Characterization and microRNA Expression Analysis of Serum-Derived Extracellular Vesicles in Severe Liver Injury from Chronic HBV Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Sample Collection
2.2. Serum EV Isolation
2.3. Characterization and Quantification of EVs
2.4. RNA Extraction from Serum EVs
2.5. RNA Sequencing and Data Analysis
2.6. RT-qPCR
2.7. Statistical Analysis
3. Results
3.1. Study Population
3.2. Characterization of Serum EVs
3.3. Serum-Derived Exosome miRNA Screening
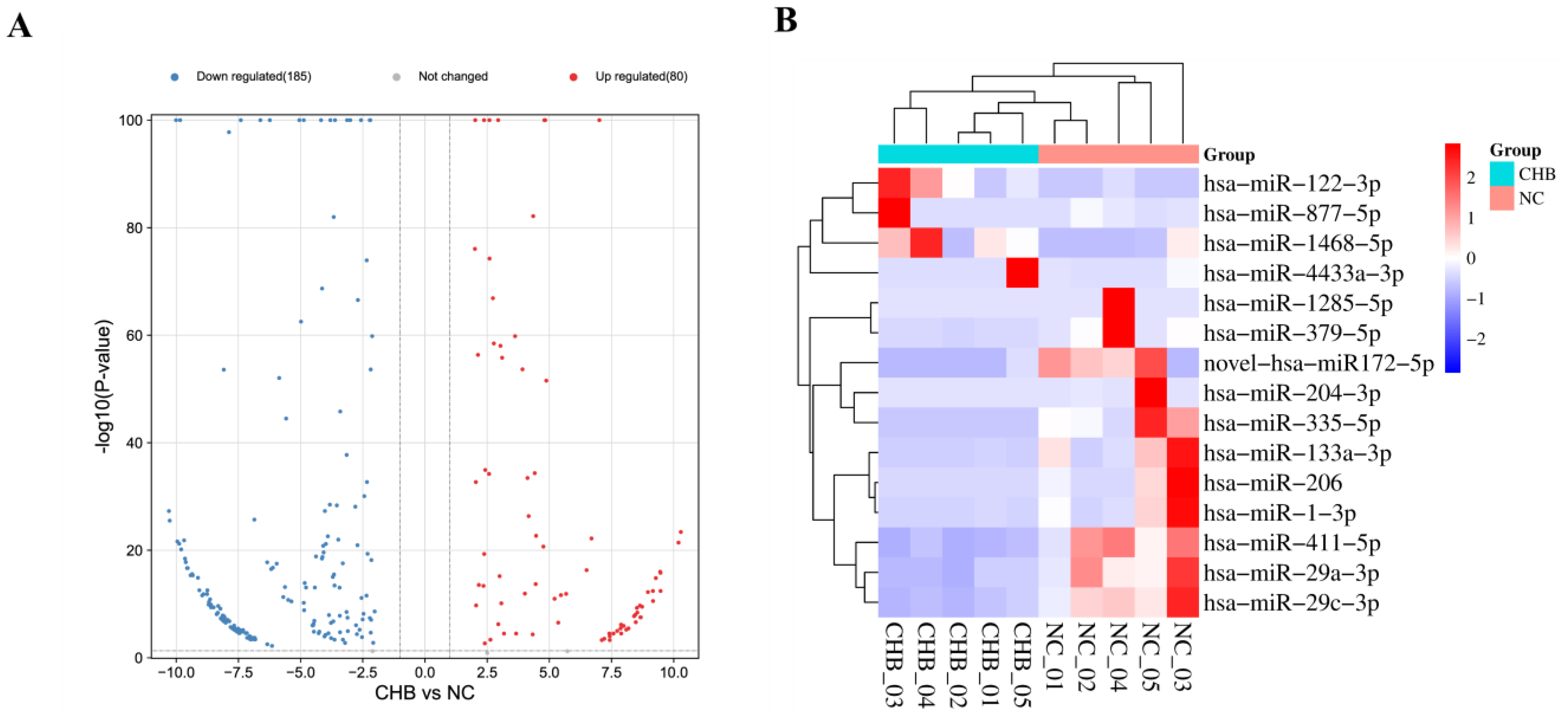
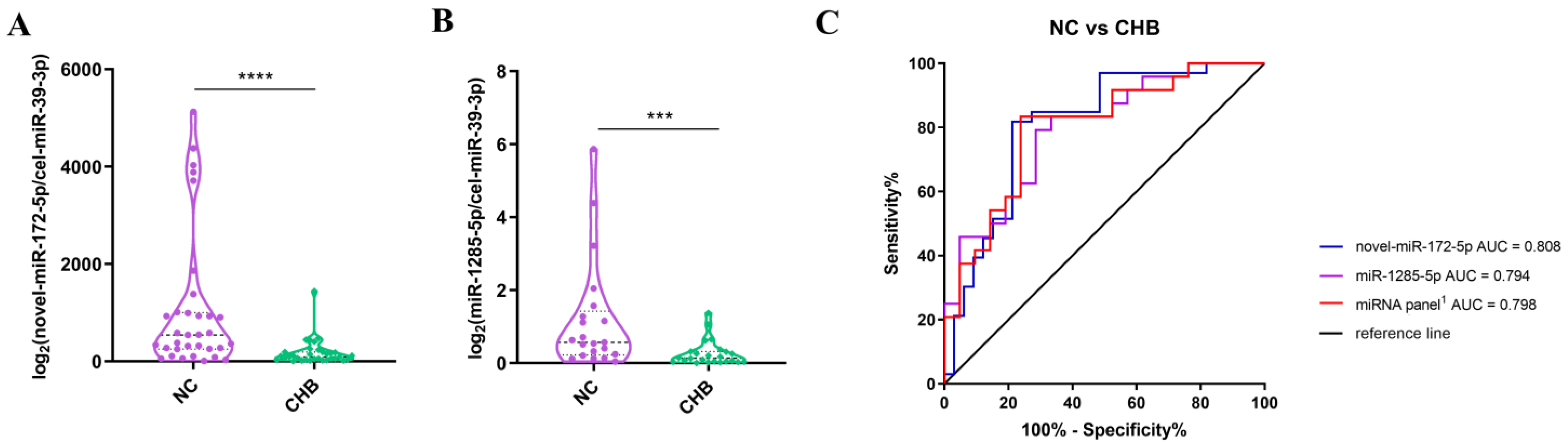
3.4. The Differences in the Expressions of miRNAs between the NC Group and the Severe Liver Injury-CHB Group
3.5. The Difference in the Expressions of miRNAs between the NC Group and the DeCi Group
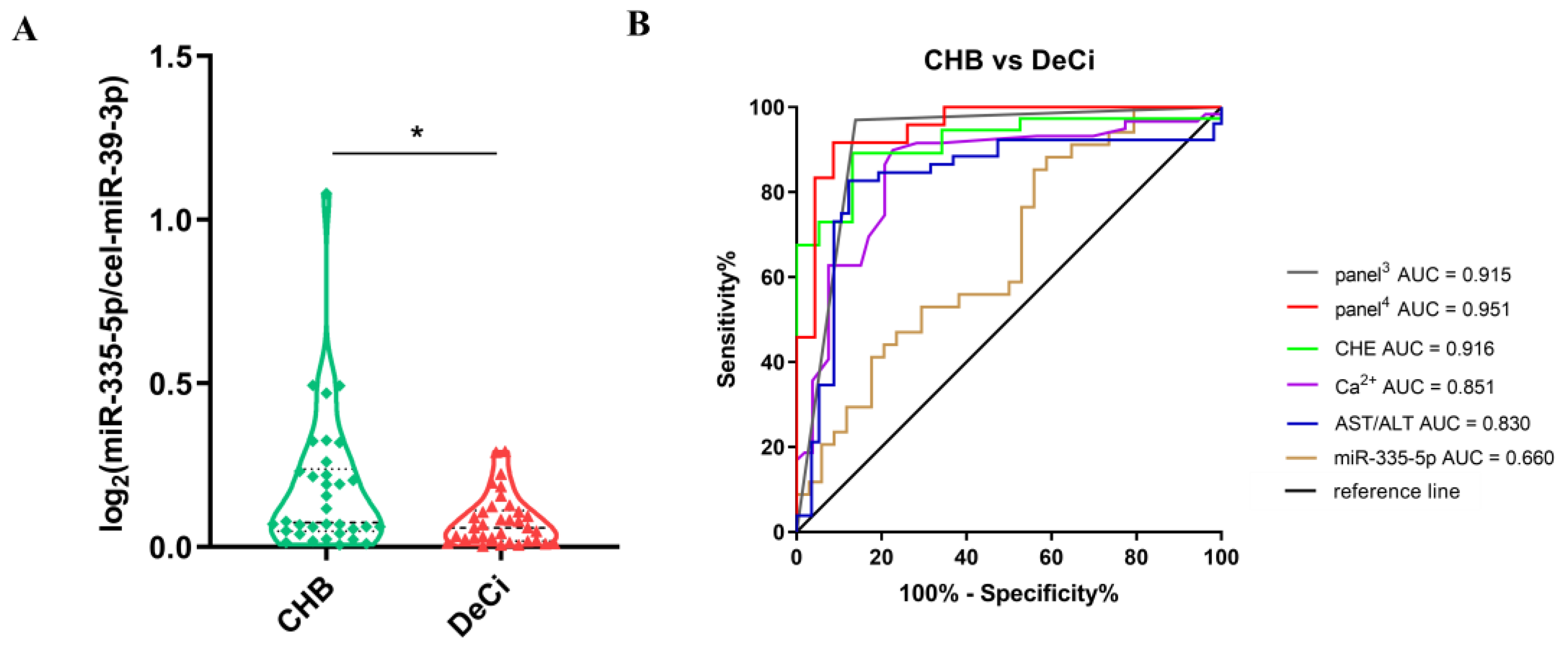
3.6. The Differences in the Expressions of miRNAs between the Severe Liver Injury-CHB Group and the DeCi Group
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asrani, S.K.; Hall, L.; Reddy, V.; Ogola, G.; Izzy, M. Comorbid Chronic Diseases and Survival in Compensated and Decompensated Cirrhosis: A Population-Based Study. Off. J. Am. Coll. Gastroenterol. | ACG 2022, 117, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhu, Y.; Zhang, J.; Chen, W.; Li, Z.; Li, L.; Zhang, L.; Cao, D. Potential Circulating Biomarkers of Circulating Chemokines CCL5, MIP-1β and HA as for Early Detection of Cirrhosis Related to Chronic HBV (Hepatitis B Virus) Infection. BMC Infect. Dis. 2019, 19, 523. [Google Scholar] [CrossRef] [PubMed]
- Lampertico, P.; Agarwal, K.; Berg, T.; Buti, M.; Janssen, H.L.A.; Papatheodoridis, G.; Zoulim, F.; Tacke, F. EASL 2017 Clinical Practice Guidelines on the Management of Hepatitis B Virus Infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of Chronic Hepatitis B Virus Infection. J. Hepatol. 2012, 57, 167–185. [Google Scholar] [CrossRef]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver Cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Marcellin, P.; Kutala, B.K. Liver Diseases: A Major, Neglected Global Public Health Problem Requiring Urgent Actions and Large-Scale Screening. Liver Int. 2018, 38, 2–6. [Google Scholar] [CrossRef]
- World Health Organisation Guideline for the Prevention, Care and Treatment of Persons with Chronic Hepatitis B Infection; World Health Organization: Geneva, Switzerland, 2015; bookshelf ID:NBK305553.
- Sonneveld, M.J.; Brouwer, W.P.; Chan, H.L.-Y.; Piratvisuth, T.; Jia, J.-D.; Zeuzem, S.; Liaw, Y.-F.; Hansen, B.E.; Choi, H.; Wat, C.; et al. Optimisation of the Use of APRI and FIB-4 to Rule out Cirrhosis in Patients with Chronic Hepatitis B: Results from the SONIC-B Study. Lancet Gastroenterol. Hepatol. 2019, 4, 538–544. [Google Scholar] [CrossRef]
- Calvopina, D.A.; Coleman, M.A.; Lewindon, P.J.; Ramm, G.A. Function and Regulation of MicroRNAs and Their Potential as Biomarkers in Paediatric Liver Disease. Int. J. Mol. Sci. 2016, 17, 1795. [Google Scholar] [CrossRef]
- Bedossa, P.; Dargère, D.; Paradis, V. Sampling Variability of Liver Fibrosis in Chronic Hepatitis C. Hepatology 2003, 38, 1449–1457. [Google Scholar] [CrossRef]
- Yang, G.; Zhuang, L.; Sun, T.; Yeo, Y.H.; Tao, L.; Zhang, W.; Ma, W.; Wu, L.; Yang, Z.; Yang, Y.; et al. Serum Glial Cell Line-Derived Neurotrophic Factor (SGDNF) Is a Novel Biomarker in Predicting Cirrhosis in Patients with Chronic Hepatitis B. 2021, 2022, 1048104. Can. J. Gastroenterol. Hepatol. 2021, 2022, 1048104. [Google Scholar] [CrossRef]
- Poynard, T.; de Ledinghen, V.; Zarski, J.P.; Stanciu, C.; Munteanu, M.; Vergniol, J.; France, J.; Trifan, A.; Le Naour, G.; Vaillant, J.C.; et al. Relative Performances of FibroTest, Fibroscan, and Biopsy for the Assessment of the Stage of Liver Fibrosis in Patients with Chronic Hepatitis C: A Step toward the Truth in the Absence of a Gold Standard. J. Hepatol. 2012, 56, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Imajo, K.; Kessoku, T.; Honda, Y.; Tomeno, W.; Ogawa, Y.; Mawatari, H.; Fujita, K.; Yoneda, M.; Taguri, M.; Hyogo, H.; et al. Magnetic Resonance Imaging More Accurately Classifies Steatosis and Fibrosis in Patients with Nonalcoholic Fatty Liver Disease than Transient Elastography. Gastroenterology 2016, 150, 626–637. [Google Scholar] [CrossRef]
- Chen, T.; Wang, C.; Yu, H.; Ding, M.; Zhang, C.; Lu, X.; Zhang, C.-Y.; Zhang, C. Increased Urinary Exosomal MicroRNAs in Children with Idiopathic Nephrotic Syndrome. EBioMedicine 2019, 39, 552–561. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic Stem Cell-Derived Microvesicles Reprogram Hematopoietic Progenitors: Evidence for Horizontal Transfer of MRNA and Protein Delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Quesenberry, P.J.; Aliotta, J.M.; Deregibus, M.C.; Camussi, G. Role of Extracellular RNA-Carrying Vesicles in Cell Differentiation and Reprogramming. Stem Cell Res. Ther. 2015, 6, 153. [Google Scholar] [CrossRef] [PubMed]
- de Paredes, A.G.G.; Manicardi, N.; Tellez, L.; Ibañez, L.; Royo, F.; Bermejo, J.; Blanco, C.; Fondevila, C.; Lanza, V.F.; Garcia-Bermejo, L.; et al. Molecular Profiling of Decompensated Cirrhosis by a Novel MicroRNA Signature. Hepatol. Commun. 2021, 5, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Lemoinne, S.; Thabut, D.; Housset, C.; Moreau, R.; Valla, D.; Boulanger, C.M.; Rautou, P.-E. The Emerging Roles of Microvesicles in Liver Diseases. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 350–361. [Google Scholar] [CrossRef]
- Thietart, S.; Rautou, P.-E. Extracellular Vesicles as Biomarkers in Liver Diseases: A Clinician’s Point of View. J. Hepatol. 2020, 73, 1507–1525. [Google Scholar] [CrossRef]
- Kornek, M.; Lynch, M.; Mehta, S.H.; Lai, M.; Exley, M.; Afdhal, N.H.; Schuppan, D. Circulating Microparticles as Disease-Specific Biomarkers of Severity of Inflammation in Patients with Hepatitis C or Nonalcoholic Steatohepatitis. Gastroenterology 2012, 143, 448–458. [Google Scholar] [CrossRef]
- Rautou, P.-E.; Bresson, J.; Sainte-Marie, Y.; Vion, A.-C.; Paradis, V.; Renard, J.; Devue, C.; Heymes, C.; Letteron, P.; Elkrief, L.; et al. Abnormal Plasma Microparticles Impair Vasoconstrictor Responses in Patients With Cirrhosis. Gastroenterology 2012, 143, 166–176.e6. [Google Scholar] [CrossRef] [PubMed]
- Payancé, A.; Silva-Junior, G.; Bissonnette, J.; Tanguy, M.; Pasquet, B.; Levi, C.; Roux, O.; Nekachtali, O.; Baiges, A.; Hernández-Gea, V.; et al. Hepatocyte Microvesicle Levels Improve Prediction of Mortality in Patients with Cirrhosis. Hepatology 2018, 68, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Campello, E.; Zanetto, A.; Spiezia, L.; Radu, C.M.; Gavasso, S.; Ferrarese, A.; Farinati, F.; Senzolo, M.; Simioni, P. Hypercoagulability Detected by Circulating Microparticles in Patients with Hepatocellular Carcinoma and Cirrhosis. Thromb. Res. 2016, 143, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Diehl, P.; Fricke, A.; Sander, L.; Stamm, J.; Bassler, N.; Htun, N.; Ziemann, M.; Helbing, T.; El-Osta, A.; Jowett, J.B.M.; et al. Microparticles: Major Transport Vehicles for Distinct MicroRNAs in Circulation. Cardiovasc. Res. 2012, 93, 633–644. [Google Scholar] [CrossRef]
- Vallabhajosyula, P.; Korutla, L.; Habertheuer, A.; Yu, M.; Rostami, S.; Yuan, C.X.; Reddy, S.; Liu, C.; Korutla, V.; Koeberlein, B.; et al. Tissue-Specific Exosome Biomarkers for Noninvasively Monitoring Immunologic Rejection of Transplanted Tissue. J. Clin. Investig. 2017, 127, 1375–1391. [Google Scholar] [CrossRef]
- Habertheuer, A.; Korutla, L.; Rostami, S.; Reddy, S.; Lal, P.; Naji, A.; Vallabhajosyula, P. Donor Tissue-Specific Exosome Profiling Enables Noninvasive Monitoring of Acute Rejection in Mouse Allogeneic Heart Transplantation. J. Thorac. Cardiovasc. Surg. 2018, 155, 2479–2489. [Google Scholar] [CrossRef]
- Szabo, G.; Bala, S. MicroRNAs in Liver Disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef]
- Farid, W.R.R.; Pan, Q.; van der Meer, A.J.P.; de Ruiter, P.E.; Ramakrishnaiah, V.; de Jonge, J.; Kwekkeboom, J.; Janssen, H.L.A.; Metselaar, H.J.; Tilanus, H.W.; et al. Hepatocyte-Derived MicroRNAs as Serum Biomarkers of Hepatic Injury and Rejection after Liver Transplantation. Liver Transplant. 2012, 18, 290–297. [Google Scholar] [CrossRef]
- Ohno, S.; Ishikawa, A.; Kuroda, M. Roles of Exosomes and Microvesicles in Disease Pathogenesis. Adv. Drug Deliv. Rev. 2013, 65, 398–401. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, S.Y.; Ko, E.; Lee, J.-H.; Yi, H.-S.; Yoo, Y.J.; Je, J.; Suh, S.J.; Jung, Y.K.; Kim, J.H.; et al. Exosomes Derived from Palmitic Acid-Treated Hepatocytes Induce Fibrotic Activation of Hepatic Stellate Cells. Sci. Rep. 2017, 7, 3710. [Google Scholar] [CrossRef] [PubMed]
- Pirola, C.J.; Gianotti, T.F.; Castaño, G.O.; Mallardi, P.; San Martino, J.; Ledesma, M.M.G.L.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating MicroRNA Signature in Non-Alcoholic Fatty Liver Disease: From Serum Non-Coding RNAs to Liver Histology and Disease Pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Akuta, N.; Kawamura, Y.; Watanabe, C.; Nishimura, A.; Okubo, M.; Mori, Y.; Fujiyama, S.; Sezaki, H.; Hosaka, T.; Kobayashi, M.; et al. Impact of Sodium Glucose Cotransporter 2 Inhibitor on Histological Features and Glucose Metabolism of Non-alcoholic Fatty Liver Disease Complicated by Diabetes Mellitus. Hepatol. Res. 2019, 49, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of Serum Exosomal MicroRNA-21 in Human Hepatocellular Carcinoma. Biomed Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef]
- Xue, X.; Zhao, Y.; Wang, X.; Qin, L.; Hu, R. Development and Validation of Serum Exosomal MicroRNAs as Diagnostic and Prognostic Biomarkers for Hepatocellular Carcinoma. J. Cell. Biochem. 2019, 120, 135–142. [Google Scholar] [CrossRef]
- Chinese Society of Infectious Diseases; Chinese Society of Hepatology. The Guidelines of Prevention and Treatment for Chronic Hepatitis B (2019 Version). Zhonghua Gan Zang Bing Za Zhi = Zhonghua Ganzangbing Zazhi = Chin. J. Hepatol. 2019, 27, 938. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering MicroRNAs from Deep Sequencing Data Using MiRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Callewaert, N.; Van Vlierberghe, H.; Van Hecke, A.; Laroy, W.; Delanghe, J.; Contreras, R. Noninvasive Diagnosis of Liver Cirrhosis Using DNA Sequencer–Based Total Serum Protein Glycomics. Nat. Med. 2004, 10, 429–434. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Zhu, J.-M.; Wu, H.; Fan, J.; Zhou, J.; Hu, J.; Yu, Q.; Liu, T.-T.; Yang, L.; Wu, C.-L.; et al. Circulating MicroRNAs as a Fingerprint for Liver Cirrhosis. PLoS ONE 2013, 8, e66577. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yao, Q.; Butt, A.M.; Guo, J.; Tian, Z.; Bao, X.; Li, H.; Meng, Q.; Lu, J. Expression Profiling of Serum MicroRNA-101 in HBV-Associated Chronic Hepatitis, Liver Cirrhosis, and Hepatocellular Carcinoma. Cancer Biol. Ther. 2014, 15, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Mölleken, C.; Sitek, B.; Henkel, C.; Poschmann, G.; Sipos, B.; Wiese, S.; Warscheid, B.; Broelsch, C.; Reiser, M.; Friedman, S.L.; et al. Detection of Novel Biomarkers of Liver Cirrhosis by Proteomic Analysis. Hepatology 2009, 49, 1257–1266. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian MicroRNAs Predominantly Act to Decrease Target MRNA Levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.-Y.; Cho, H.C.; Shim, S.G.; Paik, Y.-H. Serum Exosomal MicroRNAs as Novel Biomarkers for Hepatocellular Carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef]
- Csak, T.; Bala, S.; Lippai, D.; Satishchandran, A.; Catalano, D.; Kodys, K.; Szabo, G. MicroRNA-122 Regulates Hypoxia-Inducible Factor-1 and Vimentin in Hepatocytes and Correlates with Fibrosis in Diet-Induced Steatohepatitis. Liver Int. 2015, 35, 532–541. [Google Scholar] [CrossRef]
- Cho, H.J.; Eun, J.W.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Kim, S.S.; Cho, S.W.; Cheong, J.Y. Serum Exosomal MicroRNA, MiR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J. Clin. Med. 2020, 9, 281. [Google Scholar] [CrossRef]
- Saviano, A.; Henderson, N.C.; Baumert, T.F. Single-Cell Genomics and Spatial Transcriptomics: Discovery of Novel Cell States and Cellular Interactions in Liver Physiology and Disease Biology. J. Hepatol. 2020, 73, 1219–1230. [Google Scholar] [CrossRef]
- Daemen, S.; Gainullina, A.; Kalugotla, G.; He, L.; Chan, M.M.; Beals, J.W.; Liss, K.H.; Klein, S.; Feldstein, A.E.; Finck, B.N.; et al. Dynamic Shifts in the Composition of Resident and Recruited Macrophages Influence Tissue Remodeling in NASH. Cell Rep. 2021, 34, 108626. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef]
- Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Giuliani, A.; Maggi, M.; Sciarra, A.; Fais, S. Plasmatic Exosome Number and Size Distinguish Prostate Cancer Patients From Healthy Individuals: A Prospective Clinical Study. Front. Oncol. 2021, 11, 4258. [Google Scholar] [CrossRef] [PubMed]
- Burrello, J.; Gai, C.; Tetti, M.; Lopatina, T.; Deregibus, M.C.; Veglio, F.; Mulatero, P.; Camussi, G.; Monticone, S. Characterization and Gene Expression Analysis of Serum-Derived Extracellular Vesicles in Primary Aldosteronism. Hypertension 2019, 74, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Mizrachi, L.; Jy, W.; Jimenez, J.J.; Pastor, J.; Mauro, L.M.; Horstman, L.L.; De Marchena, E.; Ahn, Y.S. High Levels of Circulating Endothelial Microparticles in Patients with Acute Coronary Syndromes. Am. Heart J. 2003, 145, 962–970. [Google Scholar] [CrossRef]
- Amabile, N.; Guérin, A.P.; Leroyer, A.; Mallat, Z.; Nguyen, C.; Boddaert, J.; London, G.M.; Tedgui, A.; Boulanger, C.M. Circulating Endothelial Microparticles Are Associated with Vascular Dysfunction in Patients with End-Stage Renal Failure. J. Am. Soc. Nephrol. 2005, 16, 3381–3388. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, F.; Darmon, P.; Hugel, B.; Combes, V.; Sanmarco, M.; Velut, J.-G.; Arnoux, D.; Charpiot, P.; Freyssinet, J.-M.; Oliver, C.; et al. Type 1 And Type 2 Diabetic Patients Display Different Patterns of Cellular Microparticles. Diabetes 2002, 51, 2840–2845. [Google Scholar] [CrossRef] [PubMed]
- González-Quintero, V.H.; Jiménez, J.J.; Jy, W.; Mauro, L.M.; Hortman, L.; O’Sullivan, M.J.; Ahn, Y. Elevated Plasma Endothelial Microparticles in Preeclampsia. Am. J. Obstet. Gynecol. 2003, 189, 589–593. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Wang, Z.; Zhou, J.; Zhang, Y. Higher Urine Exosomal MiR-193a Is Associated with a Higher Probability of Primary Focal Segmental Glomerulosclerosis and an Increased Risk of Poor Prognosis among Children with Nephrotic Syndrome. Front. Cell Dev. Biol. 2021, 9, 727370. [Google Scholar] [CrossRef]
- Savina, A.; Furlán, M.; Vidal, M.; Colombo, M.I. Exosome Release Is Regulated by a Calcium-Dependent Mechanism in K562 Cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef]
- Prudent, M.; Crettaz, D.; Delobel, J.; Seghatchian, J.; Tissot, J.D.; Lion, N. Differences between Calcium-Stimulated and Storage-Induced Erythrocyte-Derived Microvesicles. Transfus. Apher. Sci. 2015, 53, 153–158. [Google Scholar] [CrossRef]
- Lange, S.; Gallagher, M.; Kholia, S.; Kosgodage, U.S.; Hristova, M.; Hardy, J.; Inal, J.M. Peptidylarginine DeiminasesRoles in Cancer and Neurodegeneration and Possible Avenues for Therapeutic Intervention via Modulation of Exosome and Microvesicle (EMV) Release? Int. J. Mol. Sci. 2017, 18, 1196. [Google Scholar] [CrossRef]
- Choi, D.-Y.; Park, J.-N.; Paek, S.-H.; Choi, S.-C.; Paek, S.-H. Detecting Early-Stage Malignant Melanoma Using a Calcium Switch-Enriched Exosome Subpopulation Containing Tumor Markers as a Sample. Biosens. Bioelectron. 2021, 198, 113828. [Google Scholar] [CrossRef] [PubMed]
- Bucki, R.; Bachelot-Loza, C.; Zachowski, A.; Giraud, F.; Sulpice, J.C. Calcium Induces Phospholipid Redistribution and Microvesicle Release in Human Erythrocyte Membranes by Independent Pathways. Biochemistry 1998, 37, 15383–15391. [Google Scholar] [CrossRef] [PubMed]
- Draeger, A.; Schoenauer, R.; Atanassoff, A.P.; Wolfmeier, H.; Babiychuk, E.B. Dealing with Damage: Plasma Membrane Repair Mechanisms. Biochimie 2014, 107, 66–72. [Google Scholar] [CrossRef]
- Luo, W.-J.; Cheng, T.-Y.; Wong, K.-I.; Fang, W.; Liao, K.-M.; Hsieh, Y.-T.; Su, K.-Y. Novel Therapeutic Drug Identification and Gene Correlation for Fatty Liver Disease Using High-Content Screening: Proof of Concept. Eur. J. Pharm. Sci. 2018, 121, 106–117. [Google Scholar] [CrossRef]
- Zhao, X.; Fan, H.; Chen, X.; Zhao, X.; Wang, X.; Feng, Y.; Liu, M.; Li, S.; Tang, H. Hepatitis B Virus DNA Polymerase Restrains Viral Replication Through the CREB1/HOXA Distal Transcript Antisense RNA Homeobox A13 Axis. Hepatology 2021, 73, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Allan, G.J.; Beattie, J.; Flint, D.J. Epithelial Injury Induces an Innate Repair Mechanism Linked to Cellular Senescence and Fibrosis Involving IGF-Binding Protein-5. J. Endocrinol. 2008, 199, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Lecca, M.R.; Maag, C.; Berger, E.G.; Hennet, T. Fibrotic Response in Fibroblasts from Congenital Disorders of Glycosylation. J. Cell. Mol. Med. 2011, 15, 1788–1796. [Google Scholar] [CrossRef]
- Hironaka-Mitsuhashi, A.; Otsuka, K.; Gailhouste, L.; Calle, A.S.; Kumazaki, M.; Yamamoto, Y.; Fujiwara, Y.; Ochiya, T. MiR-1285-5p/TMEM194A Axis Affects Cell Proliferation in Breast Cancer. Cancer Sci. 2020, 111, 395–405. [Google Scholar] [CrossRef]
- Wang, X.; Yan, M.; Zhao, L.; Wu, Q.; Wu, C.; Chang, X.; Zhou, Z. Low-Dose Methylmercury-Induced Genes Regulate Mitochondrial Biogenesis via MiR-25 in Immortalized Human Embryonic Neural Progenitor Cells. Int. J. Mol. Sci. 2016, 17, 2058. [Google Scholar] [CrossRef]
- Villanova, L.; Barbini, C.; Piccolo, C.; Boe, A.; De Maria, R.; Fiori, M.E. MiR-1285-3p Controls Colorectal Cancer Proliferation and Escape from Apoptosis through DAPK2. Int. J. Mol. Sci. 2020, 21, 2423. [Google Scholar] [CrossRef]
- Pao, S.I.; Lin, L.T.; Chen, Y.H.; Chen, C.L.; Chen, J.T. Repression of Smad4 by MicroRNA-1285 Moderates TGF-β-Induced Epithelial–Mesenchymal Transition in Proliferative Vitreoretinopathy. PLoS ONE 2021, 16, e0254873. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Lin, F.; Sun, W.; Zhu, W.; Fang, D.; Luo, L.; Li, S.; Zhang, W.; Jiang, L. Exosome-Transmitted MiRNA-335-5p Promotes Colorectal Cancer Invasion and Metastasis by Facilitating EMT via Targeting RASA1. Mol. Ther. Nucleic Acids 2021, 24, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lu, H.; Liang, W.; Zhao, G.; Ren, L.; Hu, D.; Chang, Z.; Liu, Y.; Garcia-Barrio, M.T.; Zhang, J.; et al. Endothelial TFEB (Transcription Factor EB) Improves Glucose Tolerance via Upregulation of IRS (Insulin Receptor Substrate) 1 and IRS2. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 783–795. [Google Scholar] [CrossRef] [PubMed]


| Variable | Severe Liver Injury-CHB (n = 58) | DeCi (n = 33) | p Value | NC (n = 58) |
|---|---|---|---|---|
| Gender (male/female) | 50/8 | 26/7 | 0.23 | 23/35 |
| Age (years) | 42 (29, 54) | 53 (48, 61) | <0.001 | 37 (30.5, 49) |
| WBC (109/L) | 5.37 (4.23, 6.92) | 3.50 (2.42, 5.57) | <0.001 | / |
| N (109/L) | 3.60 (2.66, 4.69) | 2.16 (1.39, 4.30) | <0.001 | / |
| L (109/L) | 1.22 (0.92, 1.58) | 0.63 (0.46, 1.06) | <0.001 | / |
| PLT (109/L) | 137 (113, 181) | 73.00 (39.00, 109.75) | <0.001 | / |
| ALT (U/L) | 207.80 (84.05, 454.55) | 28.10 (14.90, 45.75) | <0.001 | / |
| AST (U/L) | 110.00 (69.15, 171.50) | 39.30 (26.93, 68.05) | <0.001 | / |
| AST/ALT | 0.57 (0.35, 1.00) | 1.60 (1.29, 2.14) | <0.001 | / |
| ALB (g/L) | 34.03 ± 5.13 | 29.05 ± 5.12 | <0.001 | / |
| GLO (g/L) | 29.80 (26.95, 34.00) | 30.80 (26.70, 33.40) | 0.56 | / |
| TBIL (µmol/L) | 144.50 (20.40, 201.95) | 38.60 (19.80, 89.50) | 0.01 | / |
| DBIL (µmol/L) | 101.50 (11.45, 163.10) | 16.95 (7.90, 60.58) | 0.005 | / |
| Cr (µmol/L) | 68.20 (56.45, 80.10) | 71.10 (61.98, 110.53) | 0.13 | / |
| INR | 1.18 (1.04, 1.43) | 1.42 (1.33, 1.56) | <0.001 | / |
| Serum calcium (mmol/L) | 2.12 ± 0.09 | 1.98 ± 0.12 | <0.001 | / |
| CHE (IU/mL) | 4524.30 (3674.00, 5855.88) | 2382.30 (1336.25, 3098.20) | <0.001 | / |
| AKP (U/L) | 114.90 (75.80, 132.40) | 85.90 (61.10, 131.68) | 0.08 | / |
| GGT (U/L) | 114.10 (77.88, 186.53) | 48.15 (21.00, 63.10) | <0.001 | / |
| IgA (g/L) | 2.77 (2.62, 4.17) | 4.41 (3.29, 6.76) | 0.004 | / |
| AFP (ng/mL) | 207.00 (7.54, 294.00) | 2.35 (1.34, 6.92) | <0.001 | / |
| HBeAg (±) | 25/25 | 18/33 | 0.01 | / |
| HBeAb (±) | 28/23 | 11/39 | 0.43 | / |
| HBV DNA (copies/mL) | 513,000 (21,300, 11,150,000) | 0.07 (0, 904) | <0.001 | / |
| Variable | rs | p |
|---|---|---|
| ALT | 0.449 ** | <0.001 |
| AST | 0.412 ** | 0.001 |
| AST/ALT | −0.296 * | 0.016 |
| GGT | 0.287 * | 0.04 |
| AFP | 0.397 ** | 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Liu, X.; Pan, M.; Zhang, Y.; Tang, X.; Liu, W.; Zhao, M.; Ma, J.; Zhou, N.; Jiang, Y.; et al. Characterization and microRNA Expression Analysis of Serum-Derived Extracellular Vesicles in Severe Liver Injury from Chronic HBV Infection. Life 2023, 13, 347. https://doi.org/10.3390/life13020347
Liu M, Liu X, Pan M, Zhang Y, Tang X, Liu W, Zhao M, Ma J, Zhou N, Jiang Y, et al. Characterization and microRNA Expression Analysis of Serum-Derived Extracellular Vesicles in Severe Liver Injury from Chronic HBV Infection. Life. 2023; 13(2):347. https://doi.org/10.3390/life13020347
Chicago/Turabian StyleLiu, Min, Xionghao Liu, Mengmeng Pan, Yu Zhang, Xiangling Tang, Wanxi Liu, Mingri Zhao, Jing Ma, Ning Zhou, Yongfang Jiang, and et al. 2023. "Characterization and microRNA Expression Analysis of Serum-Derived Extracellular Vesicles in Severe Liver Injury from Chronic HBV Infection" Life 13, no. 2: 347. https://doi.org/10.3390/life13020347
APA StyleLiu, M., Liu, X., Pan, M., Zhang, Y., Tang, X., Liu, W., Zhao, M., Ma, J., Zhou, N., Jiang, Y., Wang, W., & Liu, M. (2023). Characterization and microRNA Expression Analysis of Serum-Derived Extracellular Vesicles in Severe Liver Injury from Chronic HBV Infection. Life, 13(2), 347. https://doi.org/10.3390/life13020347





