S-Nitrosylated Proteins Involved in Autophagy in Triticum aestivum Roots: A Bottom-Up Proteomics Approach and In Silico Predictive Algorithms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Extraction and Immunodetection of S-Nitrosylated Wheat Proteins
2.3. In-Gel Trypsin Digestion
2.4. Solid-Phase Extraction
2.5. Nano-LC-MS/MS
2.6. Data Analysis
2.7. Bioinformatic Analysis of the Availability of S-Nitrosylation Sites of Wheat Proteins
2.8. Protein–Ligand Docking and Protein–Protein Interactions
2.9. Statistical Analysis
3. Results
3.1. Extraction and Visualization of S-Nitrosylated Proteins during Induction of Autophagy
3.2. Protein Identification and Search for S-Nitrosylation Sites
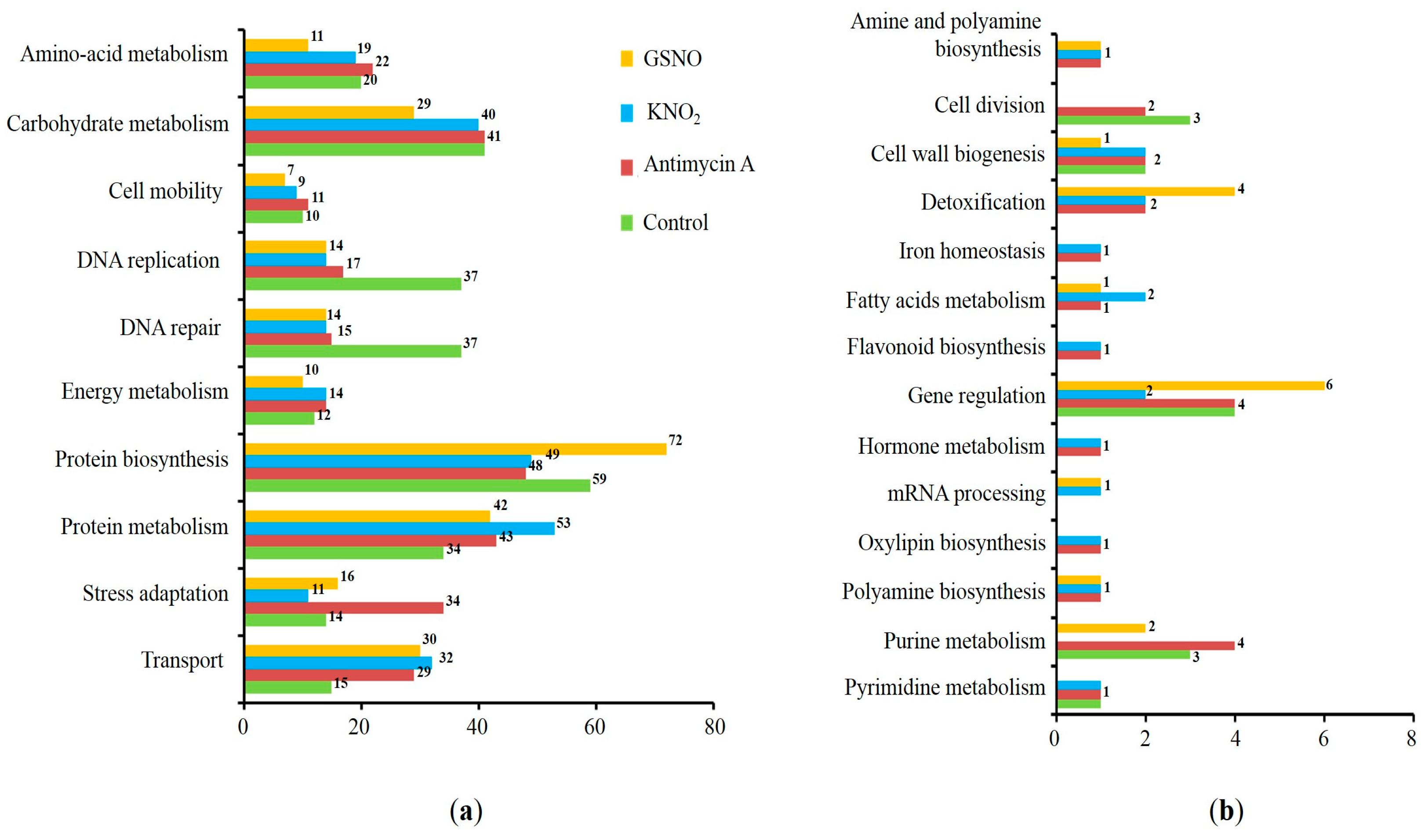
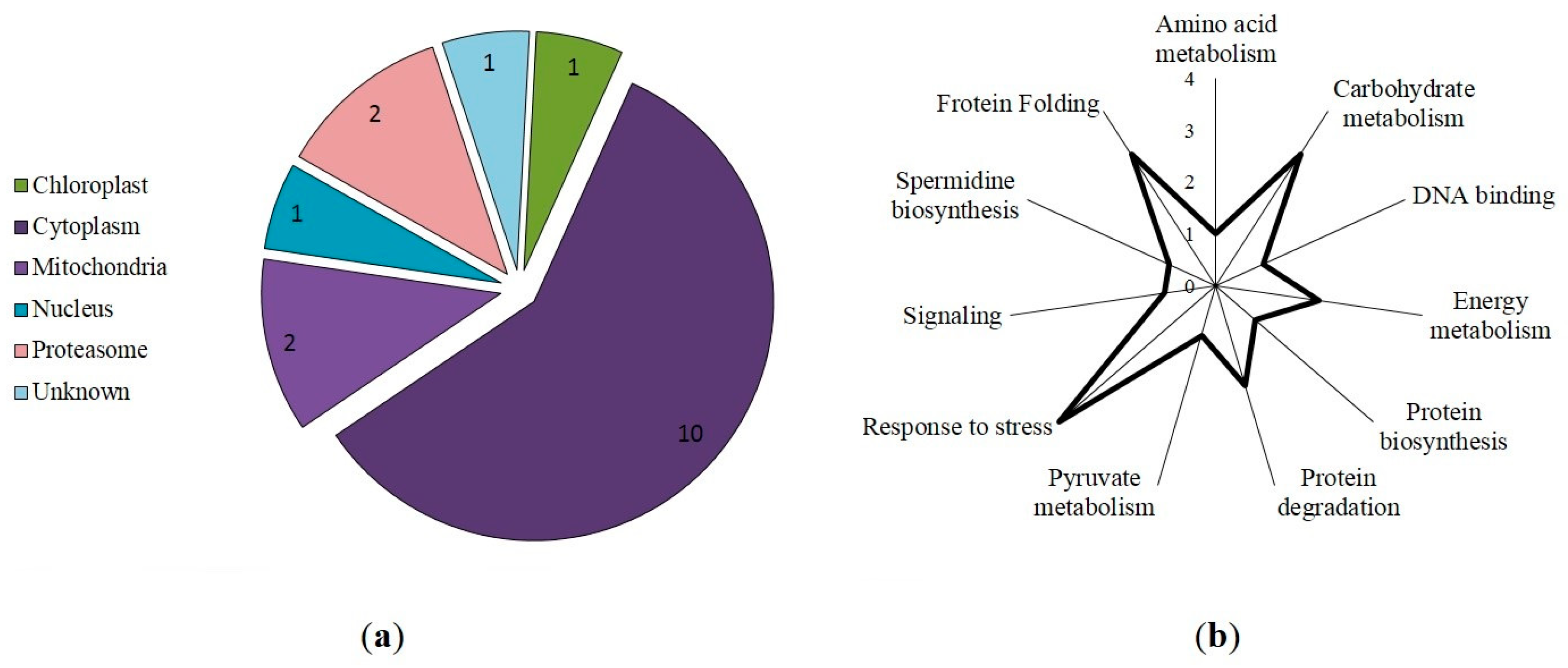
3.3. Prediction of S-Nitrosylation Sites, Localization, and Functional Annotation of S-Nitrosylated Proteins
3.4. Protein–Ligand Docking and Protein–Protein Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siyiannis, V.F.; Protonotarios, V.E.; Zechmann, B.; Chorianopoulou, S.N.; Müller, M.; Hawkesford, M.J.; Bouranis, D.L. Comparative spatiotemporal analysis of root aerenchyma formation processes in maize due to sulphate, nitrate or phosphate deprivation. Protoplasma 2012, 249, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Hanamata, S.; Kurusu, T.; Okada, M.; Suda, A.; Kawamura, K.; Tsukada, E.; Kuchitsu, K. In vivo imaging and quantitative monitoring of autophagic flux in tobacco BY-2 cells. Plant Signal. Behav. 2013, 8, e22510. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Sláviková, S.; Shy, G.; Yao, Y.; Glozman, R.; Levanony, H.; Pietrokovski, S.; Elazar, Z.; Galili, G. The autophagy-associated atg8 gene family operates both under favourable growth conditions and under starvation stresses in arabidopsis plants. J. Exp. Bot. 2005, 56, 2839–2849. [Google Scholar] [CrossRef]
- Shi, J.; Feng, H.; Lee, J.; Ning Chen, W. Comparative proteomics profile of lipid-cumulating oleaginous yeast: An ITRAQ-coupled 2-D LC-MS/MS analysis. PLoS ONE 2013, 8, e85532. [Google Scholar] [CrossRef]
- Melia, T.J.; Lystad, A.H.; Simonsen, A. Autophagosome biogenesis: From membrane growth to closure. J. Cell Biol. 2020, 219, e202002085. [Google Scholar] [CrossRef]
- Liu, B.; Huang, X.; Li, Y.; Liao, W.; Li, M.; Liu, Y.; He, R.; Feng, D.; Zhu, R.; Kurihara, H. JS-K, a nitric oxide donor, induces autophagy as a complementary mechanism inhibiting ovarian cancer. BMC Cancer 2019, 19, 645. [Google Scholar] [CrossRef]
- Sarkar, S.; Korolchuk, V.I.; Renna, M.; Imarisio, S.; Fleming, A.; Williams, A.; Garcia-Arencibia, M.; Rose, C.; Luo, S.; Underwood, B.R.; et al. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell 2011, 43, 19–32. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, L.; Tian, Z.; Wang, J.; Yang, Y.; Liu, J.; Chen, Y.; Hu, C.; Chen, T.; Zhao, Y.; et al. Nitric oxide inhibits autophagy and promotes apoptosis in hepatocellular carcinoma. Cancer Sci. 2019, 110, 1054–1063. [Google Scholar] [CrossRef]
- Dmitrieva, S.A.; Ponomareva, A.A.; Gurjanov, O.P.; Mazina, A.B.; Andrianov, V.V.; Iyudin, V.S.; Minibayeva, F.V. Spermine induces autophagy in plants: Possible role of NO and reactive oxygen species. Dokl. Biochem. Biophys. 2018, 483, 341–343. [Google Scholar] [CrossRef]
- Kuo, E.Y.; Chang, H.-L.; Lin, S.-T.; Lee, T.-M. High light-induced nitric oxide production induces autophagy and cell death in Chlamydomonas reinhardtii. Front. Plant Sci. 2020, 11, 772. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef]
- Bignon, E.; Allega, M.F.; Lucchetta, M.; Tiberti, M.; Papaleo, E. Computational structural biology of S-nitrosylation of cancer targets. Front. Oncol. 2018, 8, 272. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Kulik, A.; Koen, E.; Besson-Bard, A.; Bourque, S.; Jeandroz, S.; Lamotte, O.; Wendehenne, D. Protein S-nitrosylation: What’s going on in plants? Free Radic. Biol. Med. 2012, 53, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- León, J. Protein tyrosine nitration in plant nitric oxide signaling. Front. Plant Sci. 2022, 13, 859374. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Protein nitration: A connecting bridge between nitric oxide (NO) and plant stress. Plant Stress 2021, 2, 100026. [Google Scholar] [CrossRef]
- Montagna, C.; Rizza, S.; Maiani, E.; Piredda, L.; Filomeni, G.; Cecconi, F. To eat, or NOt to eat: S -nitrosylation signaling in autophagy. FEBS J. 2016, 283, 3857–3869. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Wang, L.; Wang, P.; Xue, Y.; Li, X.; Qiao, X.; Zhang, X.; Xu, T.; Liu, G.; et al. Autophagy impairment mediated by S-nitrosation of ATG4B leads to neurotoxicity in response to hyperglycemia. Autophagy 2017, 13, 1145–1160. [Google Scholar] [CrossRef]
- Oh, C.; Dolatabadi, N.; Cieplak, P.; Diaz-Meco, M.T.; Moscat, J.; Nolan, J.P.; Nakamura, T.; Lipton, S.A. S-nitrosylation of p62 inhibits autophagic flux to promote α-synuclein secretion and spread in Parkinson’s disease and Lewy body dementia. J. Neurosci. 2022, 42, 3011–3024. [Google Scholar] [CrossRef]
- Wright, C.; Iyer, A.K.V.; Kulkarni, Y.; Azad, N. S-nitrosylation of Bcl-2 negatively affects autophagy in lung epithelial cells. J. Cell. Biochem. 2016, 117, 521–532. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, C.; Liu, Q. PTEN S-nitrosylation by NOS1 inhibits autophagy in NPC cells. Cell Death Dis. 2019, 10, 306. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, N.; Oka, S.; Nah, J.; Wu, C.; Zhai, P.; Mukai, R.; Xu, X.; Kashyap, S.; Huang, C.-Y.; Sung, E.-A.; et al. Thioredoxin 1 promotes autophagy through transnitrosylation of Atg7 during myocardial ischemia. J. Clin. Investig. 2023, 133, e162326. [Google Scholar] [CrossRef] [PubMed]
- Guha, P.; Harraz, M.M.; Snyder, S.H. Cocaine elicits autophagic cytotoxicity via a nitric oxide-GAPDH signaling cascade. Proc. Natl. Acad. Sci. USA 2016, 113, 1417–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 2018, 71, 142–154.e6. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Dmitrieva, S.; Ponomareva, A.; Ryabovol, V. Oxidative stress-induced autophagy in plants: The role of mitochondria. Plant Physiol. Biochem. 2012, 59, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Mazina, A.; Gazizova, N.; Dmitrieva, S.; Ponomareva, A.; Rakhmatullina, D. Nitric oxide induces autophagy in Triticum aestivum roots. Antioxidants 2023, 12, 1655. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680. [Google Scholar] [CrossRef]
- Bassal, M.; Abukhalaf, M.; Majovsky, P.; Thieme, D.; Herr, T.; Ayash, M.; Tabassum, N.; Al Shweiki, M.R.; Proksch, C.; Hmedat, A.; et al. Reshaping of the Arabidopsis thaliana proteome landscape and co-regulation of proteins in development and immunity. Mol. Plant 2020, 13, 1709–1732. [Google Scholar] [CrossRef]
- Spiller, S.; Frolov, A.; Hoffmann, R. Quantification of specific glycation sites in human serum albumin as prospective type 2 diabetes mellitus biomarkers. Protein Pept. Lett. 2018, 24, 887–896. [Google Scholar] [CrossRef]
- Shumilina, J.; Kiryushkin, A.S.; Frolova, N.; Mashkina, V.; Ilina, E.L.; Puchkova, V.A.; Danko, K.; Silinskaya, S.; Serebryakov, E.B.; Soboleva, A.; et al. Integrative proteomics and metabolomics analysis reveals the role of small signaling peptide rapid alkalinization factor 34 (RALF34) in cucumber roots. Int. J. Mol. Sci. 2023, 24, 7654. [Google Scholar] [CrossRef]
- Chen, S.-J.; Liao, D.-L.; Chen, C.-H.; Wang, T.-Y.; Chen, K.-C. Construction and analysis of protein-protein interaction network of heroin use disorder. Sci. Rep. 2019, 9, 4980. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Sánchez-Calvo, B.; Mata-Pérez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Nitric oxide buffering and conditional nitric oxide release in stress response. J. Exp. Bot. 2018, 69, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Galeeva, E.I.; Trifonova, T.V.; Ponomareva, A.A.; Viktorova, L.V.; Minibayeva, F.V. Nitrate reductase from Triticum aestivum leaves: Regulation of activity and possible role in production of nitric oxide. Biochemistry 2012, 77, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.J.; Kumari, A.; Florez-Sarasa, I.; Fernie, A.R.; Igamberdiev, A.U. Interaction of nitric oxide with the components of the plant mitochondrial electron transport chain. J. Exp. Bot. 2018, 69, 3413–3424. [Google Scholar] [CrossRef]
- Antonova, K.; Vikhnina, M.; Soboleva, A.; Mehmood, T.; Heymich, M.-L.; Leonova, T.; Bankin, M.; Lukasheva, E.; Gensberger-Reigl, S.; Medvedev, S.; et al. Analysis of chemically labile glycation adducts in seed proteins: Case study of methylglyoxal-derived hydroimidazolone 1 (MG-H1). Int. J. Mol. Sci. 2019, 20, 3659. [Google Scholar] [CrossRef]
- Benhar, M.; Forrester, M.T.; Stamler, J.S. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 721–732. [Google Scholar] [CrossRef]
- Leiper, J.; Murray-Rust, J.; McDonald, N.; Vallance, P. S-nitrosylation of dimethylarginine dimethylaminohydrolase regulates enzyme activity: Further interactions between nitric oxide synthase and dimethylarginine dimethylaminohydrolase. Proc. Natl. Acad. Sci. USA 2002, 99, 13527–13532. [Google Scholar] [CrossRef]
- Sisti, G.; Kanninen, T.T.; Ramer, I.; Witkin, S.S. Interaction between the inducible 70-KDa heat shock protein and autophagy: Effects on fertility and pregnancy. Cell Stress Chaperones 2015, 20, 753–758. [Google Scholar] [CrossRef]
- Hurley, J.H.; Young, L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Jia, H.; Liang, Z.; Zhang, X.; Wang, J.; Xu, W.; Qian, H. 14-3-3 proteins: An important regulator of autophagy in diseases. Am. J. Transl. Res. 2017, 9, 4738–4746. [Google Scholar]
- Chodasiewicz, M.; Kerber, O.; Gorka, M.; Moreno, J.C.; Maruri-Lopez, I.; Minen, R.I.; Sampathkumar, A.; Nelson, A.D.L.; Skirycz, A. 2′,3′-CAMP treatment mimics the stress molecular response in Arabidopsis thaliana. Plant Physiol. 2022, 188, 1966–1978. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Chaves, S.; Alves, S.; Salin, B.; Camougrand, N.; Manon, S.; Sousa, M.J.; Côrte-Real, M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: The role of pep4 and the ADP/ATP carrier: Mitochondria degradation in apoptosis. Mol. Microbiol. 2010, 76, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Ryder, L.; Arendrup, F.S.; Martínez, J.F.; Snieckute, G.; Pecorari, C.; Shah, R.A.; Lund, A.H.; Blasius, M.; Bekker-Jensen, S. Nitric oxide-induced ribosome collision activates ribosomal surveillance mechanisms. Cell Death Dis. 2023, 14, 467. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, S.; Zhang, H.; Xu, J. Identification of nitric oxide as an endogenous inhibitor of 26S proteasomes in vascular endothelial cells. PLoS ONE 2014, 9, e98486. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Cheng, F.; Yin, L.-L.; Zhou, J.; Xia, X.-J.; Shi, K.; Yu, J.-Q.; Zhou, Y.-H.; Foyer, C.H. Interactions between 2-Cys peroxiredoxins and ascorbate in autophagosome formation during the heat stress response in Solanum lycopersicum. J. Exp. Bot. 2016, 67, 1919–1933. [Google Scholar] [CrossRef]
- Jeon, Y.-M.; Lee, M.-Y. Airborne nanoparticles (PM 0.1) induce autophagic cell death of human neuronal cells: Autophagic cell death by PM 0.1. J. Appl. Toxicol. 2016, 36, 1332–1342. [Google Scholar] [CrossRef]




| Uniprot ID | Protein | Hypothetical S-Nitrosylation Sites |
|---|---|---|
| P46077 | 14-3-3-like protein GF14 phi | 106 |
| Q9FIB6 | 26S proteasome non-ATPase regulatory subunit 12 homolog A | 397 |
| Q9LNU4 | 26S proteasome non-ATPase regulatory subunit 3 homolog A | 141 |
| P80602 | 2-Cys peroxiredoxin BAS1 chloroplastic (fragment) | 64, 185 |
| Q9LZH9 | 60S ribosomal protein L7a-2 | 193 |
| P31167 | ADP ATP carrier protein 1 mitochondrial | 130 |
| Q41629 | ADP ATP carrier protein 1 mitochondrial | 81, 206 |
| A8MS68 | Dihydrolipoyl dehydrogenase 1 chloroplastic | 400 |
| Q9S7C0 | Heat shock 70 kDa protein 14 | 268 |
| F4HQD4 | Heat shock 70 kDa protein 15 | 268 |
| Q39043 | Heat shock 70 kDa protein BIP2 | 298 |
| Q9FEF8 | rRNA 2′-O-methyltransferase fibrillarin 1 | 252 |
| Q9SVM4 | Serine hydroxymethyltransferase 5 | 324 |
| O48661 | Spermidine synthase 2 | 43 |
| P48491 | Triosephosphate isomerase cytosolic | 13, 127 |
| Q9ZUY6 | UDP-D-apiose/UDP-D-xylose synthase 1 | 187 |
| Q9SGE0 | UDP-D-apiose/UDP-D-xylose synthase 2 | 187 |
| Name | Description (UNIPROT) | UNIPROT ID |
|---|---|---|
| GRF4 | 14-3-3-like protein GF14 phi | P46077 |
| EMB2107 | 26S proteasome non-ATPase regulatory subunit 12 homolog A | Q9FIB6 |
| BAS-1 | 2-Cys peroxiredoxin BAS1 chloroplastic (fragment) | Q9C5R8 |
| RPL7AB | 60S ribosomal protein L7a-2 | Q9LZH9 |
| AAC1 | ADP ATP carrier protein 1 mitochondrial | P31167 |
| LPD1 | Dihydrolipoyl dehydrogenase 1 chloroplastic | A8MS68 |
| BIP2 | Heat shock 70 kDa protein BIP2 | Q39043 |
| FIB1 | rRNA 2′-O-methyltransferase fibrillarin 1 | Q9FEF8 |
| SHM5 | Serine hydroxymethyltransferase 5 | Q9SVM4 |
| SDS2 | Spermidine synthase 2 | O48661 |
| TPI | Triosephosphate isomerase cytosolic | P48491 |
| AXS1 | UDP-D-apiose/UDP-D-xylose synthase 1 | Q9ZUY6 |
| ATG1a | Serine/threonine-protein kinase ATG1a | Q94C95 |
| ATG2 | Autophagy-related protein 2 | F8S296 |
| ATG3 | Autophagy-related protein 3 | Q0WWQ1 |
| ATG4a | Cysteine protease ATG4a | Q8S929 |
| ATG5 | Autophagy protein 5 | Q9FFI2 |
| ATG6 | Beclin-1-like protein | Q9M367 |
| ATG7 | Ubiquitin-like modifier-activating enzyme atg7 | Q94CD5 |
| ATG8e | Autophagy-related protein 8e | Q8S926 |
| ATG9 | Autophagy-related protein 9 | Q8RUS5 |
| ATG10 | Ubiquitin-like-conjugating enzyme ATG10 | Q8VZ52 |
| ATG12b | Ubiquitin-like protein ATG12B | Q9LVK3 |
| ATG13 | Autophagy-related protein 13a | Q9SCK0 |
| ATG16 | Autophagy-related protein 16 | Q6NNP0 |
| ATG18a | Autophagy-related protein 18a | Q93VB2 |
| TOR | Serine/threonine-protein kinase TOR | Q9FR53 |
| VPS15 | Serine/threonine-protein kinase VPS15 | Q9M0E5 |
| VPS34 | Phosphatidylinositol 3-kinase VPS34 | P42339 |
| KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Q38997 |
| KIN11 | SNF1-related protein kinase catalytic subunit alpha KIN11 | P92958 |
| SNF4 | Sucrose nonfermenting 4-like protein | Q944A6 |
| GAPC2 | Glyceraldehyde-3-phosphate dehydrogenase GAPC2, cytosolic | Q9FX54 |
| GAPA-2 | Glyceraldehyde-3-phosphate dehydrogenase GAPA2, chloroplastic | Q9LPW0 |
| GAPCP2 | Glyceraldehyde-3-phosphate dehydrogenase GAPC2, cytosolic | Q9FX54 |
| PRC3 | Proteasome subunit alpha type-2-A | O23708 |
| RPT5B | 26S proteasome regulatory subunit 6A homolog B | O04019 |
| RPN3A | 26S proteasome non-ATPase regulatory subunit 3 homolog A | Q9LNU4 |
| PBC1 | Proteasome subunit beta type-3-A | Q9XI05 |
| RPN2B | 26S proteasome non-ATPase regulatory subunit 1 homolog B | Q9MAT0 |
| RPL10AA | Large ribosomal subunit protein uL1z | Q8VZB9 |
| RPL10A | Large ribosomal subunit protein uL16z | Q93VT9 |
| RPS27AA | Ubiquitin-ribosomal protein eS31z fusion protein | P59271 |
| RACK1A | Small ribosomal subunit protein RACK1z | O24456 |
| RPL6A | Large ribosomal subunit protein eL6z | Q9FZ76 |
| RPL19A | Large ribosomal subunit protein eL19x | Q9SRX2 |
| RPP1A | Large ribosomal subunit protein P1w | Q8LCW9 |
| RIG | Small ribosomal subunit protein uS19u | Q08112 |
| RPL21A | Large ribosomal subunit protein eL21z/eL21y | Q43291 |
| RPL27AB | Large ribosomal subunit protein uL15y | Q9LR33 |
| RPL34A | Large ribosomal subunit protein eL34z | Q42351 |
| PFL | Small ribosomal subunit protein uS13z/uS13y/uS13x | P34788 |
| RPS12A | Small ribosomal subunit protein eS12z | Q9S9P1 |
| BMS1 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | F4IDR3 |
| WDR | Uncharacterized protein At1g15425 | Q8L403 |
| PWP2 | Periodic tryptophan protein 2 | Q8VYZ5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazina, A.; Shumilina, J.; Gazizova, N.; Repkin, E.; Frolov, A.; Minibayeva, F. S-Nitrosylated Proteins Involved in Autophagy in Triticum aestivum Roots: A Bottom-Up Proteomics Approach and In Silico Predictive Algorithms. Life 2023, 13, 2024. https://doi.org/10.3390/life13102024
Mazina A, Shumilina J, Gazizova N, Repkin E, Frolov A, Minibayeva F. S-Nitrosylated Proteins Involved in Autophagy in Triticum aestivum Roots: A Bottom-Up Proteomics Approach and In Silico Predictive Algorithms. Life. 2023; 13(10):2024. https://doi.org/10.3390/life13102024
Chicago/Turabian StyleMazina, Anastasia, Julia Shumilina, Natalia Gazizova, Egor Repkin, Andrej Frolov, and Farida Minibayeva. 2023. "S-Nitrosylated Proteins Involved in Autophagy in Triticum aestivum Roots: A Bottom-Up Proteomics Approach and In Silico Predictive Algorithms" Life 13, no. 10: 2024. https://doi.org/10.3390/life13102024







