Streamlining the Detection of Human Thyroid Receptor Ligand Interactions with XL1-Blue Cell-Free Protein Synthesis and Beta-Galactosidase Fusion Protein Biosensors
Abstract
:1. Introduction
2. Materials and Methods
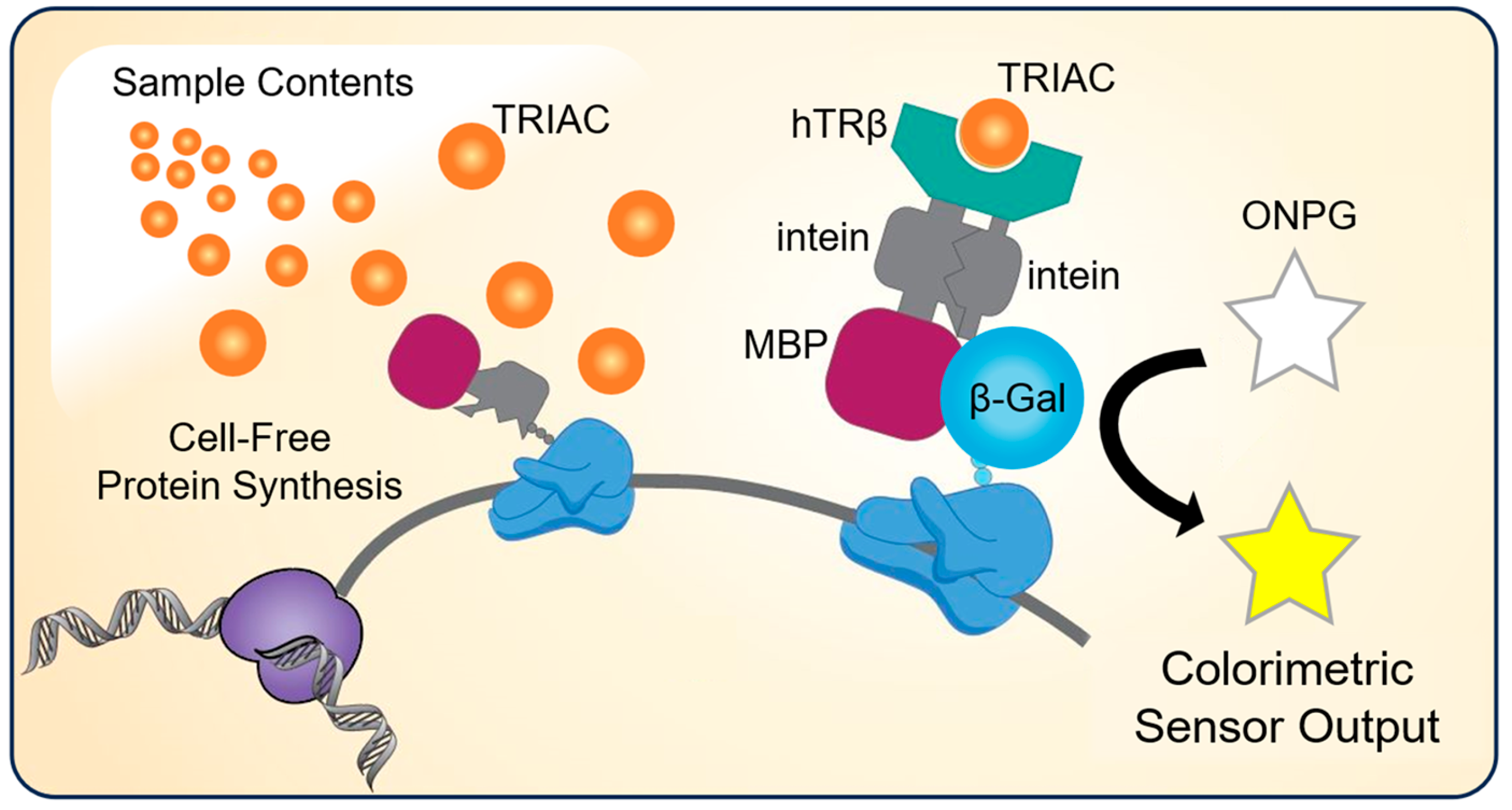
2.1. Engineering and Design of the Biosensor Constructs
2.2. Developing the XL1-Blue CFPS Platform
2.3. CFPS Reactions and Thyroid Ligand Assays
2.4. Biosensor Reagent Cost Calculations
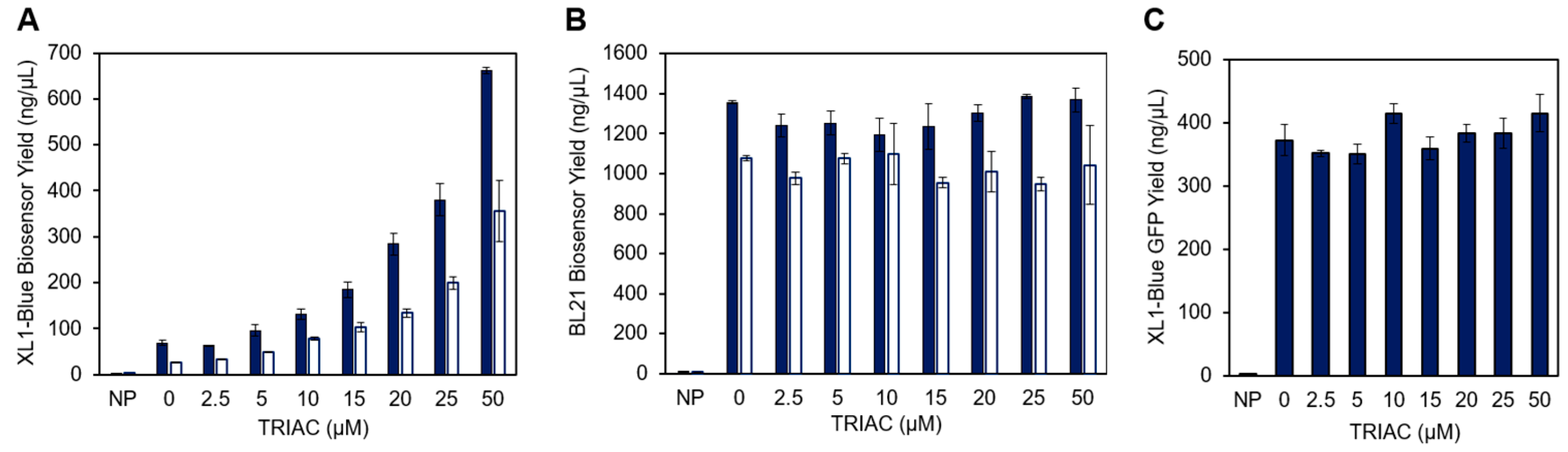
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mughal, B.B.; Fini, J.-B.; Demeneix, B.A. Thyroid-disrupting chemicals and brain development: An update. Endocr. Connect. 2018, 7, R160. [Google Scholar] [CrossRef] [PubMed]
- Alsen, M.; Sinclair, C.; Cooke, P.; Ziadkhanpour, K.; Genden, E.; van Gerwen, M. Endocrine disrupting chemicals and thyroid cancer: An overview. Toxics 2021, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.M.; Sonnenschein, C. Environmental causes of cancer: Endocrine disruptors as carcinogens. Nat. Rev. Endocrinol. 2010, 6, 363–370. [Google Scholar] [CrossRef] [PubMed]
- La Merrill, M.A.; Vandenberg, L.N.; Smith, M.T.; Goodson, W.; Browne, P.; Patisaul, H.B.; Guyton, K.Z.; Kortenkamp, A.; Cogliano, V.J.; Woodruff, T.J. Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat. Rev. Endocrinol. 2020, 16, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N. Endocrine disrupting chemicals: Strategies to protect present and future generations. Expert Rev. Endocrinol. Metab. 2021, 16, 135–146. [Google Scholar] [CrossRef]
- Saponaro, F.; Sestito, S.; Runfola, M.; Rapposelli, S.; Chiellini, G. Selective thyroid hormone receptor-beta (TRβ) agonists: New perspectives for the treatment of metabolic and neurodegenerative disorders. Front. Med. 2020, 7, 331. [Google Scholar] [CrossRef]
- Furst, A.L.; Hoepker, A.C.; Francis, M.B. Quantifying hormone disruptors with an engineered bacterial biosensor. ACS Cent. Sci. 2017, 3, 3110–3116. [Google Scholar] [CrossRef]
- Colosi, J.C.; Kney, A.D. A yeast estrogen screen without extraction provides fast, reliable measures of estrogenic activity. Environ. Toxicol. Chem. 2011, 30, 2261–2269. [Google Scholar] [CrossRef]
- Salehi, A.S.; Shakalli Tang, M.J.; Smith, M.T.; Hunt, J.M.; Law, R.A.; Wood, D.W.; Bundy, B.C. Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal. Chem. 2017, 89, 3395–3401. [Google Scholar] [CrossRef]
- Hunt, J.P.; Galiardi, J.; Free, T.J.; Yang, S.O.; Poole, D.; Zhao, E.L.; Andersen, J.L.; Wood, D.W.; Bundy, B.C. Mechanistic discoveries and simulation-guided assay optimization of portable hormone biosensors with cell-free protein synthesis. Biotechnol. J. 2022, 17, 2100152. [Google Scholar] [CrossRef]
- Salehi, A.S.; Yang, S.O.; Earl, C.C.; Tang, M.J.S.; Hunt, J.P.; Smith, M.T.; Wood, D.W.; Bundy, B.C. Biosensing estrogenic endocrine disruptors in human blood and urine: A RAPID cell-free protein synthesis approach. Toxicol. Appl. Pharmacol. 2018, 345, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.Y.; Shao, A.; Lu, Z.; Savilahti, H.; Menolascina, F.; Wang, L.; Dalchau, N.; Wang, B. A systematic approach to inserting split inteins for Boolean logic gate engineering and basal activity reduction. Nat. Commun. 2021, 12, 2200. [Google Scholar] [CrossRef] [PubMed]
- Boehle, K.E.; Carrell, C.S.; Caraway, J.; Henry, C.S. Paper-Based Enzyme Competition Assay for Detecting Falsified β-Lactam Antibiotics. ACS Sens. 2018, 3, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Hesek, D.; Mobashery, S. A practical synthesis of nitrocefin. J. Org. Chem. 2005, 70, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Lindgren, C.M.; Ebbert, L.E.; Free, T.J.; Nelson, J.A.D.; Simonson, K.M.; Hunt, J.P.; Bundy, B.C. “Just Add Small Molecules” Cell-Free Protein Synthesis: Combining DNA Template and Cell Extract Preparation into a Single Fermentation. Biotechnol. Progr. 2023, 39, e3332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guo, W.; Lu, Y. Advances in cell-free biosensors: Principle, mechanism, and applications. Biotechnol. J. 2020, 15, 2000187. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, D.M. In vitro use of cellular synthetic machinery for biosensing applications. Front. Pharmacol. 2019, 10, 1166. [Google Scholar] [CrossRef] [PubMed]
- Lopreside, A.; Wan, X.; Michelini, E.; Roda, A.; Wang, B. Comprehensive profiling of diverse genetic reporters with application to whole-cell and cell-free biosensors. Anal. Chem. 2019, 91, 15284–15292. [Google Scholar] [CrossRef] [PubMed]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A small luciferase is brightening up the field of bioluminescence. Bioconjugate Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-J.; Lee, K.-H.; Yoo, T.H.; Kim, D.-M. Interfacing a personal glucose meter with cell-free protein synthesis for rapid analysis of amino acids. Anal. Chem. 2019, 91, 2531–2535. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.P.; Barnett, R.J.; Robinson, H.; Soltani, M.; Nelson, J.A.D.; Bundy, B.C. Rapid sensing of clinically relevant glutamine concentrations in human serum with metabolically engineered E. coli-based cell-free protein synthesis. J. Biotechnol. 2020, 325, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Noireaux, V.; Bar-Ziv, R.; Libchaber, A. Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. USA 2003, 100, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. based synthetic gene networks. Cell 2014, 159, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Skretas, G.; Wood, D.W. Regulation of protein activity with small-molecule-controlled inteins. Protein Sci. 2005, 14, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, A.; Jacob, F.; Monod, J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the β-galactosidase structural gene of Escherichia coli. J. Mol. Biol. 1967, 24, 339–343. [Google Scholar] [CrossRef] [PubMed]
- McNerney, M.P.; Zhang, Y.; Steppe, P.; Silverman, A.D.; Jewett, M.C.; Styczynski, M.P. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 2019, 5, eaax4473. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Arce, A.; Guzman Chavez, F.; Gandini, C.; Puig, J.; Matute, T.; Haseloff, J.; Dalchau, N.; Molloy, J.; Pardee, K.; Federici, F. Decentralizing cell-free RNA sensing with the use of low-cost cell extracts. Front. Bioeng. Biotechnol. 2021, 9, 727584. [Google Scholar] [CrossRef]
- Ma, D.; Shen, L.; Wu, K.; Diehnelt, C.W.; Green, A.A. Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment. Synth. Biol. 2018, 3, ysy018. [Google Scholar] [CrossRef]
- Matthews, B.W. The structure of E. coli β-galactosidase. Comptes Rendus Biol. 2005, 328, 549–556. [Google Scholar] [CrossRef]
- Wood, D.W.; Wu, W.; Belfort, G.; Derbyshire, V.; Belfort, M. A genetic system yields selv-cleaving inteins for bioseparations. Nat. Biotechnol. 1999, 17, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.W.; Camarero, J.A. Intein applications: From protein purification and labeling to metabolic control methods. J. Biol. Chem. 2014, 289, 14512–14519. [Google Scholar] [CrossRef] [PubMed]
- Diamanti-Kandarakis, E.; Bourguignon, J.-P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef] [PubMed]
- Attina, T.M.; Hauser, R.; Sathyanarayana, S.; Hunt, P.A.; Bourguignon, J.-P.; Myers, J.P.; DiGangi, J.; Zoeller, R.T.; Trasande, L. Exposure to endocrine-disrupting chemicals in the USA: A population-based disease burden and cost analysis. Lancet Diabetes Endocrinol. 2016, 4, 996–1003. [Google Scholar] [CrossRef]
- Howdeshell, K.L. A model of the development of the brain as a construct of the thyroid system. Environ. Health Perspect. 2002, 110, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Schapaugh, A.W.; McFadden, L.G.; Zorrilla, L.M.; Geter, D.R.; Stuchal, L.D.; Sunger, N.; Borgert, C.J. Analysis of EPA’s endocrine screening battery and recommendations for further review. Regul. Toxicol. Pharmacol. 2015, 72, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Leusch, F.D.; Aneck-Hahn, N.H.; Cavanagh, J.-A.E.; Du Pasquier, D.; Hamers, T.; Hebert, A.; Neale, P.A.; Scheurer, M.; Simmons, S.O.; Schriks, M. Comparison of in vitro and in vivo bioassays to measure thyroid hormone disrupting activity in water extracts. Chemosphere 2018, 191, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, S.; Peeters, R.P.; Visser, T.J.; Visser, W.E. Triiodothyroacetic acid in health and disease. J. Endocrinol. 2017, 234, R99–R121. [Google Scholar] [CrossRef]
- Guo, Y.; Hui, C.-Y.; Liu, L.; Zheng, H.-Q.; Wu, H.-M. Improved monitoring of low-level transcription in Escherichia coli by a β-galactosidase α-complementation system. Front. Microbiol. 2019, 10, 1454. [Google Scholar] [CrossRef]
- Juers, D.H.; Matthews, B.W.; Huber, R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef]
- Jacobson, R.; Zhang, X.-J.; DuBose, R.; Matthews, B. Three-dimensional structure of β-galactosidase from E. coli. Nature 1994, 369, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, M.; Böcker, J.K.; Matern, J.C.; Pietrokovski, S.; Mootz, H.D. Development of a screening system for inteins active in protein splicing based on intein insertion into the LacZα-peptide. Biol. Chem. 2017, 398, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Mootz, H.D. Split inteins as versatile tools for protein semisynthesis. ChemBioChem 2009, 10, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Marx, C.J.; Lidstrom, M.E. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 2001, 147, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Swartz, J.R. Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol. Bioeng. 1999, 66, 180–188. [Google Scholar] [CrossRef]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004, 86, 19–26. [Google Scholar] [CrossRef]
- Swartz, J.R.; Jewett, M.C.; Woodrow, K.A. Cell-free protein synthesis with prokaryotic combined transcription-translation. Methods Mol. Bio. 2004, 267, 169–182. [Google Scholar]
- Calhoun, K.A.; Swartz, J.R. An economical method for cell-free protein synthesis using glucose and nucleoside monophosphates. Biotechnol. Prog. 2005, 21, 1146–1153. [Google Scholar] [CrossRef]
- Horvath, N.; Vilkhovoy, M.; Wayman, J.A.; Calhoun, K.; Swartz, J.; Varner, J.D. Toward a genome scale sequence specific dynamic model of cell-free protein synthesis in Escherichia coli. Metab. Eng. Commun. 2020, 10, e00113. [Google Scholar] [CrossRef]
- Calhoun, K.A.; Swartz, J.R. Total amino acid stabilization during cell-free protein synthesis reactions. J. Biotechnol. 2006, 123, 193–203. [Google Scholar] [CrossRef]
- Didovyk, A.; Tonooka, T.; Tsimring, L.; Hasty, J. Rapid and scalable preparation of bacterial lysates for cell-free gene expression. ACS Synth. Biol. 2017, 6, 2198–2208. [Google Scholar] [CrossRef]
- Jiang, X.; Oohira, K.; Iwasaki, Y.; Nakano, H.; Ichihara, S.; Yamane, T. Reduction of protein degradation by use of protease-deficient mutants in cell-free protein synthesis system of Escherichia coli. J. Biosci. Bioeng. 2002, 93, 151–156. [Google Scholar] [CrossRef]
- Waegeman, H.; De Lausnay, S.; Beauprez, J.; Maertens, J.; De Mey, M.; Soetaert, W. Increasing recombinant protein production in Escherichia coli K12 through metabolic engineering. New Biotechnol. 2013, 30, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Dopp, J.L.; Tamiev, D.D.; Reuel, N.F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef] [PubMed]
- UniProt: The Universal Protein knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [CrossRef] [PubMed]
- Murthy, M. Determination of protein in extracts containing interfering substances and in radioactive samples following scintillation counting. Anal. Biochem. 1975, 64, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Patel, K.G.; Wong, H.E.; Swartz, J.R. Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol. Prog. 2012, 28, 413–420. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hunt, J.P.; Free, T.J.; Galiardi, J.; Watt, K.M.; Wood, D.W.; Bundy, B.C. Streamlining the Detection of Human Thyroid Receptor Ligand Interactions with XL1-Blue Cell-Free Protein Synthesis and Beta-Galactosidase Fusion Protein Biosensors. Life 2023, 13, 1972. https://doi.org/10.3390/life13101972
Hunt JP, Free TJ, Galiardi J, Watt KM, Wood DW, Bundy BC. Streamlining the Detection of Human Thyroid Receptor Ligand Interactions with XL1-Blue Cell-Free Protein Synthesis and Beta-Galactosidase Fusion Protein Biosensors. Life. 2023; 13(10):1972. https://doi.org/10.3390/life13101972
Chicago/Turabian StyleHunt, J. Porter, Tyler J. Free, Jackelyn Galiardi, Kevin M. Watt, David W. Wood, and Bradley C. Bundy. 2023. "Streamlining the Detection of Human Thyroid Receptor Ligand Interactions with XL1-Blue Cell-Free Protein Synthesis and Beta-Galactosidase Fusion Protein Biosensors" Life 13, no. 10: 1972. https://doi.org/10.3390/life13101972
APA StyleHunt, J. P., Free, T. J., Galiardi, J., Watt, K. M., Wood, D. W., & Bundy, B. C. (2023). Streamlining the Detection of Human Thyroid Receptor Ligand Interactions with XL1-Blue Cell-Free Protein Synthesis and Beta-Galactosidase Fusion Protein Biosensors. Life, 13(10), 1972. https://doi.org/10.3390/life13101972





