Weakly Supervised Collaborative Learning for Airborne Pollen Segmentation and Classification from SEM Images
Abstract
:1. Introduction
- To eliminate the interference of noise in the classification model and improve the pollen classification performance, a weakly supervised collaborative learning method for pollen segmentation and classification is proposed in this paper. We input the segmented pollen region into the classification network to avoid the interference of impurities in the classification model, thus obtaining an accurate pollen activation region. This refined activation region was used to optimize the segmentation labels to improve the segmentation performance. The segmentation and classification models were collaboratively optimized to achieve the optimal performance simultaneously.
- To solve the problem of time-consuming and laborious annotation task, a weakly supervised pollen segmentation method is proposed with only the image-level annotation data. We combined digital image processing (based on the pollen contour and size selection) and gradient-weighted class-activation mapping to generate pollen segmentation pseudo labels. This method only requires image-level labels to obtain a segmentation network with superior performance.
- Numerous experiments on real-world SEM pollen images were conducted, and the results showed that our approach achieved satisfactory performance. An accuracy of 86.6% was achieved in the pollen classification task, and an MIoU of 92.47% was achieved in the segmentation task.
2. Materials and Methods
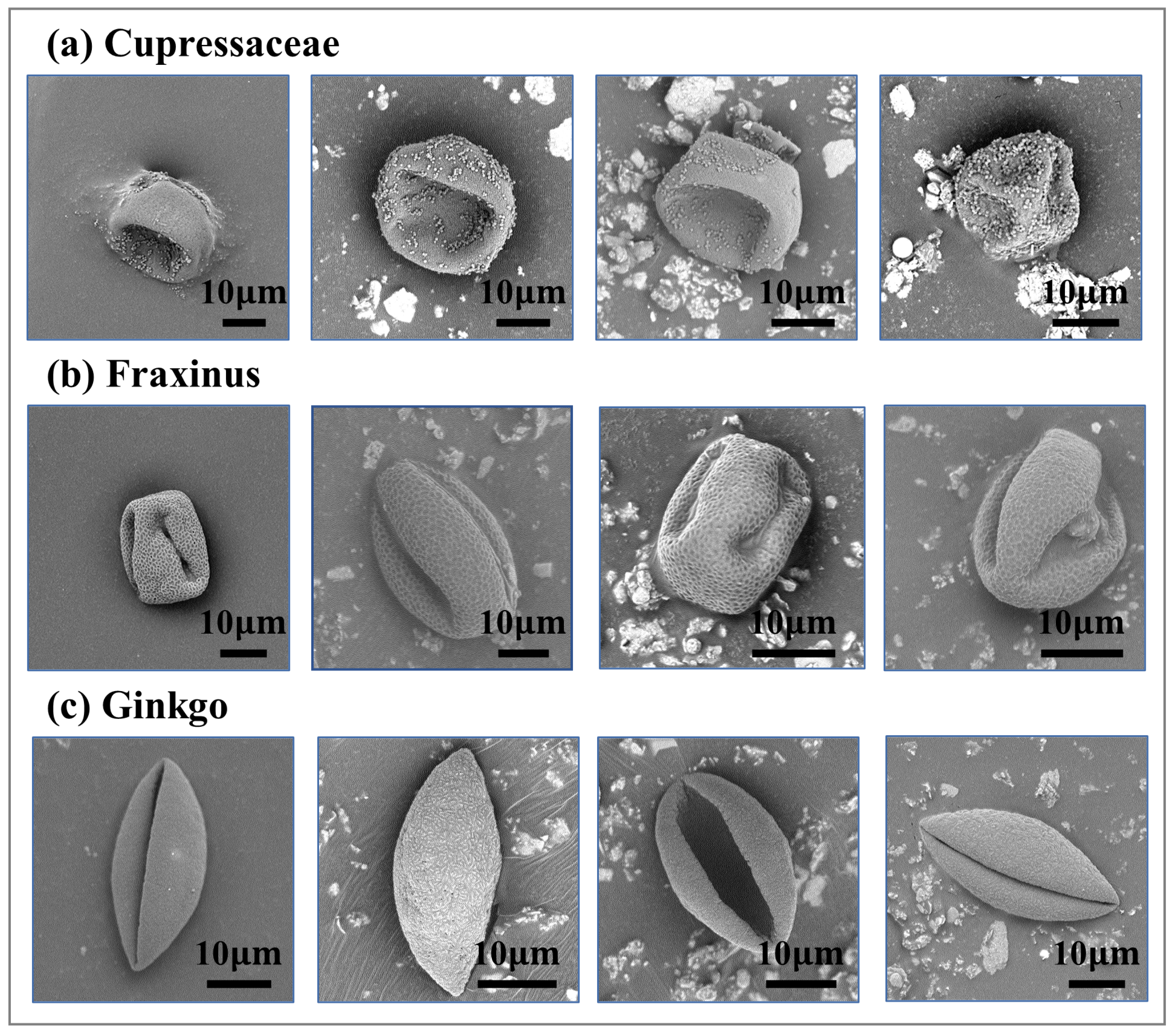
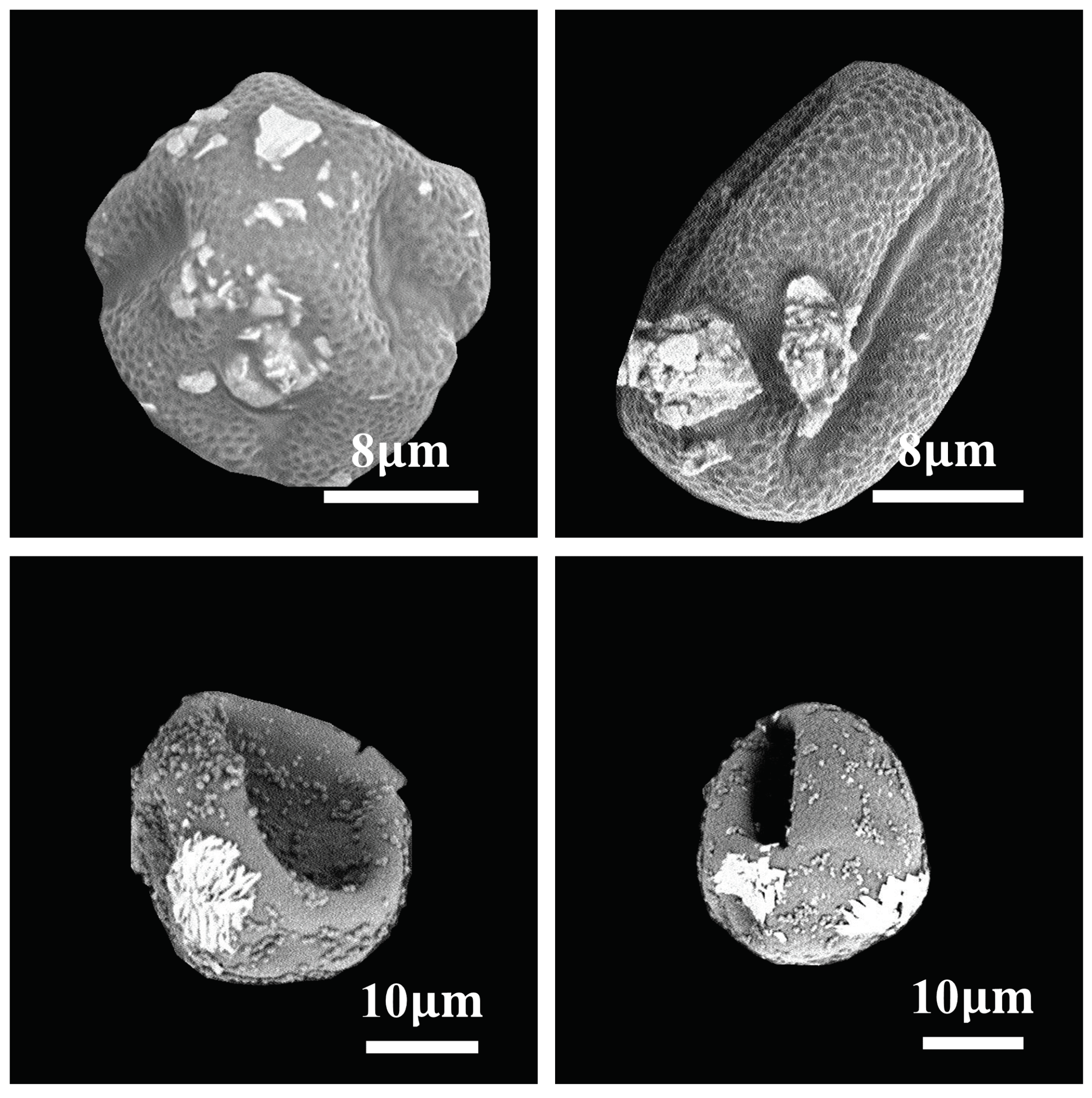
2.1. Dataset and Preprocessing
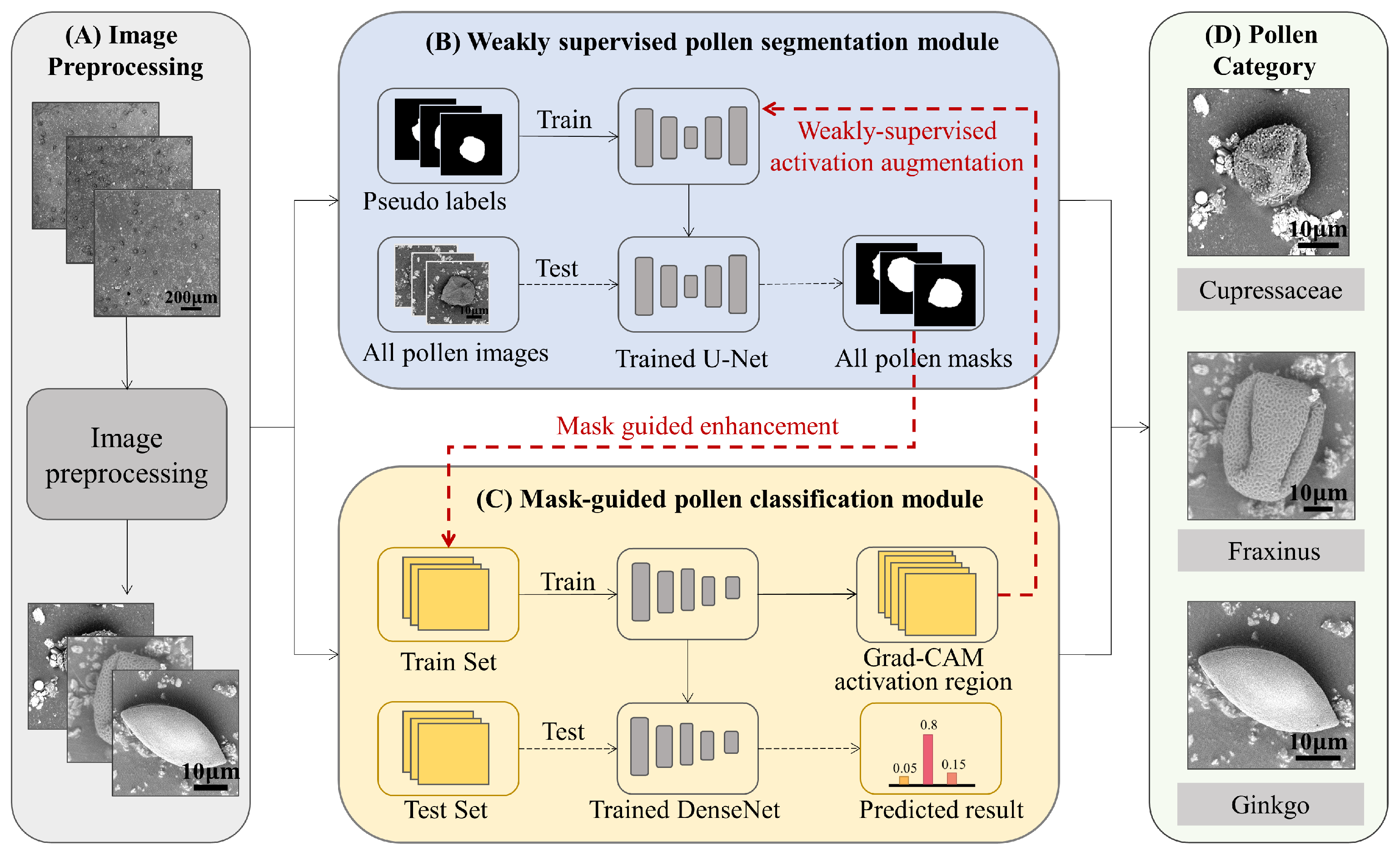
2.2. The Proposed Method
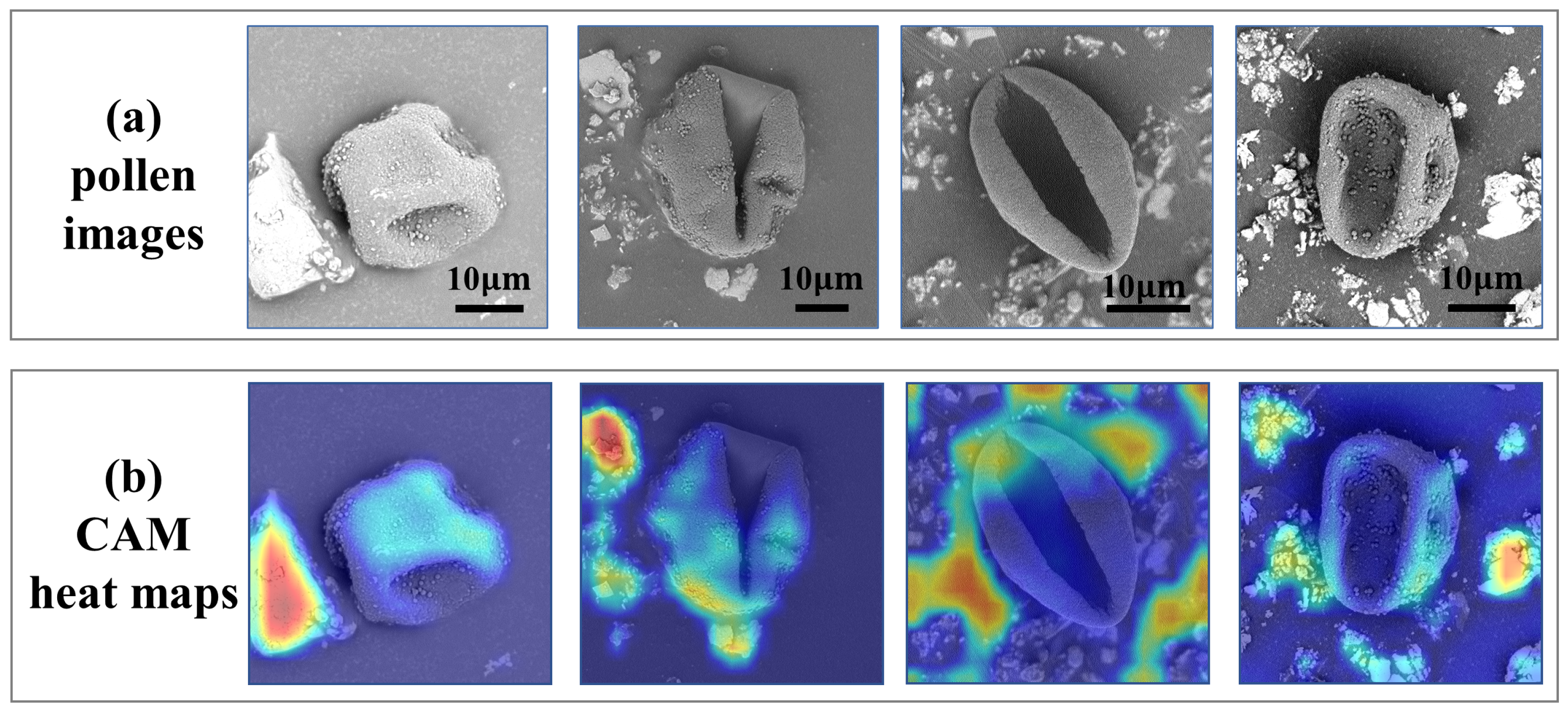
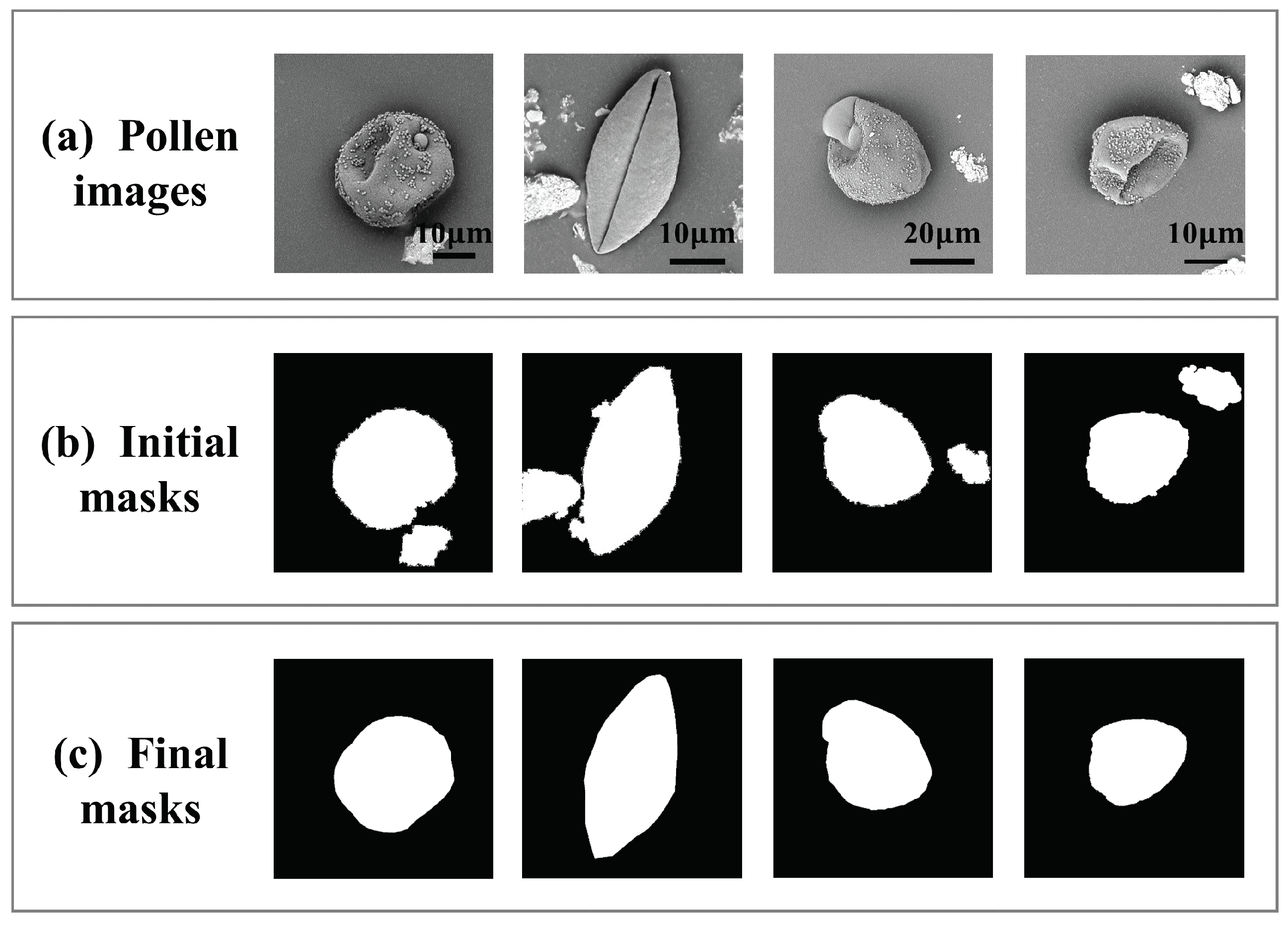
2.2.1. Weakly Supervised Pollen Segmentation Module
2.2.2. Mask-Guided Pollen Classification Module
3. Results
3.1. Experimental Settings
3.2. Comparison with Other Methods
- Precision indicates the relationship between the true positive predicted values and all positive predicted values.
- Recall refers to the proportion of true positive predicted values on all positive samples.
- Specificity denotes the ability to predict negative class.
- The F1-score provides an overall measure that combines precision and recall.
3.3. Ablation Study
- Mean intersection over union (MIoU): The MIoU is the average of the IoU for all classes. The IoU reflects the ratio of the intersection and union of the ground truth and predicted results in each pixel class.
- Mean pixel accuracy (MPA): The MPA is the average of the PA for all classes. The PA indicates the ratio of correctly classified pixels and the total pixels within each class.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Platts-Mills, T.A. The allergy epidemics: 1870–2010. J. Allergy Clin. Immunol. 2015, 136, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, H.; Morales, S.; Salmerón, C.; Cruz, A.; Calado, L.; Rodríguez-García, M.; Alché, J.; Abreu, I. Analysis of the pollen allergen content of twelve olive cultivars grown in Portugal. Aerobiologia 2013, 29, 513–521. [Google Scholar] [CrossRef]
- Fernstrom, A.; Goldblatt, M. Aerobiology and its role in the transmission of infectious diseases. J. Pathog. 2013, 2013, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Umurova, N.; Ismatova, M. Clinical course and risk factors for the development of pollinosis. World Bull. Soc. Sci. 2021, 2, 56–58. [Google Scholar]
- Sur, D.K.; Scandale, S. Treatment of allergic rhinitis. Am. Fam. Physician 2010, 81, 1440–1446. [Google Scholar] [PubMed]
- Grímsson, F.; Meller, B.; Bouchal, J.M.; Zetter, R. Combined LM and SEM study of the middle Miocene (Sarmatian) palynoflora from the Lavanttal Basin, Austria: Part III. Magnoliophyta 1–Magnoliales to Fabales. Grana 2015, 54, 85–128. [Google Scholar] [CrossRef]
- Punyasena, S.W.; Tcheng, D.K.; Wesseln, C.; Mueller, P.G. Classifying black and white spruce pollen using layered machine learning. New Phytol. 2012, 196, 937–944. [Google Scholar] [CrossRef]
- France, I.; Duller, A.; Duller, G.; Lamb, H. A new approach to automated pollen analysis. Quat. Sci. Rev. 2000, 19, 537–546. [Google Scholar] [CrossRef]
- Bonton, P.; Boucher, A.; Thonnat, M.; Tomczak, R.; Hidalgo, P.J.; Belmonte, J.; Galan, C. Colour image in 2D and 3D microscopy for the automation of pollen rate measurement. Image Anal. Stereol. 2002, 21, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Hendriks, E.A.; Duin, R.P.; Reiber, J.H.; Hiemstra, P.S.; de Weger, L.A.; Stoel, B.C. Feasibility study on automated recognition of allergenic pollen: Grass, birch and mugwort. Aerobiologia 2006, 22, 275–284. [Google Scholar] [CrossRef]
- Li, P.; Treloar, W.; Flenley, J.; Empson, L. Towards automation of palynology 2: The use of texture measures and neural network analysis for automated identification of optical images of pollen grains. J. Quat. Sci. Publ. Quat. Res. Assoc. 2004, 19, 755–762. [Google Scholar] [CrossRef]
- Daood, A.; Ribeiro, E.; Bush, M. Pollen grain recognition using deep learning. In Proceedings of the International Symposium on Visual Computing, Las Vegas, NV, USA, 12–14 December 2016; pp. 321–330. [Google Scholar]
- Sevillano, V.; Aznarte, J.L. Improving classification of pollen grain images of the POLEN23E dataset through three different applications of deep learning convolutional neural networks. PLoS ONE 2018, 13, e0201807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Li, J.; Ju, F.; Li, J.; Zu, B.; Ye, C. Automatic Pollen Detection Based on Feature Fusion and Self-attention Mechanism. In Proceedings of the International Conference on Frontier Computing, Singapore, 10–13 July 2020; pp. 93–101. [Google Scholar]
- Zhao, L.N.; Li, J.Q.; Cheng, W.X.; Liu, S.Q.; Gao, Z.K.; Xu, X.; Ye, C.H.; You, H.L. Simulation Palynologists for Pollinosis Prevention: A Progressive Learning of Pollen Localization and Classification for Whole Slide Images. Biology 2022, 11, 1841. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bao, W.; Lin, D.; Wang, Z. A local feature descriptor based on SIFT for 3D pollen image recognition. IEEE Access 2019, 7, 152658–152666. [Google Scholar] [CrossRef]
- Zhang, Y.; Fountain, D.; Hodgson, R.; Flenley, J.; Gunetileke, S. Towards automation of palynology 3: Pollen pattern recognition using Gabor transforms and digital moments. J. Quat. Sci. Publ. Quat. Res. Assoc. 2004, 19, 763–768. [Google Scholar] [CrossRef]
- Travieso, C.M.; Briceño, J.C.; Ticay-Rivas, J.R.; Alonso, J.B. Pollen classification based on contour features. In Proceedings of the 2011 15th IEEE International Conference on Intelligent Engineering Systems, Poprad, Slovakia, 23–25 June 2011; pp. 17–21. [Google Scholar]
- Park, J.H.; Seo, J.; Jackman, J.A.; Cho, N.J. Inflated sporopollenin exine capsules obtained from thin-walled pollen. Sci. Rep. 2016, 6, 28017. [Google Scholar] [CrossRef] [PubMed]
- Langford, M.; Taylor, G.; Flenley, J. Computerized identification of pollen grains by texture analysis. Rev. Palaeobot. Palynol. 1990, 64, 197–203. [Google Scholar] [CrossRef]
- Treloar, W.; Taylor, G.; Flenley, J. Towards automation of palynology 1: Analysis of pollen shape and ornamentation using simple geometric measures, derived from scanning electron microscope images. J. Quat. Sci. Publ. Quat. Res. Assoc. 2004, 19, 745–754. [Google Scholar] [CrossRef]
- Yang, J.J.; Klinkenberg, C.; Pan, J.Z.; Wyss, H.M.; den Toonder, J.M.; Fang, Q. An integrated system for automated measurement of airborne pollen based on electrostatic enrichment and image analysis with machine vision. Talanta 2022, 237, 122908. [Google Scholar] [CrossRef]
- Lu, L.L.; Jiao, B.H.; Qin, F.; Xie, G.; Lu, K.Q.; Li, J.F.; Sun, B.; Li, M.; Ferguson, D.K.; Gao, T.G.; et al. Artemisia pollen dataset for exploring the potential ecological indicators in deep time. Earth Syst. Sci. Data 2022, 14, 3961–3995. [Google Scholar] [CrossRef]
- Polling, M.; Li, C.; Cao, L.; Verbeek, F.; de Weger, L.; Belmonte, J.; De Linares, C.; Willemse, J.; de Boer, H.; Gravendeel, B. Automatic Image Classification Using Neural Networks Increases Accuracy for Allergenic Pollen Monitoring. Res. Sq. (Prepr.) 2020, 1–16. [Google Scholar] [CrossRef]
- Dhawale, V.; Dudul, S.; Tidke, J. Pollen Classification of three types of plants of the family Malvaceae using Computational Intelligence Approach. WSEAS Trans. Des. Constr. Maint. 2021, 1, 1–7. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 21–37. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]



| Attribute | SEM | LM |
|---|---|---|
| Resolution | High (usually 6000–7000×) | Low (usually 20–40×) |
| Depth of field | Large depth of field | Small depth of field |
| Imaging quality | Sharp | Blurring |
| Cost | Expensive | Low |
| Operation | Complex | Simple |
| Deployment | Difficult | Convenient |
| Method | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|
| VGG [33] | 0.747 | 0.692 | 0.842 | 0.718 |
| ResNet [34] | 0.704 | 0.685 | 0.852 | 0.694 |
| MobileNet [35] | 0.756 | 0.667 | 0.832 | 0.709 |
| Ours | 0.864 | 0.857 | 0.932 | 0.860 |
| Method | Cupressaceae | Fraxinus | Ginkgo |
|---|---|---|---|
| VGG [33] | 0.968 | 0.712 | 0.396 |
| ResNet [34] | 0.923 | 0.486 | 0.646 |
| MobileNet [35] | 0.503 | 0.568 | 0.931 |
| Ours | 0.935 | 0.775 | 0.861 |
| Method | Pollen Class | Precision | Recall | Specificity | Accuracy |
|---|---|---|---|---|---|
| Cupressaceae | 0.704 | 0.903 | 0.769 | ||
| Baseline 1 | Fraxinus | 0.615 | 0.505 | 0.833 | 0.722 |
| Ginkgo | 0.833 | 0.694 | 0.925 | ||
| Cupressaceae | 0.736 | 0.935 | 0.796 | ||
| Baseline 2 | Fraxinus | 0.802 | 0.658 | 0.940 | 0.805 |
| Ginkgo | 0.918 | 0.778 | 0.962 | ||
| Cupressaceae | 0.858 | 0.935 | 0.906 | ||
| Ours | Fraxinus | 0.843 | 0.775 | 0.946 | 0.866 |
| Ginkgo | 0.892 | 0.861 | 0.944 |
| Method | MIoU | MPA | Accuracy |
|---|---|---|---|
| Baseline 1 [30] | 0.8291 | 0.9388 | 0.9217 |
| Baseline 1 + CL | 0.9247 | 0.9671 | 0.9700 |
| Baseline 2 [36] | 0.8269 | 0.9380 | 0.9205 |
| Baseline 2 + CL | 0.9352 | 0.9663 | 0.9742 |
| Baseline 3 [37] | 0.8389 | 0.9468 | 0.9268 |
| Baseline 3 + CL | 0.9289 | 0.9629 | 0.9722 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Xu, Q.; Cheng, W.; Zhao, L.; Liu, S.; Gao, Z.; Xu, X.; Ye, C.; You, H. Weakly Supervised Collaborative Learning for Airborne Pollen Segmentation and Classification from SEM Images. Life 2023, 13, 247. https://doi.org/10.3390/life13010247
Li J, Xu Q, Cheng W, Zhao L, Liu S, Gao Z, Xu X, Ye C, You H. Weakly Supervised Collaborative Learning for Airborne Pollen Segmentation and Classification from SEM Images. Life. 2023; 13(1):247. https://doi.org/10.3390/life13010247
Chicago/Turabian StyleLi, Jianqiang, Qinlan Xu, Wenxiu Cheng, Linna Zhao, Suqin Liu, Zhengkai Gao, Xi Xu, Caihua Ye, and Huanling You. 2023. "Weakly Supervised Collaborative Learning for Airborne Pollen Segmentation and Classification from SEM Images" Life 13, no. 1: 247. https://doi.org/10.3390/life13010247
APA StyleLi, J., Xu, Q., Cheng, W., Zhao, L., Liu, S., Gao, Z., Xu, X., Ye, C., & You, H. (2023). Weakly Supervised Collaborative Learning for Airborne Pollen Segmentation and Classification from SEM Images. Life, 13(1), 247. https://doi.org/10.3390/life13010247





