SARS-CoV-2 in Animal Companions: A Serosurvey in Three Regions of Southern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Enzyme-Linked Immunosorbent Assay (ELISA)
2.3. Focus Reduction Neutralization Test (FRNT)
2.4. ELISA-Based Surrogate SARS-CoV-2 Virus Neutralization Test
2.5. Statistical Analysis
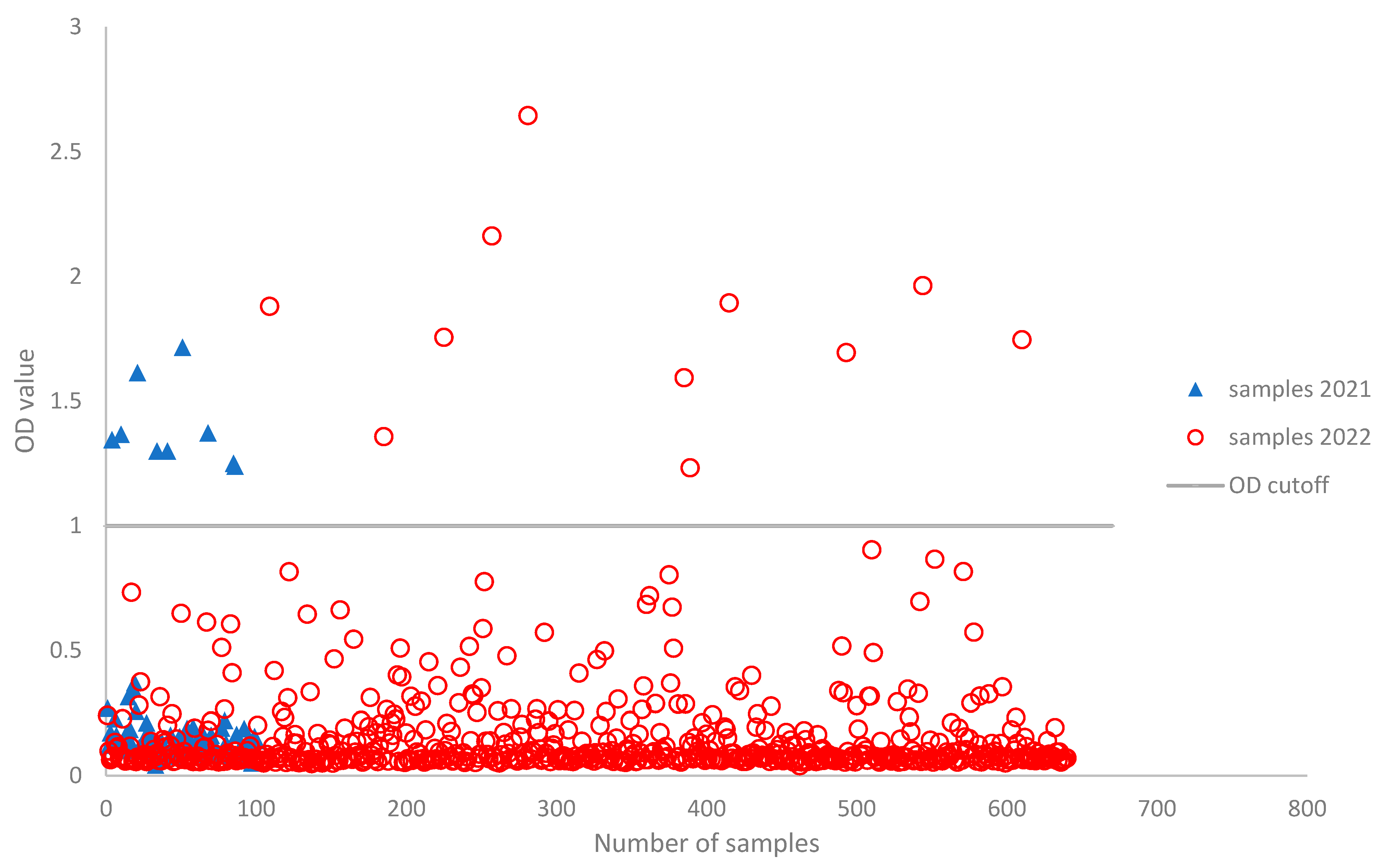
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Johnson, C.K.; Greig, D.J.; Kramer, S.; Che, X.; Wells, H.; Hicks, A.L.; Joly, D.O.; Wolfe, N.D.; Daszak, P.; et al. Global patterns in coronavirus diversity. Virus Evol. 2017, 3, vex012. [Google Scholar] [CrossRef] [PubMed]
- Leopardi, S.; Holmes, E.C.; Gastaldelli, M.; Tassoni, L.; Priori, P.; Scaravelli, D.; Zamperin, G.; De Benedictis, P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018, 58, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Sabir, J.S.M.; Lam, T.T.-Y.; Ahmed, M.M.M.; Li, L.; Shen, Y.; Abo-Aba, S.E.M.; Qureshi, M.I.; Abu-Zeid, M.; Zhang, Y.; Khiyami, M.A.; et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016, 351, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Goraichuk, I.V.; Arefiev, V.; Stegniy, B.T.; Gerilovych, A.P. Zoonotic and Reverse Zoonotic Transmissibility of SARS-CoV-2. Virus Res. 2021, 302, 198473. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- EFSA Panel on Animal Health and Welfare (AHAW); Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Rojas, J.L.G.; Gortázar, C.; et al. SARS-CoV-2 in animals: Susceptibility of animal species, risk for animal and public health, monitoring, prevention and control. EFSA J. 2023, 21, e07822. [Google Scholar] [PubMed]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.J.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F.; et al. Detection of SARS-CoV-2 in respiratory samples from cats in the UK associated with human-to-cat transmission. Vet. Rec. 2021, 188, e247. [Google Scholar] [CrossRef] [PubMed]
- Medkour, H.; Catheland, S.; Boucraut-Baralon, C.; Laidoudi, Y.; Sereme, Y.; Pingret, J.-L.; Million, M.; Houhamdi, L.; Levasseur, A.; Cabassu, J.; et al. First evidence of human-to-dog transmission of SARS-CoV-2 B.1.160 variant in France. Transbound Emerg. Dis. 2022, 69, e823–e830. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Siu, G.K.H.; Yuan, S.; Ip, J.D.; Cai, J.P.; Chu, A.W.H.; Chan, W.M.; Abdullah, S.M.U.; Luo, C.; Chan, B.P.C.; et al. Probable Animal-to-Human Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Delta Variant AY.127 Causing a Pet Shop-Related Coronavirus Disease 2019 (COVID-19) Outbreak in Hong Kong. Clin. Infect. Dis. 2022, 75, e76–e81. [Google Scholar] [CrossRef] [PubMed]
- Sila, T.; Sunghan, J.; Laochareonsuk, W.; Surasombatpattana, S.; Kongkamol, C.; Ingviya, T.; Siripaitoon, P.; Kositpantawong, N.; Kanchanasuwan, S.; Hortiwakul, T.; et al. Suspected Cat-to-Human Transmission of SARS-CoV-2, Thailand, July–September 2021–Volume 28, Number 7—July 2022. Emerg. Infect. Dis. 2022, 28, 1485–1488. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control; World Health Organization Regional Office for Europe. Methods for the Detection and Characterisation of SARS-CoV-2 Variants–Second Update; Technical Report; ECDC European Centre for Disease Prevention and Control: Stockholm, Sweden; WHO European Region: Copenhagen, Denmark, 2022. [Google Scholar]
- Chen, Y.; Huang, S.; Zhou, L.; Wang, X.; Yang, H.; Li, W. Coronavirus Disease 2019 (COVID-19): Emerging detection technologies and auxiliary analysis. J. Clin. Lab. Anal. 2022, 36, e24152. [Google Scholar] [CrossRef] [PubMed]
- Banga Ndzouboukou, J.-L.; Zhang, Y.; Fan, X.-L. Recent Developments in SARS-CoV-2 Neutralizing Antibody Detection Methods. Curr. Med. Sci. 2021, 41, 1052–1064. [Google Scholar] [CrossRef]
- Wang, W.; Lusvarghi, S.; Subramanian, R.; Epsi, N.J.; Wang, R.; Goguet, E.; Fries, A.C.; Echegaray, F.; Vassell, R.; Coggins, A.A.; et al. Antigenic cartography of well-characterized human sera shows SARS-CoV-2 neutralization differences based on infection and vaccination history. Cell Host Microbe 2022, 30, 1745–1758.e7. [Google Scholar] [CrossRef]
- Berguido, F.J.; Burbelo, P.D.; Bortolami, A.; Bonfante, F.; Wernike, K.; Hoffmann, D.; Balkema-Buschmann, A.; Beer, M.; Dundon, W.G.; Lamien, C.E.; et al. Serological Detection of SARS-CoV-2 Antibodies in Naturally-Infected Mink and Other Experimentally-Infected Animals. Viruses 2021, 13, 1649. [Google Scholar] [CrossRef]
- Padoan, A.; Bonfante, F.; Cosma, C.; Di Chiara, C.; Sciacovelli, L.; Pagliari, M.; Bortolami, A.; Costenaro, P.; Musso, G.; Basso, D.; et al. Analytical and clinical performances of a SARS-CoV-2 S-RBD IgG assay: Comparison with neutralization titers. Clin. Chem. Lab. Med. 2021, 59, 1444–1452. [Google Scholar] [CrossRef]
- Rössler, A.; Netzl, A.; Knabl, L.; Schäfer, H.; Wilks, S.H.; Bante, D.; Falkensammer, B.; Borena, W.; von Laer, D.; Smith, D.J.; et al. BA.2 and BA.5 omicron differ immunologically from both BA.1 omicron and pre-omicron variants. Nat. Commun. 2022, 13, 7701. [Google Scholar] [CrossRef] [PubMed]
- Murphy, H.L.; Ly, H. Understanding the prevalence of SARS-CoV-2 (COVID-19) exposure in companion, captive, wild, and farmed animals. Virulence 2021, 12, 2777–2786. [Google Scholar] [CrossRef] [PubMed]
- Bellinati, L.; Campalto, M.; Mazzotta, E.; Ceglie, L.; Cavicchio, L.; Mion, M.; Lucchese, L.; Salomoni, A.; Bortolami, A.; Quaranta, E.; et al. One-Year Surveillance of SARS-CoV-2 Exposure in Stray Cats and Kennel Dogs from Northeastern Italy. Microorganisms 2023, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Maggialetti, N.; Villanova, I.; Castrì, A.; Greco, C.N.; Inchingolo, F.; Virgilio, D.; Moschetta, M.; Sardaro, A.; Ianora, A.A.S.; Scardapane, A. COVID-19 in Italy: Comparison of CT Findings from Time Zero to the Delta Variant. Microorganisms 2022, 10, 796. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Iaconelli, M.; Veneri, C.; Mancini, P.; Bonanno Ferraro, G.; Brandtner, D.; Lucentini, L.; Bonadonna, L.; Rossi, M.; Grigioni, M.; et al. The rapid spread of SARS-CoV-2 Omicron variant in Italy reflected early through wastewater surveillance. Sci. Total Environ. 2022, 837, 155767. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Michelitsch, A.; Allendorf, V.; Conraths, F.J.; Beer, M.; Denzin, N.; Wernike, K. Dogs and cats are less susceptible to the omicron variant of concern of SARS-CoV-2–A field study. Transbound Emerg. Dis. 2023, 2023, 1868732. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Rissmann, M.; Kok, A.; Rosu, M.E.; Schipper, D.; Breugem, T.I.; van den Doel, P.B.; Chandler, F.; Bestebroer, T.; de Wit, M.; et al. Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Sci. Immunol. 2022, 7, eabq4450. [Google Scholar] [CrossRef]
- van der Straten, K.; Guerra, D.; van Gils, M.J.; Bontjer, I.; Caniels, T.G.; van Willigen, H.D.G.; Wynberg, E.; Poniman, M.; Burger, J.A.; Bouhuijs, J.H.; et al. Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron. Immunity 2022, 55, 1725–1731.e4. [Google Scholar] [CrossRef]
- Stefanelli, P.; Trentini, F.; Petrone, D.; Mammone, A.; Ambrosio, L.; Manica, M.; Guzzetta, G.; d’Andrea, V.; Marziano, V.; Zardini, A.; et al. Tracking the progressive spread of the SARS-CoV-2 Omicron variant in Italy, December 2021 to January 2022. Eurosurveillance 2022, 27, 2200125. [Google Scholar] [CrossRef]
- Capozzi, L.; Bianco, A.; Del Sambro, L.; Simone, D.; Lippolis, A.; Notarnicola, M.; Pesole, G.; Pace, L.; Galante, D.; Parisi, A. Genomic Surveillance of Circulating SARS-CoV-2 in South East Italy: A One-Year Retrospective Genetic Study. Viruses 2021, 13, 731. [Google Scholar] [CrossRef]
- Padoan, A.; Bonfante, F.; Pagliari, M.; Bortolami, A.; Negrini, D.; Zuin, S.; Bozzato, D.; Cosma, C.; Sciacovelli, L.; Plebani, M. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine 2020, 62, 103101. [Google Scholar] [CrossRef] [PubMed]
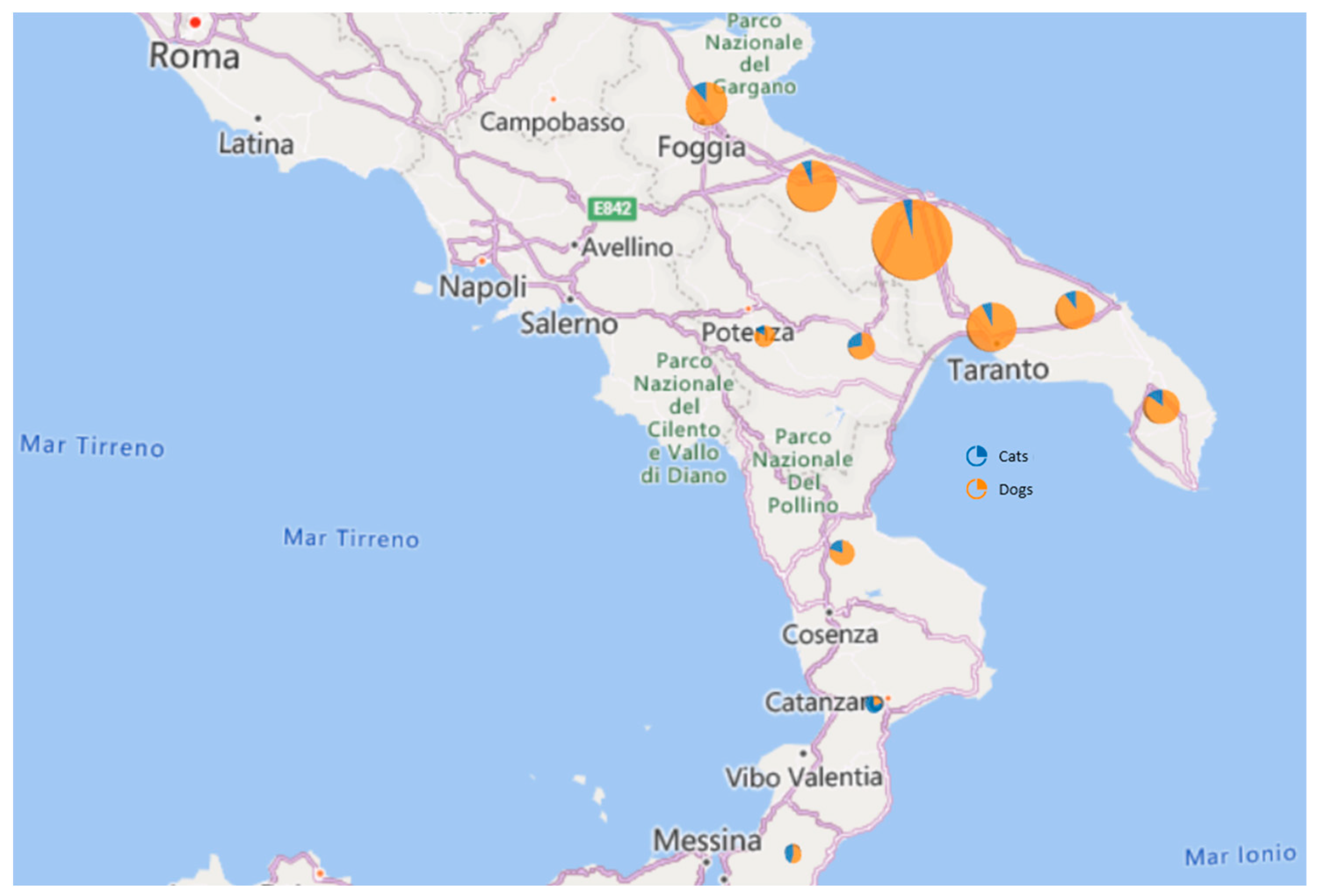

| 2021 | 2022 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples (n = 740) | Samples | ELISA | PRNT Assay | Samples | ELISA | PRNT Assay | ||||||||
| Region | Province | Dogs (n = 89) | Cats (n = 11) | Positive (D/C) | Negative | Positive | Negative | Dogs (n = 577) | Cats (n = 63) | Positive (D/C) | Negative | Positive | Negative | |
| Puglia | BA | 265 | 36 | 2 | 3(3/0) | 35 | 1 (1:40); Delta | 2 | 213 | 14 | 3 (3/0) | 259 | - | 5 |
| BT | 104 | 16 | 4 | 2(2/0) | 18 | - | 2 | 74 | 10 | 1 (1/0) | 101 | - | 3 | |
| FG | 72 | 9 | 1 | 1(1/0) | 9 | - | 1 | 55 | 7 | 2 (1/1) | 69 | - | 3 | |
| BR | 59 | 8 | 0 | 0 | 8 | - | 46 | 5 | 0 | 59 | - | - | ||
| TA | 98 | 8 | 1 | 1(1/0) | 8 | - | 1 | 80 | 9 | 2 (2/0) | 95 | - | 3 | |
| LE | 49 | 4 | 1 | 1(1/0) | 4 | - | 1 | 40 | 4 | 1 (1/0) | 47 | - | 2 | |
| Basilicata | PZ | 18 | 2 | 1 | 0 | 3 | - | 3 | 11 | 4 | 1 (1/0) | 17 | - | 1 |
| MT | 29 | 3 | 1 | 0 | 4 | - | 4 | 19 | 6 | 0 | 29 | - | - | |
| Calabria | CS | 25 | 1 | 0 | 1(1/0) | 0 | - | - | 23 | 1 | 1 (1/0) | 23 | 1 (1:20); Omicron BA.2 | 1 |
| CZ | 10 | 1 | 0 | 0 | 1 | - | 1 | 8 | 1 | 0 | 10 | - | - | |
| RC | 11 | 1 | 0 | 0 | 1 | - | 1 | 8 | 2 | 0 | 11 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, A.; Bortolami, A.; Miccolupo, A.; Sottili, R.; Ghergo, P.; Castellana, S.; Del Sambro, L.; Capozzi, L.; Pagliari, M.; Bonfante, F.; et al. SARS-CoV-2 in Animal Companions: A Serosurvey in Three Regions of Southern Italy. Life 2023, 13, 2354. https://doi.org/10.3390/life13122354
Bianco A, Bortolami A, Miccolupo A, Sottili R, Ghergo P, Castellana S, Del Sambro L, Capozzi L, Pagliari M, Bonfante F, et al. SARS-CoV-2 in Animal Companions: A Serosurvey in Three Regions of Southern Italy. Life. 2023; 13(12):2354. https://doi.org/10.3390/life13122354
Chicago/Turabian StyleBianco, Angelica, Alessio Bortolami, Angela Miccolupo, Roldano Sottili, Paola Ghergo, Stefano Castellana, Laura Del Sambro, Loredana Capozzi, Matteo Pagliari, Francesco Bonfante, and et al. 2023. "SARS-CoV-2 in Animal Companions: A Serosurvey in Three Regions of Southern Italy" Life 13, no. 12: 2354. https://doi.org/10.3390/life13122354
APA StyleBianco, A., Bortolami, A., Miccolupo, A., Sottili, R., Ghergo, P., Castellana, S., Del Sambro, L., Capozzi, L., Pagliari, M., Bonfante, F., Ridolfi, D., Bulzacchelli, C., Giannico, A., & Parisi, A. (2023). SARS-CoV-2 in Animal Companions: A Serosurvey in Three Regions of Southern Italy. Life, 13(12), 2354. https://doi.org/10.3390/life13122354





