Abstract
Macro- and microalgae are currently recognized sources of lipids with great nutritional quality and attractive bioactivities for human health promotion and disease prevention. Due to the lipidomic diversity observed among algae species, giving rise to different nutritional and functional characteristics, the mixture of macro- and microalgae has the potential to present important synergistic effects resulting from the complementarity among algae. The aim of this work was to characterize for the first time the lipidome of a blend of macro- and microalgae and evaluate the antioxidant capacity of its lipid fraction. Fatty acids were profiled by GC-MS, the polar lipidome was identified by high resolution LC-MS, and ABTS+• and DPPH• assays were used to assess the antioxidant potential. The most abundant fatty acids were oleic (18:1 n-9), α-linolenic (18:3 n-3), and linoleic (18:2 n-6) acids. The lipid extract presented a beneficial n-6/n-3 ratio (0.98) and low values of atherogenic (0.41) and thrombogenic indices (0.27). The polar lipidome revealed 462 lipid species distributed by glycolipids, phospholipids, and betaine lipids, including some species bearing PUFA and a few with reported bioactivities. The lipid extract also showed antioxidant activity. Overall, the results are promising for the valorization of this blend for food, nutraceutical, and biotechnological applications.
1. Introduction
A trend towards algae (macroalgae and microalgae) has been experienced in recent years by western countries, which is mirrored in the increase in the number of companies producing algae, particularly in Europe [1]. Algae are one of the most important marine and fresh water resources, with the potential to support carbon neutrality, the transition fostered by innovation towards healthy and sustainable food systems, and the green circular bioeconomy [2,3,4]. In this context, algae and algae-based products are progressively being used as food and food ingredients, and also the integral use and valorization of algae have been boosted, which presupposes the stepwise separation of relevant algae components for high-value applications, including functional food, cosmetics, and pharmaceutics [5]. Among its different components, algae have earned rising interest as sources of lipids, not only for biodiesel production, but also as a sustainable alternative to fish lipids, with great nutritional quality and attractive bioactivities [6,7].
The lipid fraction of macroalgae, also known as seaweeds, accounts for 1 to 8% of dry matter [8,9,10], while in microalgae, the lipid content varies within 10 to 50% of dry matter, but it can reach higher values (≥70%), depending on the growth conditions [11]. Several works have already targeted the lipid signatures of diverse algae species, including the macroalgae Ulva rigida [9,12] and Fucus vesiculosus [13], and the microalgae Chlorella vulgaris [14,15], from the fatty acid composition to the in-depth profiling of lipid classes and species. Each algae shows a specific lipidome that comprises a particular composition in fatty acids and in polar lipids—glycolipids, phospholipids, and betaine lipids [7,16]. Despite the substantial variety of profiles, lipidomic studies revealed the occurrence of noteworthy lipid species, namely, the interesting content of polyunsaturated fatty acids (PUFA) and particularly of long chain n-3 PUFA, such as α-linolenic (18:3 n-3) and eicosapentaenoic (20:5 n-3) acids [17], with well-recognized health benefits mainly against non-communicable diseases [18]. Interestingly, algae PUFA occur mainly esterified in polar lipids [12,13,14], which seems to increase the bioavailability of these healthy fatty acids [19]. Polar lipid species also display intrinsic health-promoting properties (e.g., antioxidant and anti-inflammatory activities), representing great potential for the prevention and mitigation of cardiovascular and other chronic diseases [20,21].
Algae have also drawn attention as a reservoir of antioxidant phytochemicals, in line with the demand for novel antioxidants that is driving the exploration and bioprospection of marine resources [18]. Antioxidants play a critical role in slowing or preventing oxidative stress events that underlie numerous pathological conditions, principally chronic and degenerative diseases [22]. Furthermore, they can expand the shelf life of products susceptible to oxidation, such as food and cosmetics, preserving their quality for a longer time period [22]. Algae lipids are a promising eco-friendly alternative to synthetic antioxidants, such as butylated hydroxyanisole (BHA; E-320) or butylated hydroxytoluene (BHT; E-321) [17,23], in conformity with the request of the food and healthcare industries for natural ingredients, encouraged by consumer demand [24].
Due to the lipidomic diversity observed among different algae species, giving rise to dissimilar characteristics, the mixture of several algae species has the potential to present important synergistic effects resulting from the complementarity among algae, which is not possible to obtain with only one species [25]. Regarding algae blends, the market offer is limited to products that combine only macroalgae; therefore, a blend of macro- and microalgae is a completely innovative add-value conception. The precise identification of lipids from any algae blend can elucidate its nutritional and health value, as well as encourage the development of groundbreaking products based on these lipids for multiple applications [26]. Thus, in this work, we aimed to identify for the first time the lipidome of an innovative blend of macro- and microalgae made with Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida. The polar lipidome was analyzed and characterized in detail using a lipidomic approach based on the hydrophilic interaction liquid chromatography coupled to electrospray ionization high resolution tandem mass spectrometry (HILIC-ESI-HR-MS/MS), and gas chromatography–mass spectrometry (GC-MS) allowed profiling of the total esterified fatty acids. To improve the exploitation and valorization of the blend’s total lipids, the antioxidant potential was screened using the most common spectrophotometric free-radical scavenging assays, i.e., ABTS and DPPH tests. Overall, with this work, we aim to demonstrate that the blend has additional values to be used in food, nutraceutical, and biotechnological applications, specifically at the lipid level.
2. Materials and Methods
2.1. Algae Material
Algae blend (Algaessence®; BLEND) biomass was provided by ALGAplus (Ílhavo, Portugal). Algaessence® is a microalgae and macroalgae blend composed of organic Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida. Macroalgae (F. vesiculosus and U. rigida) were produced by ALGAplus, and microalgae (C. vulgaris) was cultivated under autotrophic conditions by Allmicroalgae Natural Products (Pataias, Portugal).
2.2. Lipid Extraction
Total lipids extraction was performed according to a modified Bligh and Dyer method [25]. To 250 mg of the BLEND biomass (n = 5) was added 3.75 mL of a solvent mixture of methanol and dichloromethane (2:1, v/v), followed by homogenization in a vortex for 2 min and ultrasonication in an ultrasound bath for 1 min. After ice incubation on a rocking platform shaker (Stuart equipment, Cole-Parmer Ltd, UK) for 2 h, the suspension was centrifuged (Selecta JP Mixtasel, Abrera, Barcelona, Spain) for 10 min at 2000 rpm, and the organic phase was collected. To promote phase separation and wash the lipid extract, 1.25 mL of dichloromethane and 2.25 mL of Milli-Q water were added to the collected organic phase, centrifuged for 10 min at 2000 rpm, and the lower organic phase was recovered. Re-extraction steps using the same solvent proportions were repeated two more times. The organic phases collected were dried under a stream of nitrogen. Extracts were weighed to determine lipid content and stored at −20 °C. The results were expressed in g 100−1 of dry weight of sample (DW).
2.3. Fatty Acids Analysis by Gas Chromatography–Mass Spectrometry (GC–MS)
Fatty acid profiling was performed by gas chromatography–mass spectrometry (GC-MS) analysis of the fatty acid methyl esters (FAMEs). For that, FAMEs were prepared from the BLEND total lipid extract by alkaline transmethylation reaction using a methanolic solution of potassium hydroxide (2.0 M), according to the procedure previously detailed by Rey et al. [7]. A volume of 2.0 μL of a hexane solution containing FAMEs (n = 5) was injected in a GC-MS (Agilent Technologies 6890 N Network, Santa Clara, CA, USA) equipped with a DB-FFAP column with the following specifications: 30 m long, 0.32 mm internal diameter, and 0.25 μm film thickness (J & W Scientific, Folsom, CA, USA). The GC equipment was connected to an Agilent 5977B Mass Selective Detector operating with an electron impact mode at 70 eV and scanning range of 50–550 m/z in a one-second cycle in full scan mode acquisition. Helium was the carrier gas at a flow rate of 1.4 mL min−1. The injector and detector temperatures were 220 °C and 230 °C, respectively. The oven temperature was programmed as follows: 58 °C for 2 min, 25 °C min−1 to 160 °C, 2 °C min−1 to 210 °C, and 30 °C min−1 to 225 °C, and held for 20 min. Data acquisition was performed using GCMS5977B/Enhanced MassHunter. Data were analyzed using Agilent MassHunter Qualitative Analysis 10.0 software, and fatty acid identification was conducted by the retention time and MS spectrum comparison with the NIST chemical database library and confirmed with literature reports. The relative amount of each fatty acid was calculated by the percent relative area method, with proper normalization using internal standard methyl nonadecanoate (C19:0, Sigma-Aldrich, St. Louis, MO, USA), considering the sum of all relative areas of identified fatty acids.
Nutritional quality of the lipid fraction was assessed based on the fatty acid profile, through the PUFA/SFA and PUFA n-6/n-3 ratios, and by the atherogenicity (AI) and thrombogenicity (TI) indices, which were calculated according to the equations listed below.
2.4. Polar Lipidome Analysis by Hydrophilic Interaction Liquid Chromatography-High-Resolution Tandem Mass Spectrometry (HILIC-HR-MS/MS)
The polar lipidome was determined by hydrophilic interaction liquid chromatography (HILIC) in an Ultimate 3000 Dionex (Thermo Fisher Scientific, Bremen, Germany) with an autosampler coupled to the Q-Exactive® hybrid quadrupole Orbitrap mass spectrometer (Thermo Fisher, Scientific, Bremen, Germany), according to the methodology previously described [15]. The solvent system used consisted of two mobile phases, A and B, which were respectively composed of acetonitrile:methanol:water (2:1:1, v/v/v) and acetonitrile:methanol (3:2, v/v), both containing 5 mM ammonium acetate. The gradient program applied was as follow: 5% A (0–2 min), 5–48% A (2–10 min), 48–65% A (10–15 min), 65% A (15–17 min), returning to the initial conditions in 3 min and held for 10 min.
A mixture containing 40 μg of the BLEND total lipid extract (in 20 µL of dichloromethane), 72 μL of starting eluent (95% B and 5% A), and 8 μL of an internal standards mixture (dMPC-0.04 µg, dMPE-0.04 µg, dPPI-0.08 µg, dMPG-0.024 µg, dMPS-0.08 µg, dMPA-0.16 μg, LPC-0.04 µg, SM d18:1/17:0-0.04 µg, Cer(17:0/d18:1)-0.08 μg, CL(14:0)4-0.16 µg) was prepared. Then, a 5 μL aliquot of the sample mixture (n = 5) was injected into the microbore Ascentis® Express column (10 cm × 2.1 mm, 2.7 µm; Sigma-Aldrich) with a flow rate of 200 μL min−1 at 35 °C. The mass spectrometer was operated simultaneously in positive (electrospray voltage 3.0 kV) and negative (electrospray voltage—2.7 kV) modes, configured as follows: high resolution of 70,000, capillary temperature of 350 °C, sheath gas flow of 20 U, automatic gain control (AGC) target of 1 × 106, m/z range of 400–1600, 2 microscans, and maximum inject time (IT) of 100 ms. In MS/MS experiments, the following configuration was set: high resolution of 17,500, AGC target of 1 × 105, 1 microscan, and maximum IT of 50 ms. Cycles of one full-scan mass spectrum and ten data-dependent MS/MS scans were continuously repeated throughout the experiments, with a dynamic exclusion of 60 s, intensity threshold of 2 × 104, and normalized collision energy ranged between 25, 30, and 35 eV. Data acquisition was accomplished using the Xcalibur v3.3 software (Thermo Fisher Scientific, USA), which was also used to analyze the acquired spectra in order to identify the polar lipid species.
The identification was performed based on the approach previously described [27]. Briefly, lipid species was assigned according to the retention time of internal standards and accurate mass measurements (error of <5 ppm), and the interpretation of well-known fragmentation patterns provided structural information and validated the identification of the most lipid species. To assist in the identification, MZmine v2.42 software was used to filter the raw LC-MS data, peak detection (intensity threshold of 1 × 104), peak processing, and assignment against an in-house database.
2.5. ABTS+• Scavenging Assay
The antioxidant scavenging activity against the 2,2′-azinobis-3-ethylbenzthiazoline-6-sulphonic acid radical cation (ABTS+•) was evaluated as performed in other studies [8,17]. A volume of 150 μL of the lipid extract (31.25–250 μg mL−1 in ethanol, n = 3) or 150 μL of the Trolox standard solution (5–37.5 μmol L−1 in ethanol) were mixed with 150 μL of an ABTS+• working solution in ethanol (3.5 mmol L−1; absorbance ≈ 0.9, 734 nm). The mixture was incubated for 120 min, and the absorbance was measured at 734 nm every 5 min (Multiskan GO 1.00.38, Thermo Scientific, Hudson, NH, USA). Control lipid extracts were prepared by replacing the ABTS+• solution with ethanol. The antioxidant activity, expressed as a percentage of inhibition of the ABTS+•, was determined according to Equation (3), where is the absorbance of the ABTS+• and is the difference between the absorbance of the sample and the absorbance of the control.
The lipid extract concentration capable of scavenging 50% (IC50) of ABTS+• after 120 min of reaction was calculated by linear regression plotting of the concentration of lipid extract versus the percentage of the inhibition curve. The activity was expressed as Trolox Equivalents (TE, μmol Trolox g−1 of sample), according to Equation (4).
Calibration curve for ABTS test is provided in Supplementary Figure S1.
2.6. DPPH• Scavenging Assay
The antioxidant scavenging activity against the 2,2-diphenyl-1-picrylhydrazyl radical (DPPH•) was determined using a previously described method [8,17]. A volume of 150 μL of the lipid extracts (31.25–250 μg mL−1 in ethanol, n = 3) or 150 μL of the Trolox standard solution (5–37.5 μmol L−1 in ethanol) were mixed with 150 μL of a DPPH• working solution in ethanol (250 μmol L−1; absorbance ≈ 0.9, 517 nm). After incubating the mixture for 120 min, the absorbance was measured at 517 nm every 5 min (Multiskan GO 1.00.38, Thermo Scientific, Hudson, NH, USA). Control lipid extracts were prepared by replacing the DPPH• solution with ethanol. The antioxidant activity, expressed as a percentage of inhibition of the DPPH•, was determined according to Equation (5), where is the absorbance of the ABTS+• and is the difference between the absorbance of the sample and the absorbance of the control.
The lipid extract concentration capable of scavenging 35% (IC35) and 20% (IC20) of DPPH• after 120 min of reaction was calculated by linear regression plotting of the concentration of lipid extract versus the percentage of the inhibition curve. The activity was expressed as Trolox Equivalents (TE, μmol Trolox g−1 of sample), according to Equation (6) for IC35 and Equation (7) for IC20
Calibration curve for DPPH test is provided in Supplementary Figure S2.
3. Results and Discussion
3.1. Total Lipid Content, Fatty Acid Profile, and Lipid Quality Indicators of the Algae Blend
The total lipid content of the BLEND was estimated gravimetrically after the lipid extraction procedure as 4.02 ± 0.20 g 100 g–1 of DW biomass. The analysis of the fatty acid composition of the BLEND total lipid extract by GC-MS revealed the presence of 31 fatty acids (Table 1). The monounsaturated fatty acid (MUFA) 18:1 n-9 was detected as the most abundant fatty acid (30.10 ± 0.51%), followed by the saturated fatty acid (SFA) 16:0 (14.74 ± 0.31%) and the polyunsaturated fatty acids (PUFA) 18:3 n-3 (11.73 ± 0.33%) and 18:2 n-6 (11.58 ± 0.13%). The other identified fatty acids had relative abundances of less than 5%. The algae blend was characterized by containing a lower content of SFA (24.58 ± 0.63%), followed by MUFA (37.07 ± 0.74%) and PUFA (39.99 ± 0.29%). The total n-3 and n-6 fatty acid content reached, respectively, 19.42 ± 0.10% and 19.02 ± 0.32%.

Table 1.
Fatty acid profile of the algae blend (BLEND) determined by GC-MS expressed in relative abundance (%). Values are expressed as the mean ± SD (n = 5).
The determined fatty acid profile of the BLEND has a particular composition when compared to those reported in the literature for the individual algae species that compose the mixture, i.e., Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida. The FA 18:1 n-9 (oleic acid, OA) was described as the main fatty acid in F. vesiculosus (20–28%) [13,17,28], while in U. rigida, it can be presented only in relevant amounts (≈9%) [9,28]. The FA 16:0 was the most abundant SFA in all species [7,9,13,17,29,30], but some studies reported that it was the major fatty acid in U. rigida, reaching 42% [28,29,31]. Lastly, the essential fatty acids FA 18:3 n-3 (α-linolenic acid, ALA) and FA 18:2 n-6 (linoleic acid, LA) were identified at high levels in C. vulgaris (ALA: 24–26% and LA: 15–18%) [7,30], in contrast to the minor abundances reported for the seaweeds F. vesiculosus (ALA: 4–7% and LA: 7–9%) and U. rigida (ALA: 5–11% and LA: 1–2%).
Thus, in the BLEND, we were able to obtain a fatty acid profile characterized by a high content of OA, complemented by the presence of both essential fatty acids ALA and LA in interesting contents, therefore, a specific composition that was not achieved in the isolated algae species. OA, which is the main fatty acid in olive oil (>70% of the total fatty acids), has exhibited different health-promoting properties for the management and prevention of NCDs [32]. LA and ALA are essential fatty acids that need to be obtained through diet, as the human body is unable to synthesize them [33]. From LA and ALA, humans can synthesize, respectively, n-6 long chain PUFA, as 20:4 n-6 (arachidonic acid, AA), and n-3 long chain PUFA, as 20:5 n-3 (eicosapentaenoic acid, EPA) and 22:6 n-3 (docosahexaenoic acid, DHA) [33]. These fatty acids are valuable structural components of lipids in membranes and can have a substantial biological role because they are precursors of lipid mediators. Generally, AA is metabolized to lipid pro-inflammatory mediators, and EPA and DHA are precursors of anti-inflammatory signaling lipids, making these PUFA that play a crucial role in an extensive spectrum of biological processes, including inflammation and blood clotting [34].
To give new insight into the beneficial impact on health and assess the nutritional lipid quality of the BLEND, the PUFA/SFA and n-6/n-3 PUFA ratios, as well as the atherogenicity (AI) and thrombogenicity (TI) indices, were calculated (Table 2). The PUFA/SFA ratio was 1.63 ± 0.05 and the n-6/n-3 PUFA ratio was 0.98 ± 0.02, while the AI and TI were 0.41 ± 0.01 and 0.27 ± 0.01, respectively.

Table 2.
Lipid quality indicators values obtained for the algae blend (BLEND), considering the 31 fatty acids identified, in comparison with the calculated values reported in the literature for the algae species—Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida.
The ratio PUFA/SFA is the elementary index to estimate the impact of diet on cardiovascular health, and the higher this ratio, the more beneficial the effect [35]. The n-6/n-3 PUFA ratio is suggested as a relevant factor for brain development and decreasing the risk of developing non-communicable diseases, including autoimmune and neurodegenerative diseases [33]. The recommendations for healthy diets prioritize the ingestion of foods with n-6/n-3 PUFA ≤ 1; however, this recommended value is still controversial, and the absolute amounts of the fatty acids must be taken into account, too [33]. In addition, the indices AI and TI are the most commonly used theoretical calculations to measure the probability of reducing the risk of atherogenic plaques and blood clot formation, respectively [35]. The lower the AI and TI of a product, the higher the nutritional quality and the greater the potential to contribute to mitigating the prevalence of cardiovascular diseases. Our results are within the range described in the literature for the individual algae species (Table 2), other algae, fish, and shellfish [35]. Remarkably, some of the calculated values showed better results than those reported for seaweed blends [25]. Overall, the profitable values found support the nutritional potential of the BLEND total lipid extract, which could be useful to improve human diet quality and sustainability, and also to help prevent inflammatory, cardiovascular, and brain disorders.
3.2. Polar Lipidome of the Algae Blend
The polar lipidome of the BLEND total lipid extract was profiled by HILIC–ESI–HR-MS and MS/MS spectra analysis. Lipid species were identified based on the retention time information and accurate mass identification of ions detected in LC–MS data, and the comprehensive interpretation of MS/MS spectra allowed confirmation of the structural features of the polar head and fatty acid composition of lipid species, as described in detail by Rey et al. [27]. A total of 462 polar lipid species, distributed among glycolipids (GL, 118 species), phospholipids (PL, 206 species), and betaine lipids (BL, 138 species), were identified in the BLEND total lipid extract (Table 3).

Table 3.
Polar lipid profile of the BLEND total lipid extract identified by HILIC–ESI-MS/MS with the indication of the number of lipid species identified and the most abundant lipid species by the class of polar lipids, in comparison with polar lipid profiles of the individual algae species—Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida—from literature. The number of lipid species is related to the molecular weight determined by MS and corresponds to the number of identified lipid molecular ions.
Digalactosylmonoacylglycerol (DGMG), digalactosyldiacylglycerol (DGDG), diacylglyceryl-hydroxymethyl-N,N,N-trimethyl-β-alanine (DGTA), diacylglycerol-trimethyl homoserine (DGTS), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), lysophosphati-dylglycerol (LPG), lysophosphatidylinositol (LPI), monogalactosylmonoacylglycerol (MGMG), monogalactosyldiacylglycerol (MGDG), monoacylglyceryl-hydroxymethyl-N,N,N-trimethyl-β-alanine (MGTA), monoacylglycerol-trimethyl homoserine (MGTS), phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), sulfoquinovosyl diacylglycerol (SQDG), sulfoquinovosyl monoacylglycerol (SQMG).
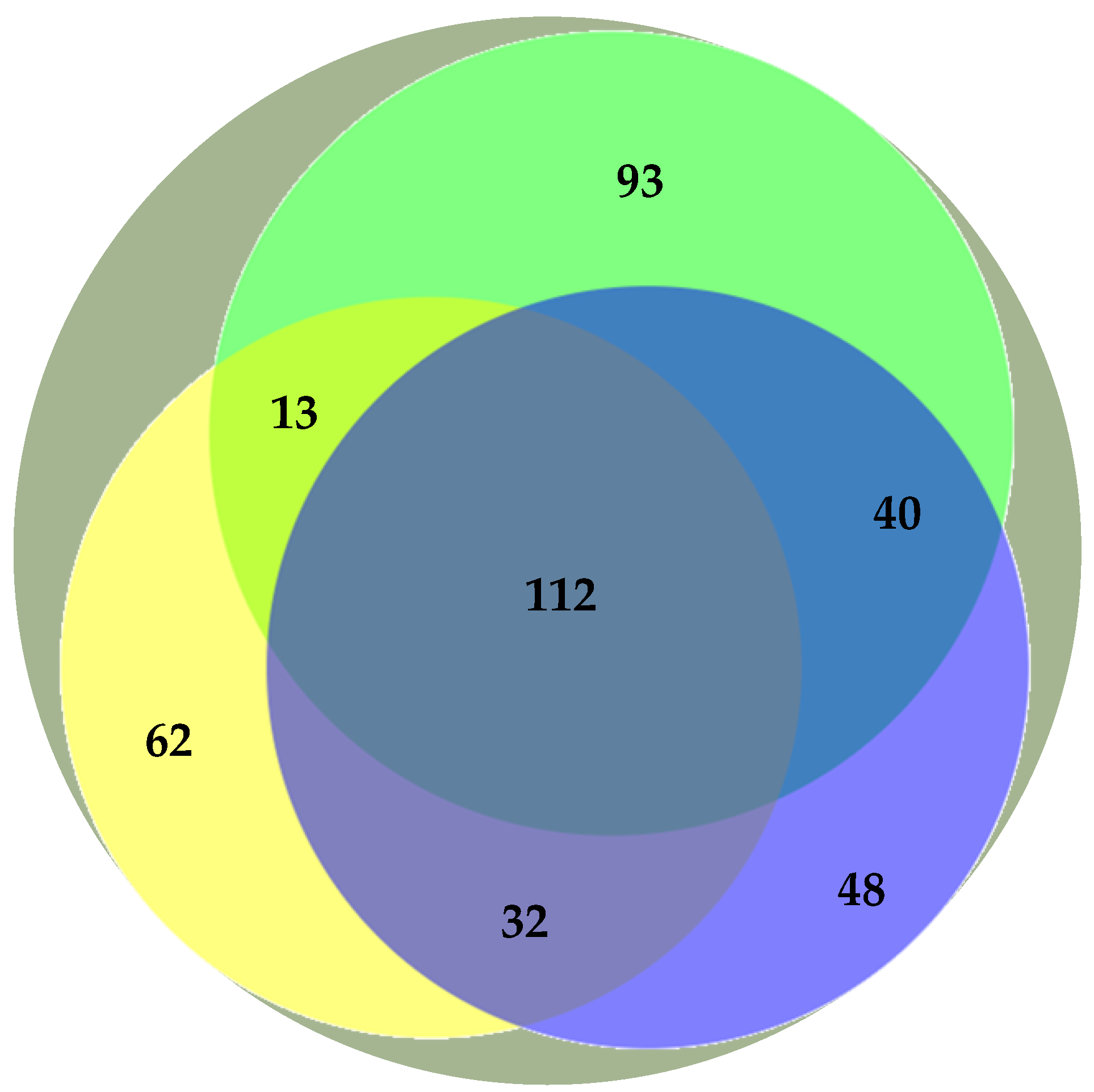
Previous studies have addressed the polar lipidome of the individual algae species present in the BLEND (e.g., Refs. [13,15,29]). Actually, the BLEND lipid extract showed a greater diversity of lipid species in comparison with C. vulgaris, F. vesiculosus, and U. rigida (Table 3). To describe the relationship between the polar lipid profile of the BLEND and the three algae, the polar lipid species identified in the BLEND and reported until now for C. vulgaris, F. vesiculosus, and U. rigida were plotted on a Venn diagram (Figure 1). From the total of 462 lipid species identified in the BLEND lipid extract, only 112 were common to the three algae, while 93, 62, and 48 lipids were derived exclusively from C. vulgaris, F. vesiculosus, and U. rigida, respectively.
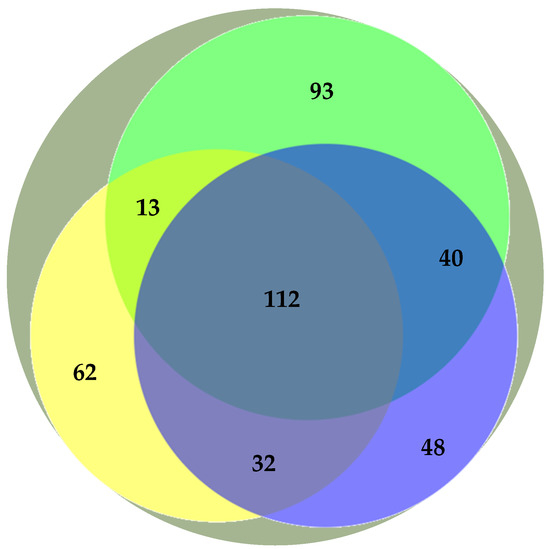
Figure 1.
Venn diagram representation of the number of common polar lipid species identified in the algae blend (BLEND,  ) and reported in the literature for Chlorella vulgaris (
) and reported in the literature for Chlorella vulgaris ( ), Fucus vesiculosus (
), Fucus vesiculosus ( ), and Ulva rigida (
), and Ulva rigida ( ).
).
 ) and reported in the literature for Chlorella vulgaris (
) and reported in the literature for Chlorella vulgaris ( ), Fucus vesiculosus (
), Fucus vesiculosus ( ), and Ulva rigida (
), and Ulva rigida ( ).
).
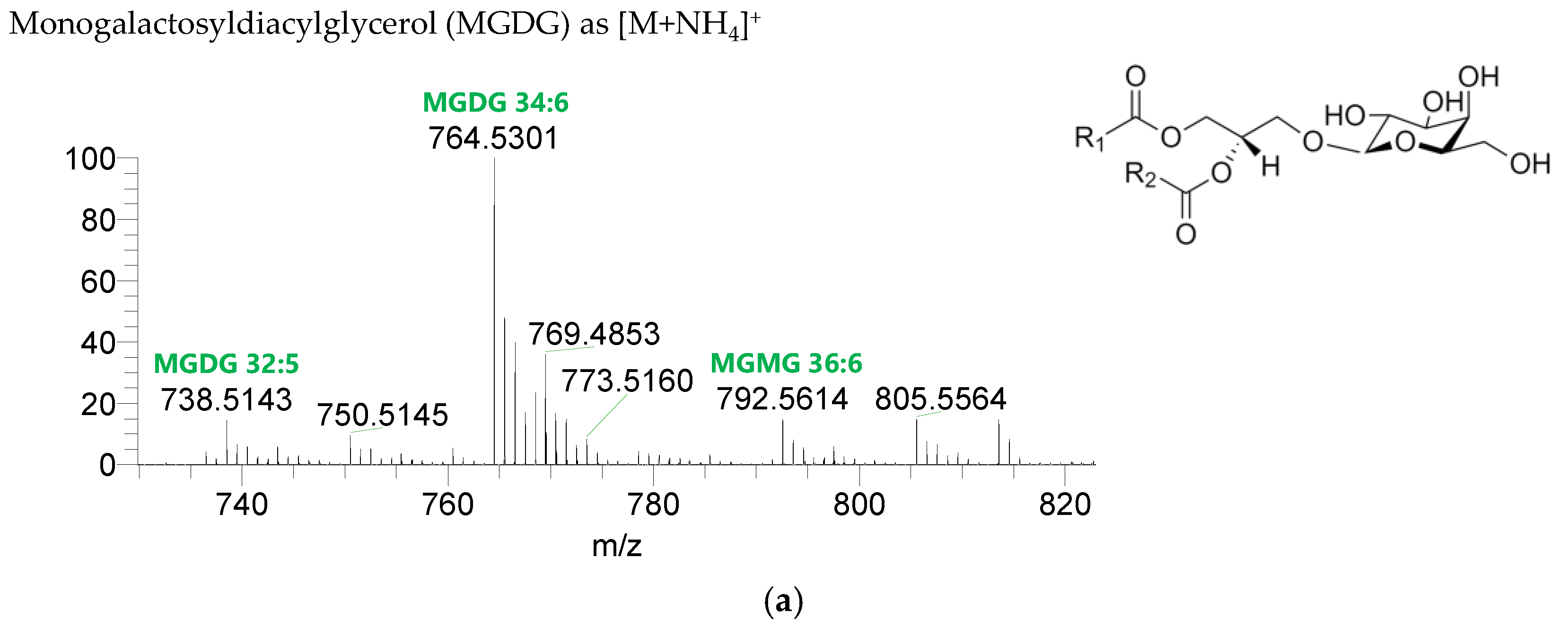
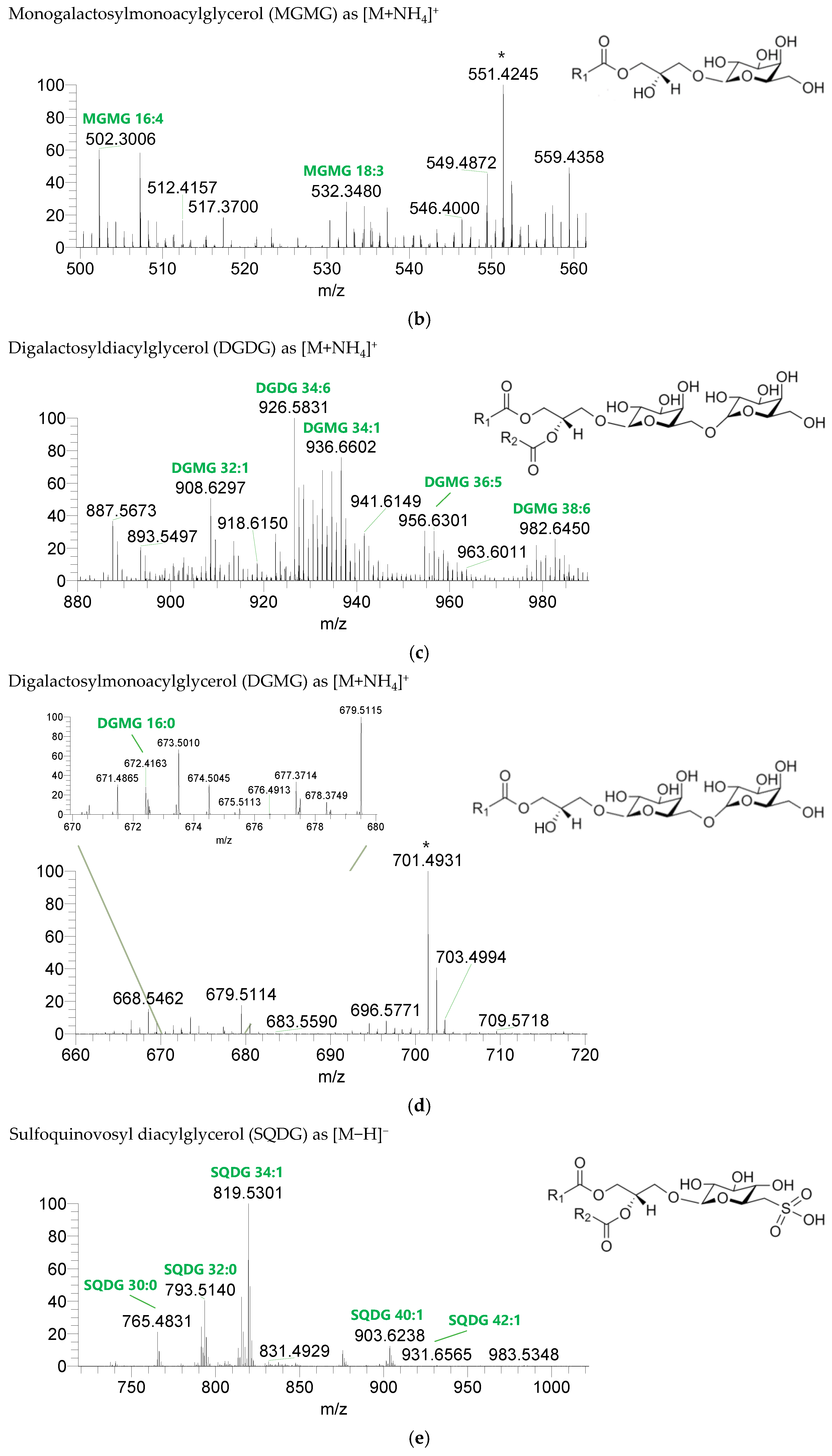
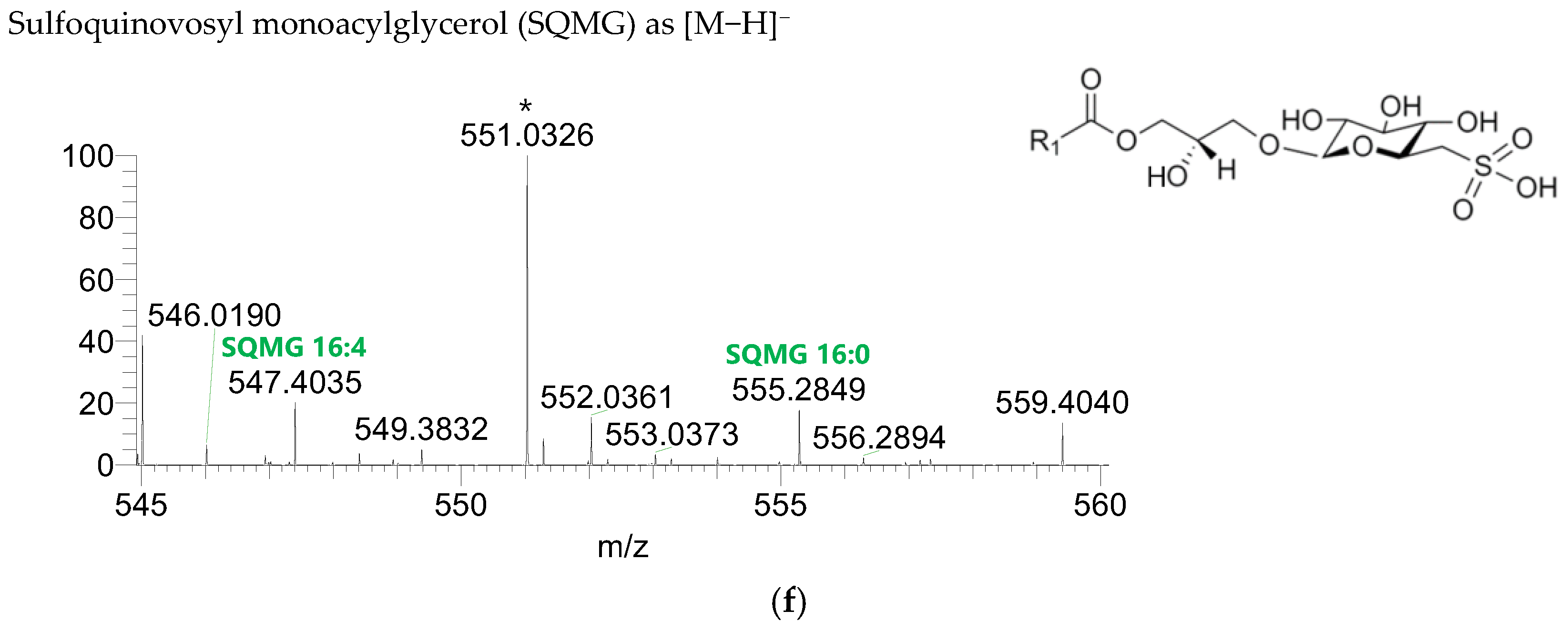
The GL identified included four classes of galactolipids—monogalactosyldiacylglycerol (MGDG), monogalactosylmonoacylglycerol (MGMG), digalactosyldiacylglycerol (DGDG), and digalactosylmonoacylglycerol (DGMG)—identified as [M+NH4]+ ions (Table 4, Supplementary Table S1), and two classes of sulfolipids—sulfoquinovosyl diacylglycerol (SQDG) and sulfoquinovosyl monoacylglycerol (SQMG)—identified as [M−H]− ions (Table 3) (Figure 2). A total of 29 lipid species of MGDG, 11 of MGMG, 33 of DGDG, 8 of DGMG, 34 of SQDG, and 3 of SQMG were recognized. The most abundant species in each class of glycolipids were: MGDG 34:6, assigned as MGDG (16:2_18:4), MGDG (16:3_18:3), and MGDG (16:4_18:2); MGMG 16:4; DGDG 34:6, assigned as DGDG (16:3_18:3); DGMG 16:0; SQDG 34:1, assigned as SQDG (16:0_18:1); and SQMG 16:0. GL species included mostly C16- and C18-fatty acyl chains, but also C14-, C15-, C17-, C19-, C22-, and C24-fatty acyl chains.

Table 4.
Glycolipids identified in the polar lipidome of the algae blend (BLEND) by high resolution HILIC–ESI–MS and MS/MS. C carbons, N number of double bonds. * Lipid species identified based on the polar head fragment, calculated mass, and retention time. ** Lipid species identified based on calculated mass and retention time. The presence of the lipid species in the individual algae species—Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida—is indicated by a cross (×), according to the reports in the literature.
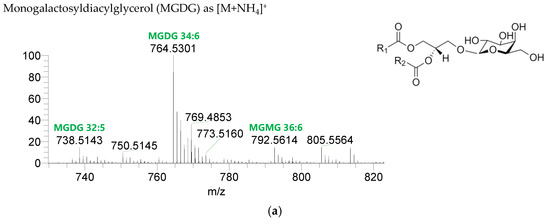
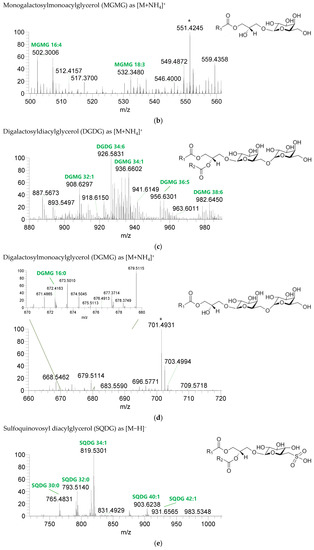
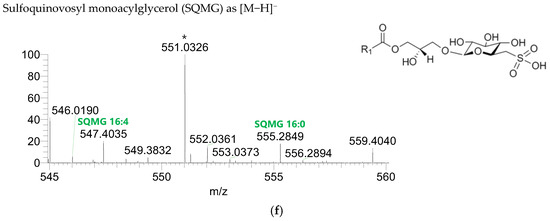
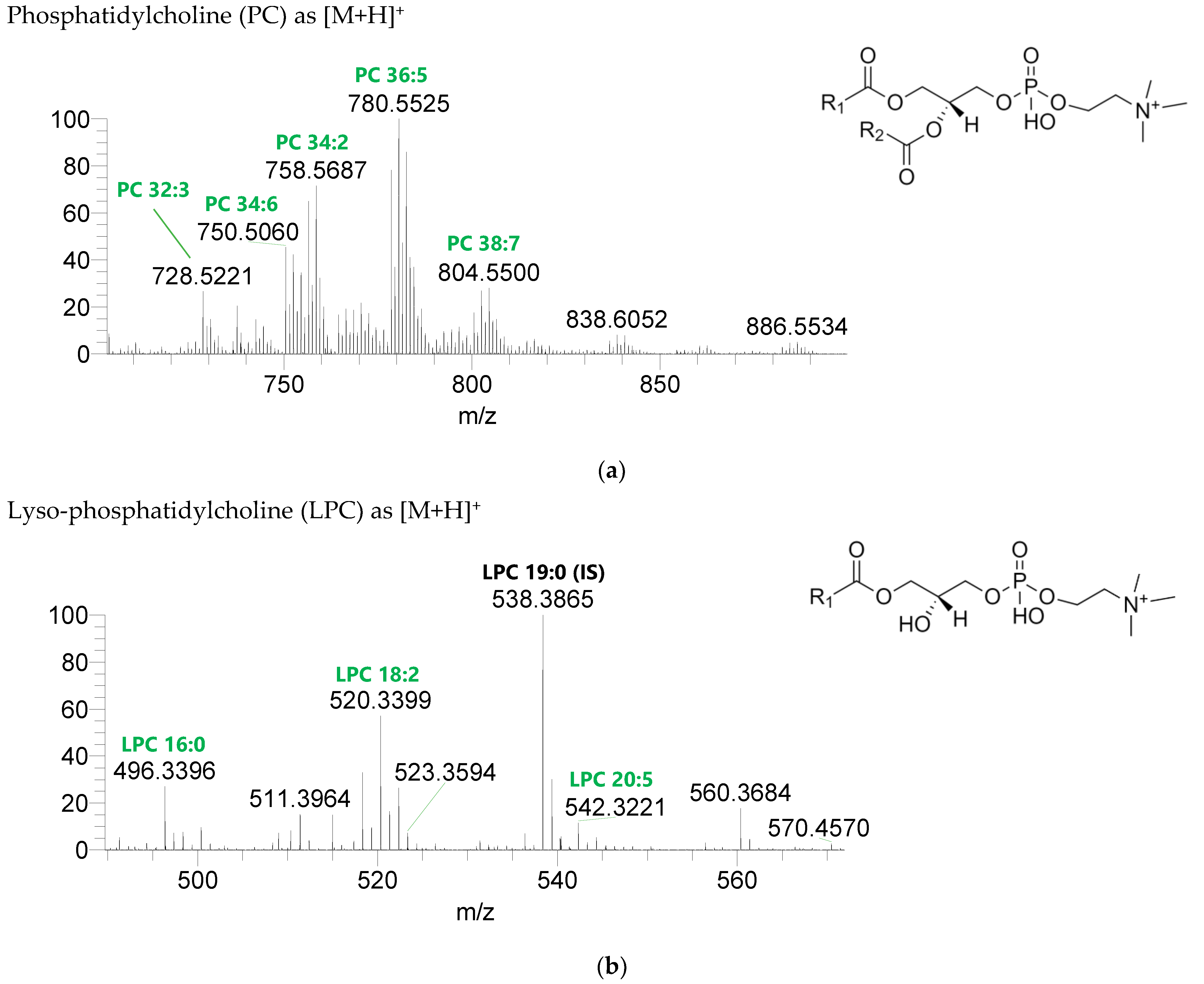
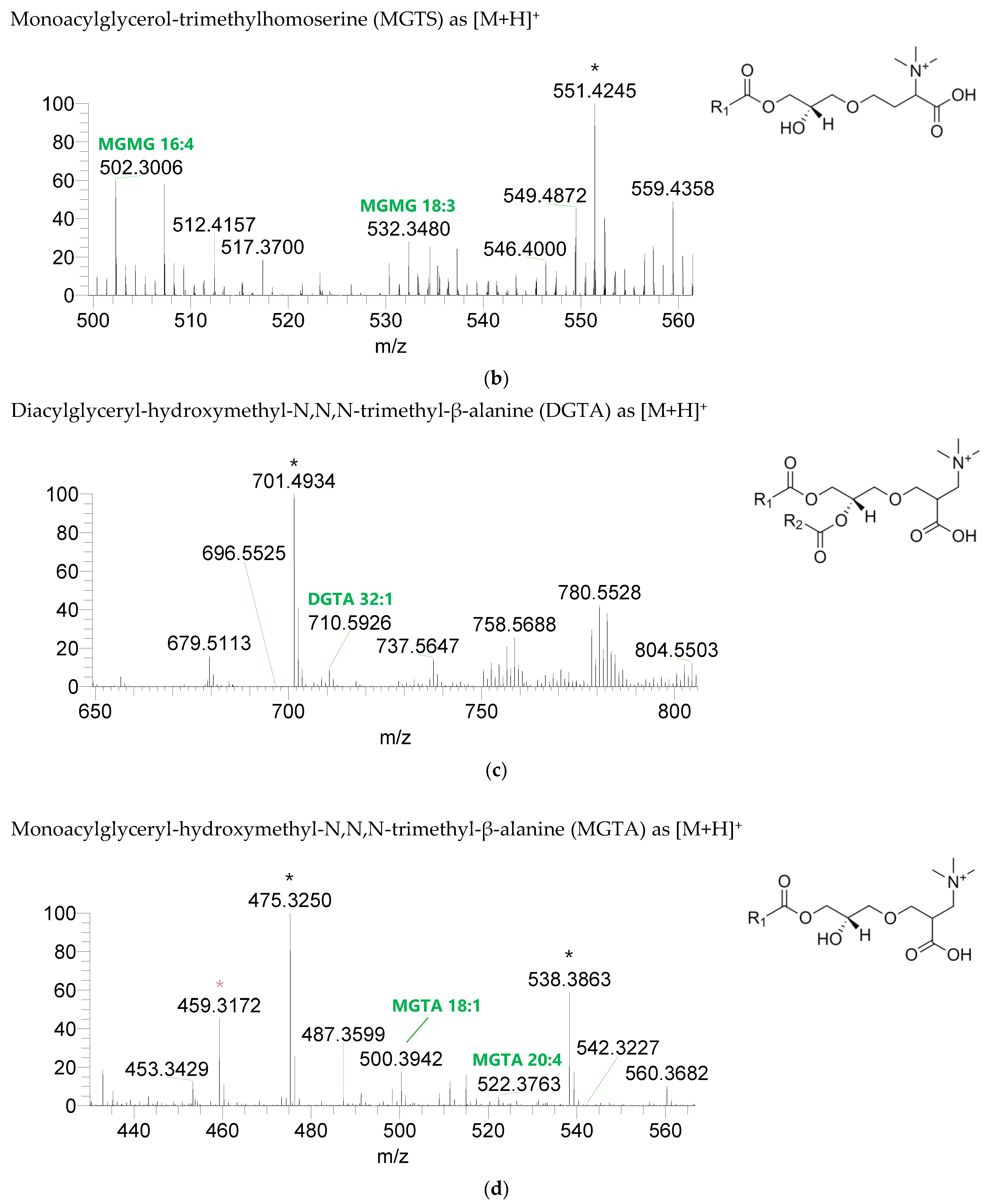
Figure 2.
LC–MS spectra of glycolipid classes identified in the polar lipidome of the algae blend. (a) MGDG, (b) MGMG, (c) DGDG, and (d) DGMG were identified in the positive mode as [M+NH4]+ ions, and (e) SQDG and (f) SQMG were identified in the negative mode as [M−H]− ions. The ions group assigned with symbol (asterisk) are a background.
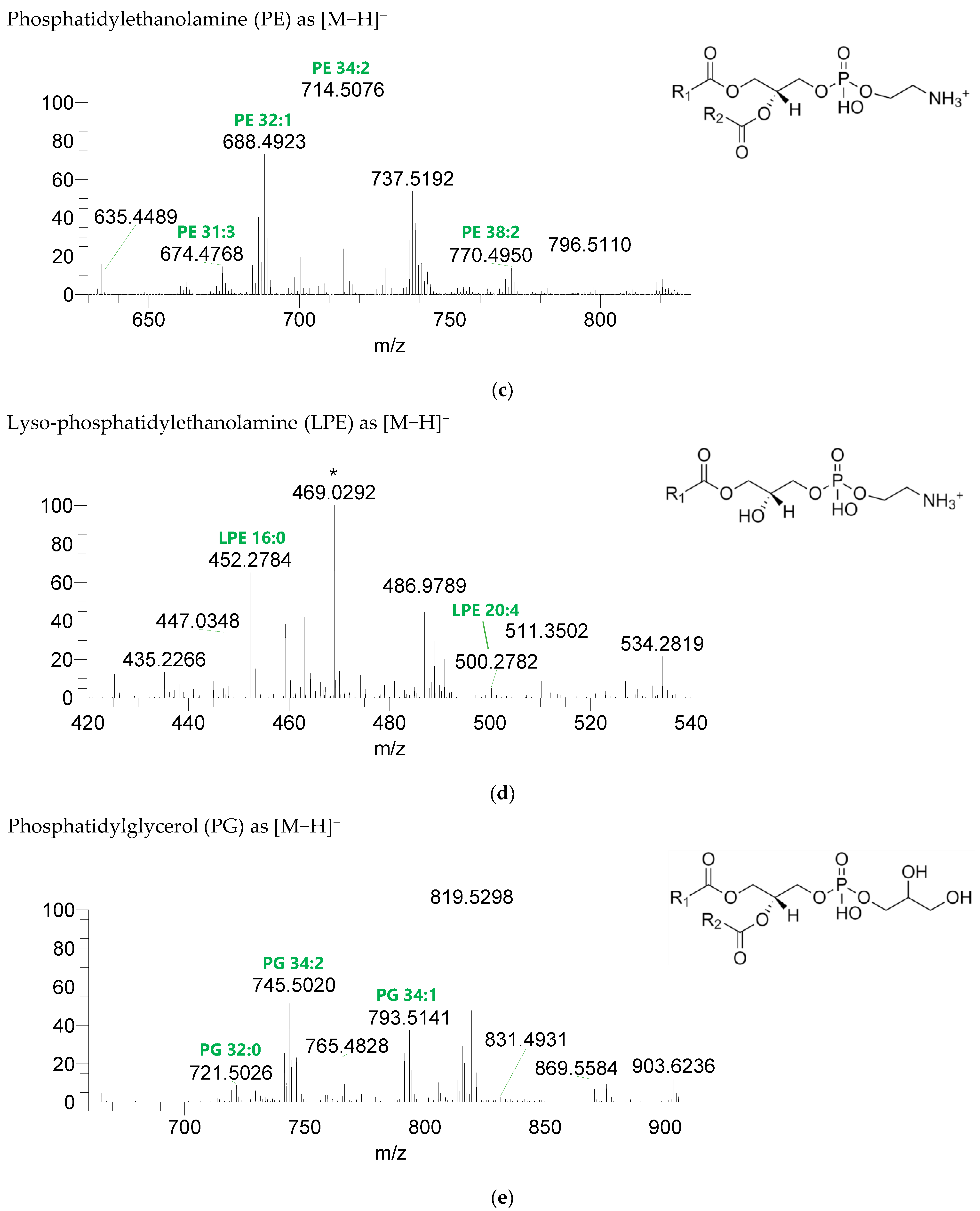
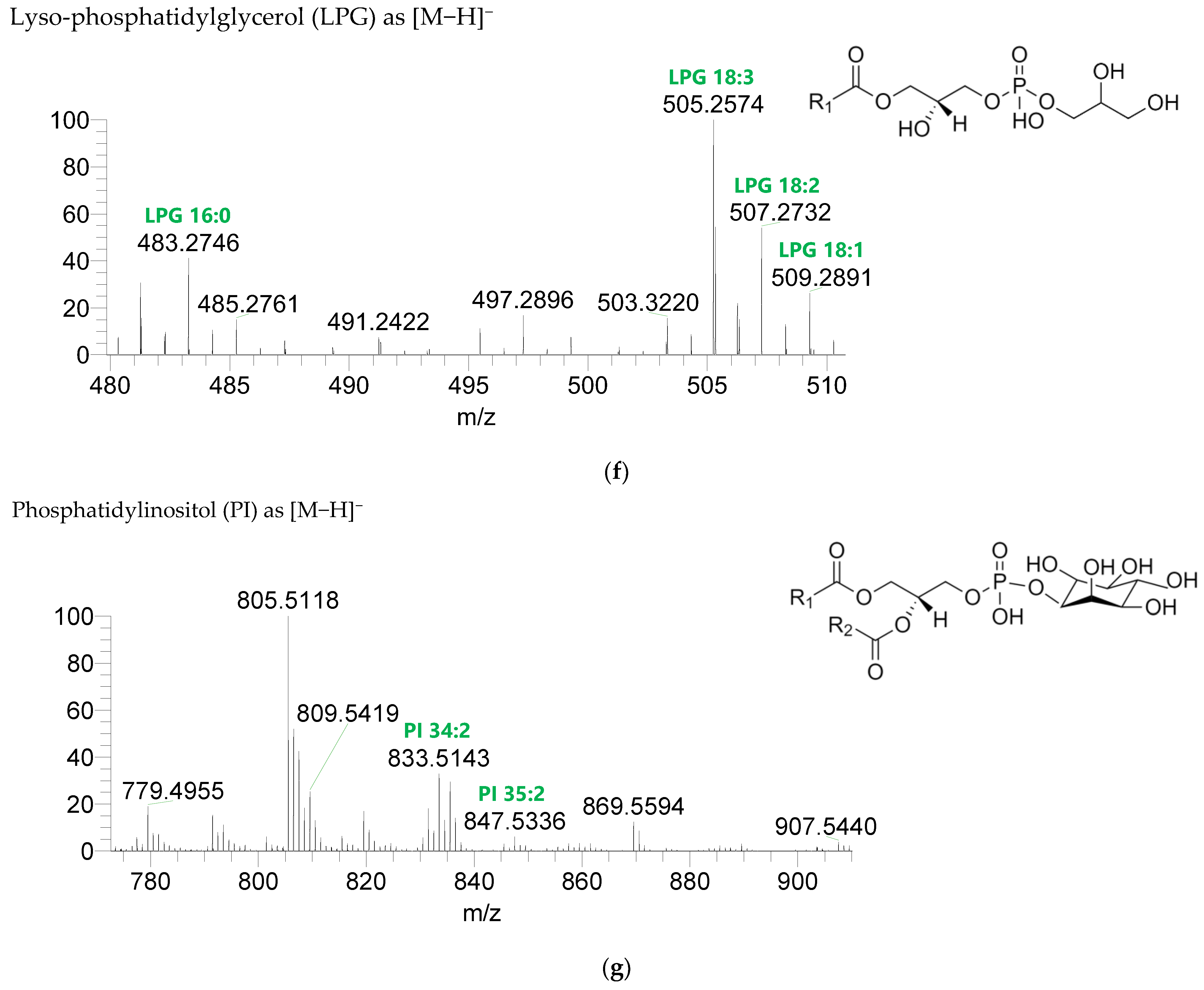
Seven classes of PL were detected in the BLEND lipid extract: phosphatidylcholine (PC), lysophosphatidylcholine (LPC), phosphatidylethanolamine (PE), lysophosphatidylethanolamine (LPE), phosphatidylglycerol (PG), lysophosphatidylglycerol (LPG), and phosphatidylinositol (PI) (Figure 3, Table 5, Supplementary Table S2). PC and LPC were identified in the positive mode as [M+H]+ ions, and the remaining classes were identified in the negative mode as [M-H]- ions. A total of 60 lipid species of PC, 21 of LPC, 47 of PE, 18 of LPE, 34 of PG, 5 of LPG, and 21 of PI were recognized. The most abundant species in each class of PL were: PC 36:5, assigned as PC (16:0_20:5), PC (16:1_20:4), and PC (18:2_18:3); LPC 18:2; PE 34:2, assigned as PE (16:0_18:2) and PE (16:1_18:1); LPE 16:0; PG 34:2, assigned as PG (16:0_18:2) and PG (16:1_18:1); LPG 18:3; and PI 34:2, assigned as PI (16:0_18:2) and PI (16:1_18:1). PL species were mainly esterified with C16-, C18-, and C20-fatty acyl chains, but also C14-, C15-, C17-, C19-, C21-, and C22-fatty acyl chains can be found esterified.
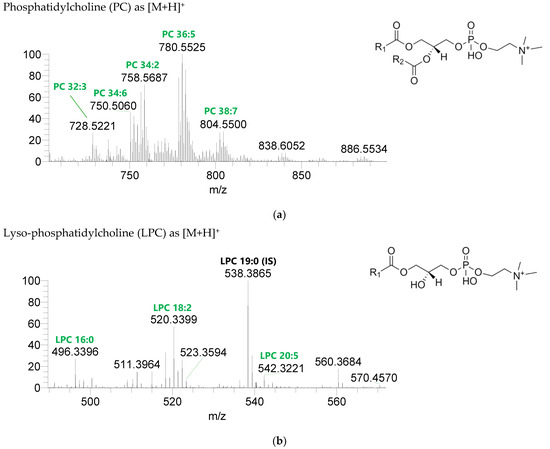
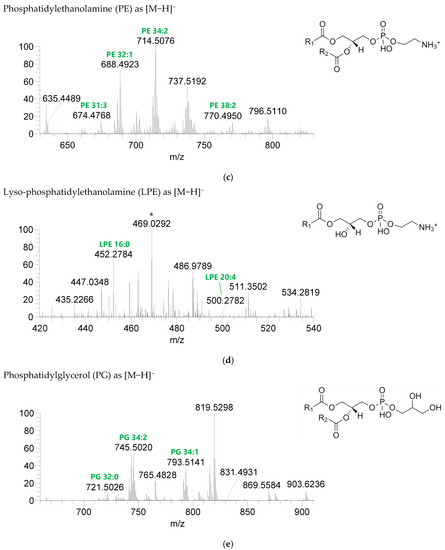
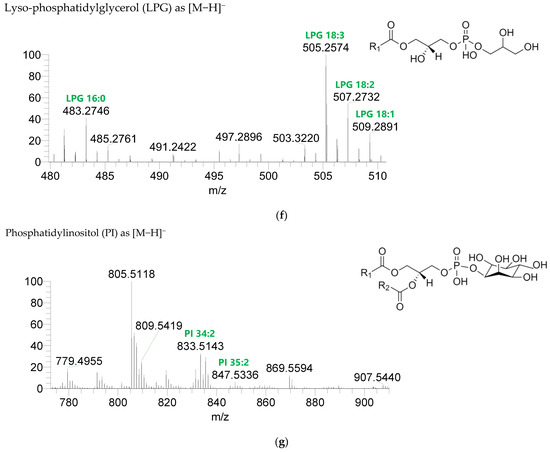
Figure 3.
LC-MS spectra of phospholipids classes identified the polar lipidome of the algae blend. (a) PC and (b) LPC were identified in the positive mode as [M+H]+ ions, and (c) PE, (d) LPE, (e) PG, (f) LPG, and (g) PI were identified in the negative mode as [M-H]- ions. The ions group assigned with symbol (asterisk) are a background.

Table 5.
Phospholipids identified in the polar lipidome of the algae blend (BLEND) by high resolution HILIC-ESI-MS and MS/MS. C carbons, N number of double bonds. * Lipid species identified based on the polar head fragment, calculated mass, and retention time. ** Lipid species identified based on calculated mass and retention time. The presence of the lipid species in the individual algae species—Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida—is indicated by a cross (×), according to the reports in the literature.
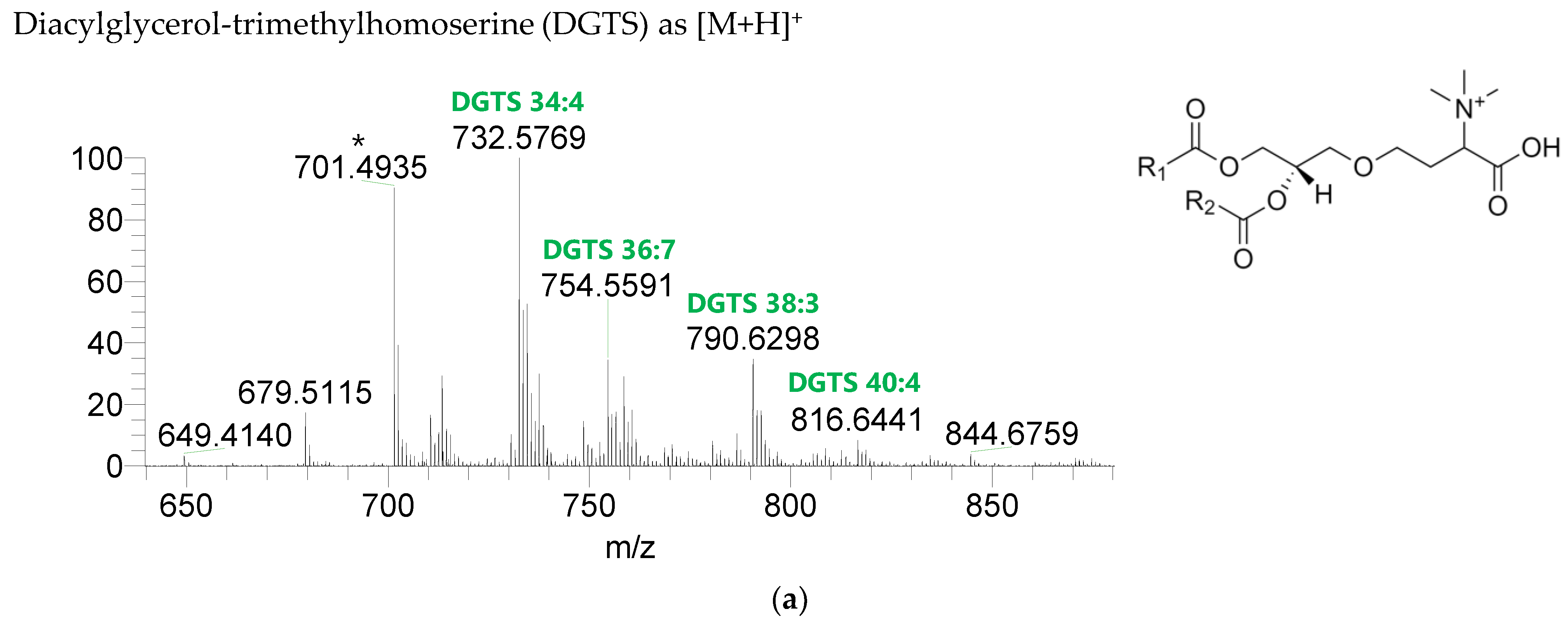
The BL found in the BLEND lipid extract comprised four classes, including diacylglycerol-trimethyl homoserine (DGTS), monoacylglycerol-trimethyl homoserine (MGTS), diacylglyceryl-hydroxymethyl-N,N,N-trimethyl-β-alanine (DGTA), and monoacylglyceryl-hydroxymethyl-N,N,N-trimethyl-β-alanine (MGTA), which were identified in the positive mode as [M+H]+ ions (Figure 4, Table 6, Supplementary Table S3). A total of 61 lipid species of DGTS, 28 of MGTS, 36 of DGTA, and 13 of MGTA were identified. The most abundant species in each class of BL were: DGTS 34:4, assigned as DGTS (16:0_18:4); MGTS 16:0; DGTA 32:1, assigned as DGTS (14:0_18:1) and DGTS (16:0_16:1); and MGTA 18:1. Although C14-, C15-, C17-, C19-, C22-, and C24-fatty acyl chains can be detected esterified in BL species, they were primarily esterified with C16-, C18-, C20-, and C22-fatty acyl chains.
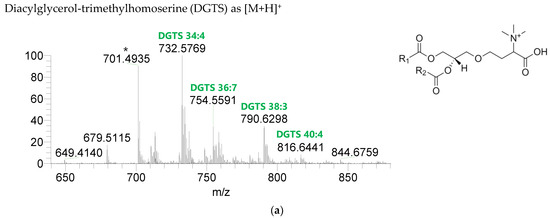
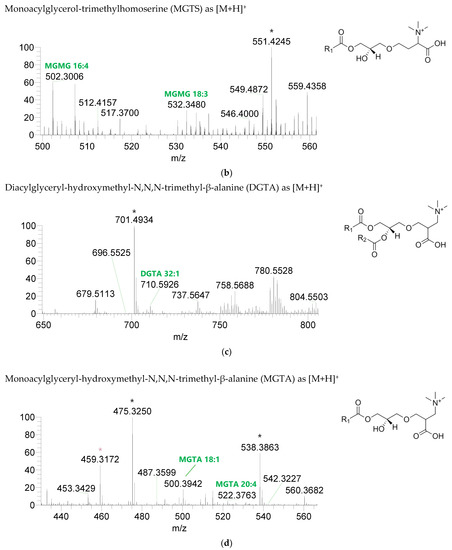
Figure 4.
LC-MS spectra of betaine lipids classes identified the polar lipidome of the algae blend. (a) DGTS, (b) MGTS, (c) DGTA, and (d) MGTA were identified in the positive mode as [M+H]+ ions. The ions group assigned with symbol (asterisk) are a background.

Table 6.
Betaine lipids identified in the polar lipidome of the algae blend (BLEND) by high resolution HILIC–ESI–MS and MS/MS. C carbons, N number of double bonds. * Lipid species identified based on the polar head fragment, calculated mass, and retention time. ** Lipid species identified based on calculated mass and retention time. The presence of the lipid species in the individual algae species—Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida—is indicated by a cross (×), according to the reports in the literature.
The GL species identified in the BLEND lipid extract were already described in C. vulgaris, F. vesiculosus, and/or U. rigida [13,29,30], with a few exceptions (Table 4). The PL fraction showed a vast diversity of phosphoglycerol-containing lipid species, mainly in comparison with the individual algae species (Table 3), with a strong contribution of lipid species from C. vulgaris (Table 5). However, we found a considerable number of PL species that were not described in lipidomic studies targeting C. vulgaris, F. vesiculosus, and U. rigida [9,13,15,16,29,30]. Finally, the arrangement of the three algae gave rise to a more complex BL profile, where DGTS and DGTA are present, as well as their lyso forms (Table 3). DGTS and MGTS are characteristically present in green seaweeds, such as U. rigida [16], and in microalgae only is the occurrence of DGTS more prominent [37]. The majority of the DGTS and MGTS species have been identified in at least one of the isolated algae species constituting the BLEND [9,12,13,14,16,29], mainly U. rigida (Table 6). The presence of DGTA and MGTA in the BLEND lipid extract is clearly derived from F. vesiculosus (Table 6), as these lipids are found most exclusively in Ochrophyta phylum [13,16]. Contrary to what was observed for the PL profile, the BL fraction benefits from the contribution of macroalgae rather than microalgae. Interestingly, BL are non-phosphorous zwitterionic polar glycerolipids analogous of PC and, together with PL, are considered the major polar lipids of cell membranes contributing to the maintenance of membrane architecture and functions due to the positively charged polar heads [38]. Thus, to conserve the charge and properties of membranes, in algae with lower levels of PL, BL are present in a higher amount (as noticed for F. vesiculosus and U. rigida), and vice-versa (as observed for C. vulgaris) [16,37]. Regarding the most abundant lipid species of each lipid class distinguished in the BLEND lipid extract, Table 3 shows that there is a correlation with what was reported for C. vulgaris, F. vesiculosus, and U. rigida.
In fact, some GL, PL, and BL species undetected in the individual algae were present in the BLEND. These lipid species were detected in very low abundances, which could be below the threshold used for lipid analysis in previous studies that identified the individual algae lipidomes. In addition, this could be related to changes in the lipid composition, namely, in terms of relative abundance, that may occur due to the adaptation of each algae to the growth conditions, as micro- and macroalgae samples analyzed in these other studies were not exactly the same ones used to formulate the blend sample under investigation here [13,30]. This incidence can be corroborated by other studies, as in the case of U. rigida, where minority species are not always likely to be identified [9,16,29]. Bear in mind that fluctuations in the lipidome can be observed in both wild and cultivated algae, in the latter to a lesser extent [16,29]. Another influencing factor was the approach to the analysis of the LC-MS data, because in the first studies that focused on the algae lipidome, which were carried out in our laboratory, lipid species with odd carbon numbers were not considered, such as MGDG 35:6, PC 31:4, and SQDG 31:1 [29,39]. Additionally, in the BLEND lipid extract, it was not possible to detect lipid species belonging to the PL classes phosphatidic acid (PA), phosphatidylserine (PS), and lysophosphatidylnositol (LPI). These classes have been identified before in F. vesiculosus (Table 3), but as lower abundant classes. The non-identification of these classes in the BLEND lipid extract could be due to a dilution effect or a suppression effect [27], which could mean that F. vesiculosus represents a minor percentage in the mixture.
It Is also important to point out that some polar lipid species found in C. vulgaris, F. vesiculosus, and/or U. rigida were not identified in the BLEND lipid extract. These lipid species were detected in very low abundance in the individual algae species [16]; therefore, in our study, signal suppression may have occurred, and the detection limit may not have been reached [27].
To sum up, the combination of C. vulgaris, F. vesiculosus, and U. rigida provides a single polar lipid profile, with a greater diversity of lipid species. The present identification of the polar lipids contained in the BLEND lipid extract is crucial to suit specific applications, as the numerous combinations of different acyl chains with the polar head give rise to GL, PL, and BL species with distinct properties and functions [26,37].
Marine GL have been exploited and characterized for being natural compounds with an extensive variety of molecular structures and biological activities, such as anti-inflammatory, antiproliferative, and antimicrobial [6]. The potential of the BLEND total lipid extract for biotechnological purposes is enhanced by the occurrence of several GL molecular species previously associated with bioactivity and potential health benefits of lipids. For instance, in the case of galactolipids, MGDG 34:4 (18:4_16:0) [40], MGDG 34:8 (18:4_16:4) [41], and MGMG 16:3 [42] displayed strong and dose-dependent nitric oxide inhibition, indicative of their potential as anti-inflammatory agents. MGDG 34:4 (18:4_16:0) and DGDG 34:4 (18:4_16:0) tested in two in vivo mouse models of inflammation showed anti-inflammatory activities and substantially lower toxicities than that of the reference treatments [43]. Antitumor properties have been unveiled in MGDG 32:4 (16:2/16:2), MGDG 32:5 (16:2_16:3), MGDG 34:4 (18:2_16:2), MGDG 34:6 (18:3_16:3), MGDG 36:4 (18:2/18:2), DGDG 34:4 (16:2_18:2), and DGDG 34:6 (16:3_18:3) from C. vulgaris [44], in MGDG 38:9 (18:4_20:5) from Fucus evanescens [45], and in MGDG 36:4 (20:4_16:0) from Hydrolithon reinboldii [46]. MGDG species with LA at the sn-2 position, as MGDG 32:2 (14:0/18:2) and MGDG 34:2 (16:0/18:2), have been suggested to play an important role in the inhibition of triglyceride accumulation in 3T3-L1 adipocytes [47]. Of particular note are the most abundant species of each sulfolipids class SQDG 34:1 (16:0_18:1) and SQMG 16:0, which have shown great antitumor activity against human carcinomas and antibacterial effect against Bacillus subtilis and Escherichia coli [48]. In addition, a study on Chlorococcum sp. suggested that SQDG bearing the bioactive fatty acid ALA or OA at the sn-2 position of their structure, as SQDG 34:1 (16:0/18:1) and SQDG 32:1 (14:0/18:1), are associated with strong anti-inflammatory and antithrombotic properties [20]. Meanwhile, SQDG 32:0 (16:0/16:0) isolated from a brown seaweed demonstrated potential as an antiviral agent against human herpesviruses [49]. The GL fraction of the BLEND appears to be a strong contender for a healthy diet, namely, as a more efficient bioavailable carrier of beneficial fatty acids, contributing to functional foods and ingredients, and a promising source of therapeutic agents intended for nutraceutical applications [6,19]. Alternatively, these bio-based and biodegradable food-grade compounds may be of interest as biosurfactants to prepare emulsions in food products or to develop drug delivery systems [50].
PL include a variety of versatile lipid species that are required in a wide range of biological processes. Studies on bioactivities of PL from algae are quite scarce [6]. Among the lipid species identified in the BLEND total lipid extract, LPC 16:0 and PG 34:2 (16:0_18:2) have already shown potential anti-inflammatory action [43,51]. Moreover, a body of evidence suggests that dietary marine PL have numerous health benefits, with a positive impact in several diseases—for example, age-related, cardiovascular, cognitive, and neurological disorders—especially PL esterified to n-3 PUFA [26,52]. The PL fraction of the BLEND is enriched in lipid species esterified to healthy fatty acids, such as 18:1, 18:2, and 18:3, and because marine PL products showed remarkably high stability against oxidation and better bioavailability [52,53], the application of this lipid fraction as ingredients for functional food, namely, food fortification, is promising [53]. Effectively, PL have been regularly and extensively used in a large set of industries. In short, the PL of the BLEND could be explored in the food (emulsifiers and fat-replacements), pharmaceutical (drug delivery systems), and cosmetic (emulsifiers and rehydrating agents for skincare) industries [26], as well as in neutron scattering research (mimics cell membranes) [54].
Although little is known about the BL bioactive potential, the molecular species MGTS 20:5 and DGTS 40:10 (20:5/20:5), detected in the BLEND lipid extract, were respectively described with antiatherogenic and anti-inflammatory activity [55,56]. More studies are needed to understand the bioactive potential of these abundant algae lipids.
3.3. Antioxidant Activity of the Algae Blend Total Lipid Extract
The antioxidant activity of the BLEND total lipid extract was evaluated using the ABTS•+ and DPPH• scavenging assays (Table 7). The percentage of radical inhibition in the presence of the lipid extract was calculated after 120 min. An inhibition of 87.64 ± 2.29% of ABTS•+ and 37.98 ± 0.65% of DPPH• was reached with the maximum tested concentration (250 μg mL−1). The concentration that provided 50% of ABTS•+ inhibition (IC50) was 140.01 ± 4.40 μg mL−1 with a TE of 126.56 ± 3.91 μmol Trolox g−1, while for DPPH• assay, 35% of inhibition (IC35) was attained at a concentration of 230.62 ± 3.30 μg mL−1, representing a TE of 74.09 ± 1.07 μmol Trolox g−1.

Table 7.
Lipid extract concentration (μg mL−1) of the algae blend (BLEND) that provided 50% (IC50) inhibition of the ABTS•+, and 35% (IC35) and 20% (IC20) inhibition of the DPPH•, in comparison with the values reported in the literature for the individual algae species—Chlorella vulgaris, Fucus vesiculosus, and Ulva rigida. Values are expressed as the mean ± SD (n = 3).
Natural compounds with antioxidant activity are currently of interest mainly in the food and pharmaceutical industries [22]. Algae total lipid extracts contain lipids that have already been described as powerful antioxidants, such as GL, PL, pigments, and PUFA [15,57,58]. Therefore, recent studies have been focused on the antioxidant potential of total lipid extracts from different algae species [7,8,17,59,60]. The ABTS•+ and DPPH• scavenging assays are chemical methods widely used to measure the antioxidant capacity of natural extracts, including algae and derived compounds, due to their low cost, operational simplicity, and radical stability, in spite of their biological irrelevance [60,61,62]. In our work, the BLEND lipid extract showed higher ability to scavenge the ABTS•+ than the DPPH•, as reported for natural extracts from fruits, vegetables, and algae [17,61], including C. vulgaris, F. vesiculosus, and U. rigida (Table 6). Effectively, our results are in agreement with those obtained for the individual algae species and with previous studies on other algae [7,59,60]. Overall, the BLEND total lipid extract showed antioxidant activity and can be used as a natural antioxidant, valued for promoting environmental and economic sustainability [2].
4. Conclusions
In this study, the fatty acid profile and the polar lipidome of an innovative blend of macro- and microalgae (BLEND) were determined for the first time. The essential fatty acids LA (18:2 n-6) and ALA (18:3 n-3) were among the most abundant fatty acids in the lipid pool, and the calculated lipid indicators included a well-balanced n-6/n-3 ratio and low AI and TI, which betoken a great nutritional value. Through a HILIC–ESI–MS/MS lipidomic approach, we identified 462 species of polar lipids, including glycolipids, phospholipids, and betaine lipids, some of which have already been recognized for their bioactive potential. The mixture of different macro- and microalgae species resulted in a blend characterized by a unique lipid profile, which could not be achieved by a single algae species. In addition, the BLEND total lipid extract displayed antioxidant activity. Ultimately, the mixture of different algae species can be an approach to achieve products that comprise more benefits privileging health, nutrition, and environmental sustainability. However, further studies are required. Nonetheless, this study acknowledges the BLEND as a promising sustainable source of natural bioactive lipid compounds with potential application, namely, in the food, nutraceutical, and cosmetic industries.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life13010231/s1, Figure S1. Calibration curve for the ABTS•+ scavenging assay as generated by measuring the absorbance of the reaction medium at 734 nm after 120 min. Trolox was used as standard. Figure S2. Calibration curve for the DPPH• scavenging assay as generated by measuring the absorbance of the reaction medium at 517 nm after 120 min. Trolox was used as standard. Table S1: Glycolipids identified in the polar lipidome of the algae blend (BLEND) by high resolution HILIC-ESI-MS and MS/MS. C carbons, N number of double bonds. * Lipid species identified based on the polar head fragment, calculated mass and retention time. ** Lipid species identified based on calculated mass and retention time. Table S2: Phospholipids identified in the polar lipidome of the algae blend (BLEND) by high resolution HILIC–ESI–MS and MS/MS. C carbons, N number of double bonds. * Lipid species identified based on the polar head fragment, calculated mass and retention time. ** Lipid species identified based on calculated mass and retention time. Table S3: Betaine lipids identified in the polar lipidome of the algae blend (BLEND) by high resolution HILIC-ESI-MS and MS/MS. C carbons, N number of double bonds. * Lipid species identified based on the polar head fragment, calculated mass and retention time. ** Lipid species identified based on calculated mass and retention time.
Author Contributions
Conceptualization, F.M. and M.R.D.; methodology, T.M., P.D. and M.R.D.; validation, D.L., T.M. and M.R.D.; formal analysis, F.M. and M.R.D.; investigation, F.M., D.L., T.C. and T.M.; resources, J.S., M.H.A., P.D. and M.R.D.; writing-original draft preparation, F.M.; writing-review and editing, all authors; supervision, M.R.D.; project administration, M.R.D.; funding acquisition, M.R.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundação para a Ciência e Tecnologia: 2021.12131.BD; Fundação para a Ciência e Tecnologia: 2020.05678.BD; Fundação para a Ciência e Tecnologia: CESAM- UIDB/50017/2020 + UIDP/50017/2020 + LA/P/0094/2020; Fundação para a Ciência e Tecnologia: REQIMTE-LAQV UIDP/50006/2020 + UIDB/50006/2020; FCT - Fundação para a Ciência e a Tecnologia, within the PT2020 Partnership Agreement and Compete 2020 co-funded by the FEDER—Fundo Europeu de Desenvolvimento Regional: RNEM- LISBOA-01-0145-FEDER-402-022125.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Thanks are due to LAQV/REQUIMTE (UIDP/50006/2020 + UIDB/50006/2020), CESAM (UIDB/50017/2020 + UIDP/50017/2020 + LA/P/0094/2020), RNEM, the Portuguese Mass Spectrometry Network, (LISBOA-01-0145-FEDER-402-022125) through national funds and, where applicable, co-financed by the FEDER, within the PT2020 Partnership Agreement and Compete 2020. The authors are also thankful to the COST Action EpiLipidNET, CA19105—Pan-European Network in Lipidomics and EpiLipidomics and to the COST Action CA20106—TOMORROW’S ‘WHEAT OF THE SEA’: ULVA, A MODEL FOR AN INNOVATIVE MARICULTURE (SEAWHEAT). Francisca Marques (2021.12131.BD) and Tiago Alexandre Conde (2020.05678.BD) are grateful to FCT for their grants. Tânia Melo thanks the Junior Researcher contract in the scope of the Individual Call to Scientific Employment Stimulus 2020 [CEECIND/01578/2020].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Araújo, R.; Vázquez Calderón, F.; Sánchez López, J.; Azevedo, I.C.; Bruhn, A.; Fluch, S.; Garcia Tasende, M.; Ghaderiardakani, F.; Ilmjärv, T.; Laurans, M.; et al. Current Status of the Algae Production Industry in Europe: An Emerging Sector of the Blue Bioeconomy. Front. Mar. Sci. 2021, 7, 1–24. [Google Scholar] [CrossRef]
- Ullmann, J.; Grimm, D. Algae and Their Potential for a Future Bioeconomy, Landless Food Production, and the Socio-Economic Impact of an Algae Industry. Org. Agric. 2021, 11, 261–267. [Google Scholar] [CrossRef]
- Slegers, P.M.; Helmes, R.J.K.; Draisma, M.; Broekema, R.; Vlottes, M.; van den Burg, S.W.K. Environmental Impact and Nutritional Value of Food Products Using the Seaweed Saccharina latissima. J. Clean. Prod. 2021, 319, 1–11. [Google Scholar] [CrossRef]
- European Commission Directorate General for Maritime Affairs and Fisheries; Addamo, A.M.; Calvo Santos, A.; Guillén, J.; Neehus, S.; Peralta Baptista, A.; Quatrini, S.; Telsnig, T.; Petrucco, G. The EU Blue Economy Report 2022; Publications Office of the European Union, 2022. [Google Scholar]
- Torres, M.D.; Flórez-Fernández, N.; Domínguez, H. Integral Utilization of Red Seaweed for Bioactive Production. Mar. Drugs 2019, 17, 314. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.; Rey, F.; Leal, M.C.; Lillebø, A.I.; Calado, R.; Domingues, M.R. Bioactivities of Lipid Extracts and Complex Lipids from Seaweeds: Current Knowledge and Future Prospects. Mar. Drugs 2021, 19, 686. [Google Scholar] [CrossRef]
- Conde, T.A.; Neves, B.F.; Couto, D.; Melo, T.; Neves, B.; Costa, M.; Silva, J.; Domingues, P.; Domingues, M.R. Microalgae as Sustainable Bio-Factories of Healthy Lipids: Evaluating Fatty Acid Content and Antioxidant Activity. Mar. Drugs 2021, 19, 357. [Google Scholar] [CrossRef]
- Rey, F.; Cartaxana, P.; Melo, T.; Calado, R.; Pereira, R.; Abreu, H.; Domingues, P.; Cruz, S.; Domingues, M.R. Domesticated Populations of Codium tomentosum Display Lipid Extracts with Lower Seasonal Shifts than Conspecifics from the Wild—Relevance for Biotechnological Applications of This Green Seaweed. Mar. Drugs 2020, 18, 188. [Google Scholar] [CrossRef]
- Moreira, A.S.P.; da Costa, E.; Melo, T.; Lopes, D.; Pais, A.; Santos, S.; Pitarma, B.; Mendes, M.; Abreu, M.H.; Collén, P.N.; et al. Polar Lipids of Commercial Ulva spp. of Different Origins: Profiling and Relevance for Seaweed Valorization. Foods 2021, 10, 914. [Google Scholar] [CrossRef]
- Susanto, E.; Fahmi, A.S.; Hosokawa, M.; Miyashita, K. Variation in Lipid Components from 15 Species of Tropical and Temperate Seaweeds. Mar. Drugs 2019, 17, 630. [Google Scholar] [CrossRef]
- Nascimento, I.A.; Marques, S.S.I.; Cabanelas, I.T.D.; Pereira, S.A.; Druzian, J.I.; de Souza, C.O.; Vich, D.V.; de Carvalho, G.C.; Nascimento, M.A. Screening Microalgae Strains for Biodiesel Production: Lipid Productivity and Estimation of Fuel Quality Based on Fatty Acids Profiles as Selective Criteria. BioEnergy Res. 2013, 6, 1–13. [Google Scholar] [CrossRef]
- da Costa, E.; Ricardo, F.; Melo, T.; Mamede, R.; Abreu, M.H.; Domingues, P.; Domingues, M.R.; Calado, R. Site-Specific Lipidomic Signatures of Sea Lettuce (Ulva spp., Chlorophyta) Hold the Potential to Trace Their Geographic Origin. Biomolecules 2020, 10, 489. [Google Scholar] [CrossRef]
- da Costa, E.; Domingues, P.; Melo, T.; Coelho, E.; Pereira, R.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomic Signatures Reveal Seasonal Shifts on the Relative Abundance of High-Valued Lipids from the Brown Algae Fucus vesiculosus. Mar. Drugs 2019, 17, 335. [Google Scholar] [CrossRef]
- White, D.; Rooks, P.; Kimmance, S.; Tait, K.; Jones, M.; Tarran, G.; Cook, C.; Llewellyn, C. Modulation of Polar Lipid Profiles in Chlorella sp. in Response to Nutrient Limitation. Metabolites 2019, 9, 39. [Google Scholar] [CrossRef]
- Couto, D.; Melo, T.; Conde, T.A.; Moreira, A.S.P.; Ferreira, P.; Costa, M.; Silva, J.; Domingues, R.; Domingues, P. Food Grade Extraction of Chlorella vulgaris Polar Lipids: A Comparative Lipidomic Study. Food Chem. 2022, 375, 1–10. [Google Scholar] [CrossRef]
- Lopes, D.; Melo, T.; Rey, F.; Costa, E.; Moreira, A.S.P.; Abreu, M.H.; Domingues, P.; Lillebø, A.I.; Calado, R.; Domingues, M.R. Insights of Species-Specific Polar Lipidome Signatures of Seaweeds Fostering Their Valorization in the Blue Bioeconomy. Algal Res. 2021, 55, 1–16. [Google Scholar] [CrossRef]
- Lopes, D.; Melo, T.; Rey, F.; Meneses, J.; Monteiro, F.L.; Helguero, L.A.; Abreu, M.H.; Lillebø, A.I.; Calado, R.; Domingues, M.R. Valuing Bioactive Lipids from Green, Red and Brown Macroalgae from Aquaculture, to Foster Functionality and Biotechnological Applications. Molecules 2020, 25, 3883. [Google Scholar] [CrossRef]
- Ghosh, S.; Sarkar, T.; Pati, S.; Kari, Z.A.; Edinur, H.A.; Chakraborty, R. Novel Bioactive Compounds From Marine Sources as a Tool for Functional Food Development. Front. Mar. Sci. 2022, 9, 1–28. [Google Scholar] [CrossRef]
- Kagan, M.L.; Levy, A.; Leikin-Frenkel, A. Comparative Study of Tissue Deposition of Omega-3 Fatty Acids from Polar-Lipid Rich Oil of the Microalgae Nannochloropsis oculata with Krill Oil in Rats. Food Funct. 2015, 6, 185–191. [Google Scholar] [CrossRef]
- Shiels, K.; Tsoupras, A.; Lordan, R.; Nasopoulou, C.; Zabetakis, I.; Murray, P.; Saha, S.K. Bioactive Lipids of Marine Microalga Chlorococcum sp. SABC 012504 with Anti-Inflammatory and Anti-Thrombotic Activities. Mar. Drugs 2021, 19, 28. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.P.; Craigie, J.S.; Hafting, J.T.; Critchley, A.T. Polar Lipids from the Marine Macroalga Palmaria palmata Inhibit Lipopolysaccharide-Induced Nitric Oxide Production in RAW264.7 Macrophage Cells. Phytochemistry 2014, 101, 101–108. [Google Scholar] [CrossRef]
- Mendonça, J.d.S.; Guimarães, R.d.C.A.; Zorgetto-Pinheiro, V.A.; Fernandes, C.D.P.; Marcelino, G.; Bogo, D.; Freitas, K.d.C.; Hiane, P.A.; de Pádua Melo, E.S.; Vilela, M.L.B.; et al. Natural Antioxidant Evaluation: A Review of Detection Methods. Molecules 2022, 27, 3563. [Google Scholar] [CrossRef] [PubMed]
- Abd El Baky, H.; El-Baroty, G.S.A.; Ibrahim, E.A.; El-Baz, F.K. Cytotoxicity, Antioxidants and Antimicrobial Activities of Lipids Extracted from Some Marine Algae. J. Aquac. Res. Dev. 2014, 5, 7. [Google Scholar] [CrossRef]
- Losada-Lopez, C.; Dopico, D.C.; Faína-Medín, J.A. Neophobia and Seaweed Consumption: Effects on Consumer Attitude and Willingness to Consume Seaweed. Int. J. Gastron. Food Sci. 2021, 24, 1–9. [Google Scholar] [CrossRef]
- Marques, F.; Lopes, D.; da Costa, E.; Conde, T.; Rego, A.; Ribeiro, A.I.; Abreu, M.H.; Domingues, M.R. Seaweed Blends as a Valuable Source of Polyunsaturated and Healthy Fats for Nutritional and Food Applications. Mar. Drugs 2021, 19, 684. [Google Scholar] [CrossRef] [PubMed]
- Haq, M.; Suraiya, S.; Ahmed, S.; Chun, B.-S. Phospholipids from Marine Source: Extractions and Forthcoming Industrial Applications. J. Funct. Foods 2021, 80, 1–13. [Google Scholar] [CrossRef]
- Rey, F.; Melo, T.; Lopes, D.; Couto, D.; Marques, F.; Rosário, M. Applications of Lipidomics in Marine Organisms: Progresses, Challenges and Future Perspectives. Mol. Omics 2022, 18, 357–386. [Google Scholar] [CrossRef]
- Neto, R.; Marçal, C.; Queirós, A.; Abreu, H.; Silva, A.; Cardoso, S. Screening of Ulva rigida, Gracilaria sp., Fucus vesiculosus and Saccharina latissima as Functional Ingredients. Int. J. Mol. Sci. 2018, 19, 2987. [Google Scholar] [CrossRef]
- Lopes, D.; Moreira, A.S.P.; Rey, F.; da Costa, E.; Melo, T.; Maciel, E.; Rego, A.; Abreu, M.H.; Domingues, P.; Calado, R.; et al. Lipidomic Signature of the Green Macroalgae Ulva rigida Farmed in a Sustainable Integrated Multi-Trophic Aquaculture. J. Appl. Phycol. 2019, 31, 1369–1381. [Google Scholar] [CrossRef]
- Couto, D.; Melo, T.; Conde, T.A.; Costa, M.; Silva, J.; Domingues, M.R.M.; Domingues, P. Chemoplasticity of the Polar Lipid Profile of the Microalgae Chlorella vulgaris Grown under Heterotrophic and Autotrophic Conditions. Algal Res. 2021, 53, 1–15. [Google Scholar] [CrossRef]
- Echave, J.; Lourenço-Lopes, C.; Carreira-Casais, A.; Chamorro, F.; Fraga-Corral, M.; Otero, P.; Garcia-Perez, P.; Baamonde, S.; Fernández-Saa, F.; Cao, H.; et al. Nutritional Composition of the Atlantic Seaweeds Ulva rigida, Codium tomentosum, Palmaria palmata and Porphyra purpurea. Chem. Proc. 2021, 5, 67. [Google Scholar] [CrossRef]
- Farag, M.A.; Gad, M.Z. Omega-9 Fatty Acids: Potential Roles in Inflammation and Cancer Management. J. Genet. Eng. Biotechnol. 2022, 20, 1–11. [Google Scholar] [CrossRef]
- Monnard, C.R.; Dulloo, A.G. Polyunsaturated Fatty Acids as Modulators of Fat Mass and Lean Mass in Human Body Composition Regulation and Cardiometabolic Health. Obes. Rev. 2021, 22, 1–18. [Google Scholar] [CrossRef]
- Hiki, M.; Miyazaki, T.; Shimada, K.; Sugita, Y.; Shimizu, M.; Aikawa, T.; Ouchi, S.; Shiozawa, T.; Takasu, K.; Takahashi, S.; et al. Significance of Serum Polyunsaturated Fatty Acid Level Imbalance in Patients with Acute Venous Thromboembolism. J. Atheroscler. Thromb. 2017, 24, 1016–1022. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H. Nutritional Indices for Assessing Fatty Acids: A Mini-Review. Int. J. Mol. Sci. 2020, 21, 5695. [Google Scholar] [CrossRef]
- Couto, D.; Conde, T.A.; Melo, T.; Neves, B.; Costa, M.; Silva, J.; Domingues, M.R.; Domingues, P. The Chemodiversity of Polar Lipidomes of Different Taxa of Commercial Microalgae. Algal Res. 2023; under review. [Google Scholar]
- Kato, M.; Sakai, M.; Adachi, K.; Ikemoto, H.; Sano, H. Distribution of Betaine Lipids in Marine Algae. Phytochemistry 1996, 42, 1341–1345. [Google Scholar] [CrossRef]
- Riekhof, W.R.; Andre, C.; Benning, C. Two Enzymes, BtaA and BtaB, Are Sufficient for Betaine Lipid Biosynthesis in Bacteria. Arch. Biochem. Biophys. 2005, 441, 96–105. [Google Scholar] [CrossRef]
- Lopes, D.; Melo, T.; Meneses, J.; Abreu, M.H.; Pereira, R.; Domingues, P.; Lillebø, A.I.; Calado, R.; Domingues, M.R. A New Look for the Red Macroalga Palmaria palmata: A Seafood with Polar Lipids Rich in EPA and with Antioxidant Properties. Mar. Drugs 2019, 17, 533. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.; Craigie, J.S.; Hafting, J.T. Lipids Isolated from the Cultivated Red Alga Chondrus crispus Inhibit Nitric Oxide Production. J. Appl. Phycol. 2014, 26, 1565–1571. [Google Scholar] [CrossRef]
- Banskota, A.H.; Gallant, P.; Stefanova, R.; Melanson, R.; O’Leary, S.J.B. Monogalactosyldiacylglycerols, Potent Nitric Oxide Inhibitors from the Marine Microalga Tetraselmis chui. Nat. Prod. Res. 2013, 27, 1084–1090. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Gallant, P.; Osborne, J.A.; Melanson, R.; O’Leary, S.J.B. Nitric Oxide Inhibitory Activity of Monogalactosylmonoacylglycerols from a Freshwater Microalgae Chlorella sorokiniana. Nat. Prod. Res. 2013, 27, 1028–1031. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Rossi, C.; Marcolongo, G.; Di Lena, A.; Venzo, A.; Berrie, C.P.; Corda, D. Selective In Vivo Anti-Inflammatory Action of the Galactolipid Monogalactosyldiacylglycerol. Eur. J. Pharmacol. 2005, 524, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, T.; Nagatsu, A.; Murakami, N.; Sakakibara, J.; Tokuda, H.; Nishino, H.; Iwashima, A. Anti-Tumour-Promoting Glyceroglycolipids from the Green Alga, Chlorella vulgaris. Phytochemistry 1995, 40, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Imbs, T.I.; Ermakova, S.P.; Fedoreyev, S.A.; Anastyuk, S.D.; Zvyagintseva, T.N. Isolation of Fucoxanthin and Highly Unsaturated Monogalactosyldiacylglycerol from Brown Alga Fucus evanescens C Agardh and In Vitro Investigation of Their Antitumor Activity. Mar. Biotechnol. 2013, 15, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.-W.; Hay, M.E.; Fairchild, C.R.; Prudhomme, J.; Roch, K.L.; Aalbersberg, W.; Kubanek, J. Antineoplastic Unsaturated Fatty Acids from Fijian Macroalgae. Phytochemistry 2008, 69, 2495–2500. [Google Scholar] [CrossRef]
- Ma, A.-C.; Chen, Z.; Wang, T.; Song, N.; Yan, Q.; Fang, Y.-C.; Guan, H.-S.; Liu, H.-B. Isolation of the Molecular Species of Monogalactosyldiacylglycerols from Brown Edible Seaweed Sargassum horneri and Their Inhibitory Effects on Triglyceride Accumulation in 3T3-L1 Adipocytes. J. Agric. Food Chem. 2014, 62, 11157–11162. [Google Scholar] [CrossRef]
- Baz, F.K.E.; Baroty, G.S.E.; Baky, H.H.A.E.; El-Salam, O.I.A.; Ibrahim, E.A. Structural Characterization and Biological Activity of Sulfolipids from Selected Marine Algae. Grasas Aceites 2013, 64, 561–571. [Google Scholar] [CrossRef]
- Plouguerné, E.; De Souza, L.M.; Sassaki, G.L.; Cavalcanti, J.F.; Villela Romanos, M.T.; Da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Antiviral Sulfoquinovosyldiacylglycerols (SQDGs) from the Brazilian Brown Seaweed Sargassum vulgare. Mar. Drugs 2013, 11, 4628–4640. [Google Scholar] [CrossRef]
- Azevedo, M.A.; Cerqueira, M.A.; Fuciños, P.; Silva, B.F.B.; Teixeira, J.A.; Pastrana, L. Rhamnolipids-Based Nanostructured Lipid Carriers: Effect of Lipid Phase on Physicochemical Properties and Stability. Food Chem. 2021, 344, 1–9. [Google Scholar] [CrossRef]
- Lauritano, C.; Helland, K.; Riccio, G.; Andersen, J.H.; Ianora, A.; Hansen, E.H. Lysophosphatidylcholines and Chlorophyll-Derived Molecules from the Diatom Cylindrotheca closterium with Anti-Inflammatory Activity. Mar. Drugs 2020, 18, 166. [Google Scholar] [CrossRef]
- Tang, X.; Li, Z.-J.; Xu, J.; Xue, Y.; Li, J.-Z.; Wang, J.-F.; Yanagita, T.; Xue, C.-H.; Wang, Y.-M. Short Term Effects of Different Omega-3 Fatty Acid Formulation on Lipid Metabolism in Mice Fed High or Low Fat Diet. Lipids Health Dis. 2012, 11, 1–8. [Google Scholar] [CrossRef]
- Lu, F.S.H.; Nielsen, N.S.; Baron, C.P.; Jacobsen, C. Marine Phospholipids: The Current Understanding of Their Oxidation Mechanisms and Potential Uses for Food Fortification. Crit. Rev. Food Sci. Nutr. 2017, 57, 2057–2070. [Google Scholar] [CrossRef]
- Holderer, O.; Koutsioubas, A.; Garvey, C.J.; Nylander, T. Editorial: Membrane Structure and Dynamics Studied With Neutron Scattering. Front. Chem. 2021, 9, 1–2. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; McGinn, P.J. New Diacylglyceryltrimethylhomoserines from the Marine Microalga Nannochloropsis granulata and Their Nitric Oxide Inhibitory Activity. J. Appl. Phycol. 2013, 25, 1513–1521. [Google Scholar] [CrossRef]
- Khatib, S.; Artoul, F.; Paluy, I.; Boluchevsky, L.; Kvitnitsky, E.; Vaya, J. Nannochloropsis Sp. Ethanol Extract Prevents Macrophage and LDL Oxidation and Enhances PON1 Activity through the Principal Active Compound Lyso-Diacylglyceryltrimethylhomoserine (Lyso-DGTS). J. Appl. Phycol. 2018, 30, 1679–1689. [Google Scholar] [CrossRef]
- Terme, N.; Boulho, R.; Kucma, J.-P.; Bourgougnon, N.; Bedoux, G. Radical Scavenging Activity of Lipids from Seaweeds Isolated by Solid-Liquid Extraction and Supercritical Fluids. OCL 2018, 25, 1–6. [Google Scholar] [CrossRef]
- Richard, D.; Kefi, K.; Barbe, U.; Bausero, P.; Visioli, F. Polyunsaturated Fatty Acids as Antioxidants. Pharmacol. Res. 2008, 57, 451–455. [Google Scholar] [CrossRef]
- da Costa, E.; Melo, T.; Reis, M.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingues, M.R. Polar Lipids Composition, Antioxidant and Anti-Inflammatory Activities of the Atlantic Red Seaweed Grateloupia turuturu. Mar. Drugs 2021, 19, 414. [Google Scholar] [CrossRef]
- Santos, F.; Monteiro, J.P.; Duarte, D.; Melo, T.; Lopes, D.; da Costa, E.; Domingues, M.R. Unraveling the Lipidome and Antioxidant Activity of Native Bifurcaria bifurcata and Invasive Sargassum muticum Seaweeds: A Lipid Perspective on How Systemic Intrusion May Present an Opportunity. Antioxidants 2020, 9, 642. [Google Scholar] [CrossRef]
- Floegel, A. Comparison of ABTS/DPPH Assays to Measure Antioxidant Capacity in Popular Antioxidant-Rich US Foods. J. Food Compos. Anal. 2011, 24, 1043–1048. [Google Scholar] [CrossRef]
- Monteiro, M.; Santos, R.A.; Iglesias, P.; Couto, A.; Serra, C.R.; Gouvinhas, I.; Barros, A.; Oliva-Teles, A.; Enes, P.; Díaz-Rosales, P. Effect of Extraction Method and Solvent System on the Phenolic Content and Antioxidant Activity of Selected Macro- and Microalgae Extracts. J. Appl. Phycol. 2020, 32, 349–362. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).