meng, F.; Ji, C.H.; Xiao, R.; Li, J.P.; Dai, Z.T.; Liao, X.H. ALDH2 promotes uterine corpus endometrial carcinoma proliferation and construction of clinical survival prognostic model. Aging (Albany NY) 2021, 13, 23588. [Google Scholar] [CrossRef]
- Baek, S.H.; Jang, Y.K. AMBRA1 Negatively Regulates the Function of ALDH1B1, a Cancer Stem Cell Marker, by Controlling Its Ubiquitination. Int. J. Mol. Sci. 2021, 22, 2079. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Zhou, D.; Li, X.; Jia, S.; Qi, S.; Huang, J. Aldehyde dehydrogenase 1B1 is a potential marker of colorectal tumors. Histol. Histopathol. 2021, 36, 183–194. [Google Scholar] [CrossRef]
- Golla, J.P.; Kandyliari, A.; Tan, W.Y.; Chen, Y.; Orlicky, D.J.; Thompson, D.C.; Shah, Y.M.; Vasiliou, V. Interplay between APC and ALDH1B1 in a newly developed mouse model of colorectal cancer. Chem. Biol. Interact. 2020, 331, 109274. [Google Scholar] [CrossRef]
- Matsumoto, A.; Arcaroli, J.; Chen, Y.; Gasparetto, M.; Neumeister, V.; Thompson, D.C.; Singh, S.; Smith, C.; Messersmith, W.; Vasiliou, V. Aldehyde dehydrogenase 1B1: A novel immunohistological marker for colorectal cancer. Br. J. Cancer 2017, 117, 1537–1543. [Google Scholar] [CrossRef]
- Langan, R.C.; Mullinax, J.E.; Ray, S.; Raiji, M.T.; Schaub, N.; Xin, H.W.; Koizumi, T.; Steinberg, S.M.; Anderson, A.; Wiegand, G.; et al. A pilot study assessing the potential role of non-CD133 colorectal cancer stem cells as biomarkers. J. Cancer 2012, 3, 231–240. [Google Scholar] [CrossRef]
- Charepalli, V.; Reddivari, L.; Radhakrishnan, S.; Vadde, R.; Agarwal, R.; Vanamala, J.K.P. Anthocyanin-containing purple-fleshed potatoes suppress colon tumorigenesis via elimination of colon cancer stem cells. J. Nutr. Biochem. 2015, 26, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Mameishvili, E.; Serafimidis, I.; Iwaszkiewicz, S.; Lesche, M.; Reinhardt, S.; Bölicke, N.; Büttner, M.; Stellas, D.; Papadimitropoulou, A.; Szabolcs, M.; et al. Aldh1b1 expression defines progenitor cells in the adult pancreas and is required for Kras-induced pancreatic cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 20679–20688. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.K.; Wang, X.K.; Liao, X.W.; Han, C.Y.; Yu, T.D.; Qin, W.; Zhu, G.Z.; Su, H.; Yu, L.; Liu, X.G.; et al. Aldehyde dehydrogenase 1 (ALDH1) isoform expression and potential clinical implications in hepatocellular carcinoma. PLoS ONE 2017, 12, e0182208. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Guo, X.; Wang, Z.; Li, X.; Bu, Y.; Bai, X.; Zheng, L.; Huang, Y. The prognostic roles of ALDH1 isoenzymes in gastric cancer. Onco. Targets. Ther. 2016, 9, 3405–3414. [Google Scholar] [CrossRef] [PubMed]
- Brunner, C.; Davies, N.M.; Martin, R.M.; Eeles, R.; Easton, D.; Kote-Jarai, Z.; Al Olama, A.A.; Benlloch, S.; Muir, K.; Giles, G.; et al. Alcohol consumption and prostate cancer incidence and progression: A Mendelian randomisation study. Int. J. Cancer 2017, 140, 75–85. [Google Scholar] [CrossRef]
- Li, C.; Wang, Q.; Ma, J.; Shi, S.; Chen, X.; Yang, H.; Han, J. Integrative pathway analysis of genes and metabolites reveals metabolism abnormal subpathway regions and modules in esophageal squamous cell carcinoma. Molecules 2017, 22, 1599. [Google Scholar] [CrossRef]
- He, J.; Song, X.; Yu, L.; Li, J.; Qiao, Z.; Jiu, R.; Yu, B.; Liu, X. [Increased expression of acetaldehyde dehydrogenase in cisplatin-resistant human lung adenocarcinoma A549/DDP cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2015, 31, 625–629. [Google Scholar]
- Leung, Y.K.; Govindarajah, V.; Cheong, A.; Veevers, J.; Song, D.; Gear, R.; Zhu, X.; Ying, J.; Kendler, A.; Medvedovic, M.; et al. Gestational high-fat diet and bisphenol A exposure heightens mammary cancer risk. Endocr. Relat. Cancer 2017, 24, 365–378. [Google Scholar] [CrossRef]
- Wang, Q.-E. DNA damage responses in cancer stem cells: Implications for cancer therapeutic strategies. World J. Biol. Chem. 2015, 6, 57. [Google Scholar] [CrossRef]
- Lin, H.; Ma, X.; Yang, X.; Chen, Q.; Wen, Z.; Yang, M.; Fu, J.; Yin, T.; Lu, G.; Qi, J.; et al. Natural shikonin and acetyl-shikonin improve intestinal microbial and protein composition to alleviate colitis-associated colorectal cancer. Int. Immunopharmacol. 2022, 111, 109097. [Google Scholar] [CrossRef]
- Hartomo, T.B.; Van Huyen Pham, T.; Yamamoto, N.; Hirase, S.; Hasegawa, D.; Kosaka, Y.; Matsuo, M.; Hayakawa, A.; Takeshima, Y.; Iijima, K.; et al. Involvement of aldehyde dehydrogenase 1A2 in the regulation of cancer stem cell properties in neuroblastoma. Int. J. Oncol. 2015, 46, 1089–1098. [Google Scholar] [CrossRef]
- Wang, X.; Li, C.; Yao, W.; Tian, Z.; Liu, Z.; Ge, H. MicroRNA-761 suppresses tumor progression in osteosarcoma via negatively regulating ALDH1B1. Life Sci. 2020, 262, 118544. [Google Scholar] [CrossRef]
- Chen, X.; Huang, J.; Yu, C.; Liu, J.; Gao, W.; Li, J.; Song, X.; Zhou, Z.; Li, C.; Xie, Y.; et al. A noncanonical function of EIF4E limits ALDH1B1 activity and increases susceptibility to ferroptosis. Nat. Commun. 2022, 13, 6318. [Google Scholar] [CrossRef]

| Type of Cancer | Techniques Used | Samples Used | Ref. |
|---|---|---|---|
| Colorectal Cancer | Computational analysis | Colon and rectum adenocarcinoma (TCGA samples) | [202] |
| Western blot/Flow cytometry/PCR/Comet assay | Cell line/TCGA samples | [134] | |
| shRNA/Immunoblot/qPCR | Cell line | [203] | |
| Immunohistochemistry/Computational analysis | Patient and tissue samples | [204] | |
| Macroscopic evaluation/Immunohistochemistry | Xenografts | [205] | |
| shRNA/Western blot | Cell lines | [145] | |
| qPCR/Western blot/Flow Cytometry | Cell line | [146] | |
| qPCR/Western blot/Aldefluor assay | Cell lines/Tissue | [206] | |
| Immunohistochemistry | Tisue | [192] | |
| Colorectal Tissue Micro Array/Immunohistochemistry | Patients | [207] | |
| p53 lentiviral shRNA | Colon cancer stem cells | [208] | |
| Pancreatic Cancer | FACS/Immunofluorescence | In vivo | [209] |
| shRNA/Immunohistochemistry/RT-PCR | Cell lines/Tissue | [153] | |
| Osteosarcoma | siRNA/Flow cytometry/Western blot | Osteosarcoma patients/Cell lines/In vivo | [162] |
| Nasopharyngeal Carcinoma (NPC) | Immunohistochemistry/Bioinformatic analysis | Patients | [158] |
| Hepatocellular Carcinoma | Kaplan–Meier survival analysis | Patients | [210] |
| Gastric Cancer | Kaplan–Meier survival analysis | Patients | [211] |
| qPCR | MKN-45, SGC-7901 and MKN-45, SGC-7901 spheres | [160] | |
| Prostate cancer | SNPs | Prostate cancer patients | [212] |
| Esophageal Squamous Cell Carcinoma (ESCC) | Computational analysis | Patients | [213] |
| Lung Adenocarcinoma/Lung Squamous Cell Carcinoma | Computational analysis | Lung adenocarcinoma/Lung Squamous Cell Carcinoma (TCGA samples) | [202] |
| Flow cytometry/Western blot/qPCR | Cell line | [214] | |
| Breast Cancer | Computational analysis | Breast cancer (TCGA samples) | [202] |
| RNA-seq/survival data | Breast cancer patients(TCGA) | [215] | |
| Glioblastoma | Computational analysis | Glioblastoma (TCGA samples) | [202] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsochantaridis, I.; Roupas, A.; Mohlin, S.; Pappa, A.; Voulgaridou, G.-P. The Concept of Cancer Stem Cells: Elaborating on ALDH1B1 as an Emerging Marker of Cancer Progression. Life 2023, 13, 197. https://doi.org/10.3390/life13010197
Tsochantaridis I, Roupas A, Mohlin S, Pappa A, Voulgaridou G-P. The Concept of Cancer Stem Cells: Elaborating on ALDH1B1 as an Emerging Marker of Cancer Progression. Life. 2023; 13(1):197. https://doi.org/10.3390/life13010197
Chicago/Turabian StyleTsochantaridis, Ilias, Angelos Roupas, Sofie Mohlin, Aglaia Pappa, and Georgia-Persephoni Voulgaridou. 2023. "The Concept of Cancer Stem Cells: Elaborating on ALDH1B1 as an Emerging Marker of Cancer Progression" Life 13, no. 1: 197. https://doi.org/10.3390/life13010197
APA StyleTsochantaridis, I., Roupas, A., Mohlin, S., Pappa, A., & Voulgaridou, G.-P. (2023). The Concept of Cancer Stem Cells: Elaborating on ALDH1B1 as an Emerging Marker of Cancer Progression. Life, 13(1), 197. https://doi.org/10.3390/life13010197










