Clinical Biomarker of Sterile Inflammation, HMGB1, in Patients with Chronic Non-Specific Low Back Pain: A Pilot Cross-Sectional Study
Abstract
1. Introduction
2. Methods
2.1. Participating Subjects
2.2. Laboratory Studies
2.2.1. Blood Collection and Processing
2.2.2. Immunophenotype Staining
2.2.3. HMGB1 Determination in Plasma and TC Supernatants
2.2.4. Preparation of Adherent Cell Cultures and Immunofluorescent Staining
2.2.5. Quantitative RT-PCR (qRT-PCR)
3. Statistics
4. Results
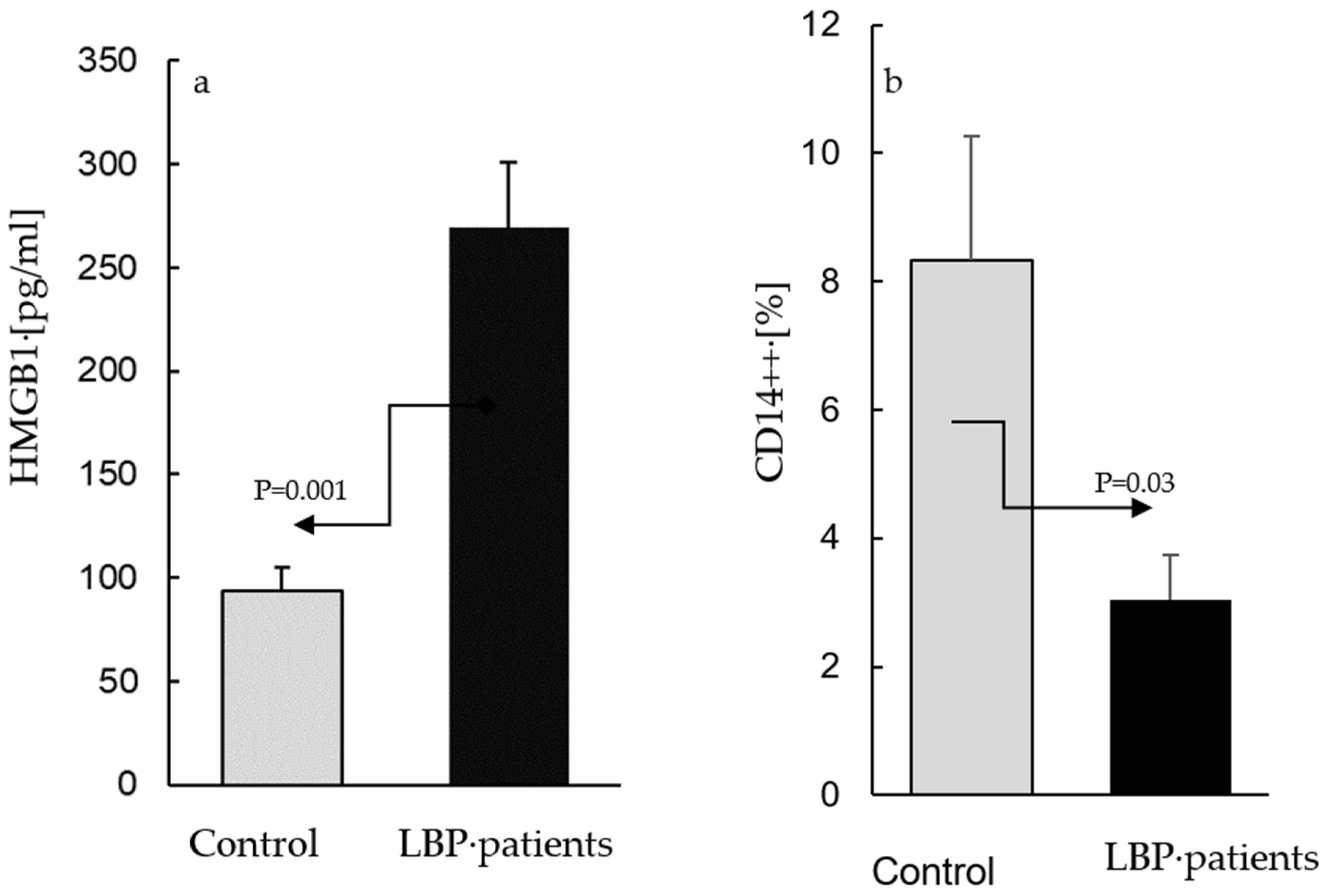
4.1. Determinations of Extracellular HMGB1 Levels
4.1.1. Plasma
4.1.2. AC Culture Supernatants
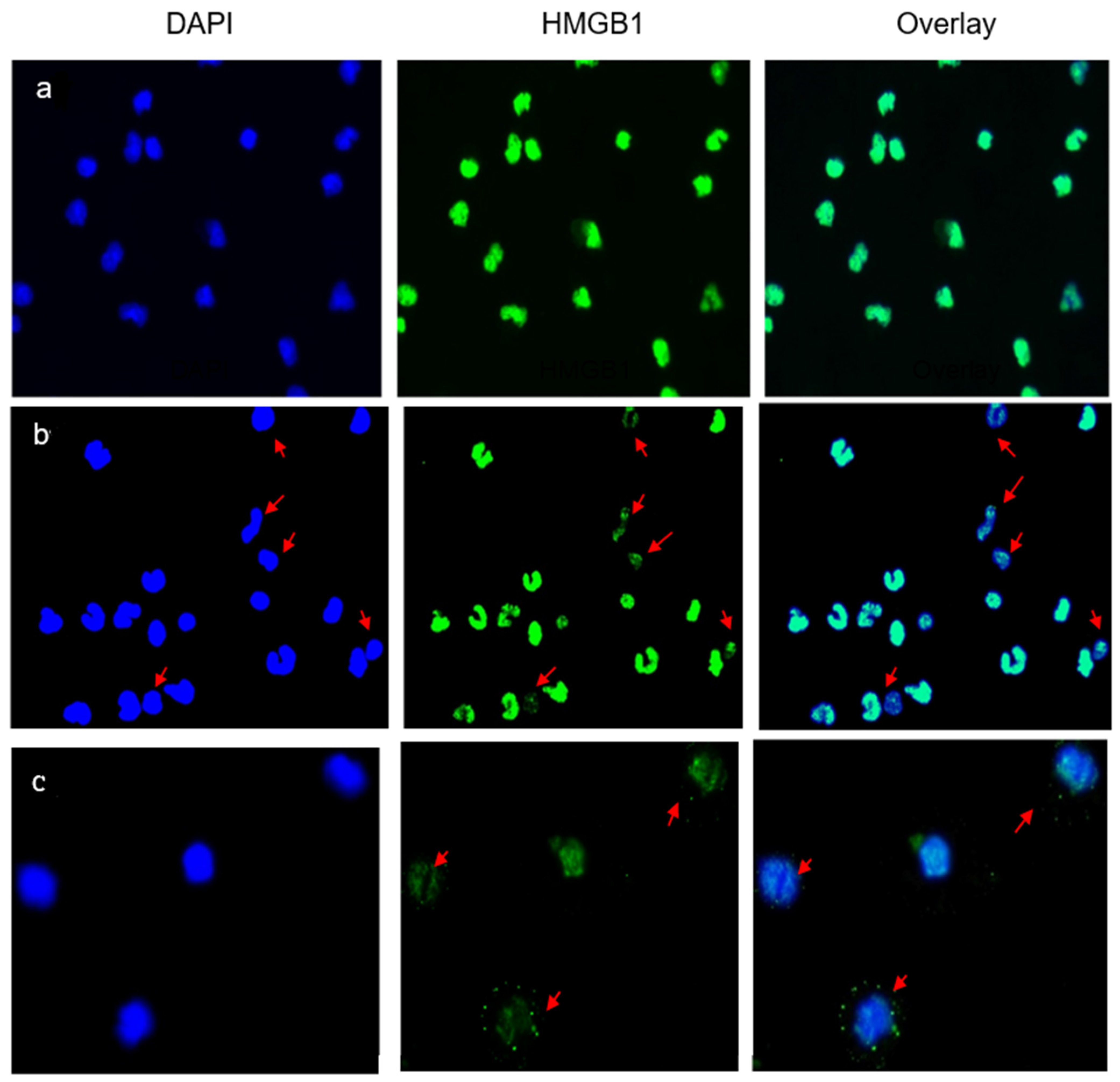
4.2. Immunofluorescent Staining of Adherent Cells (AC)
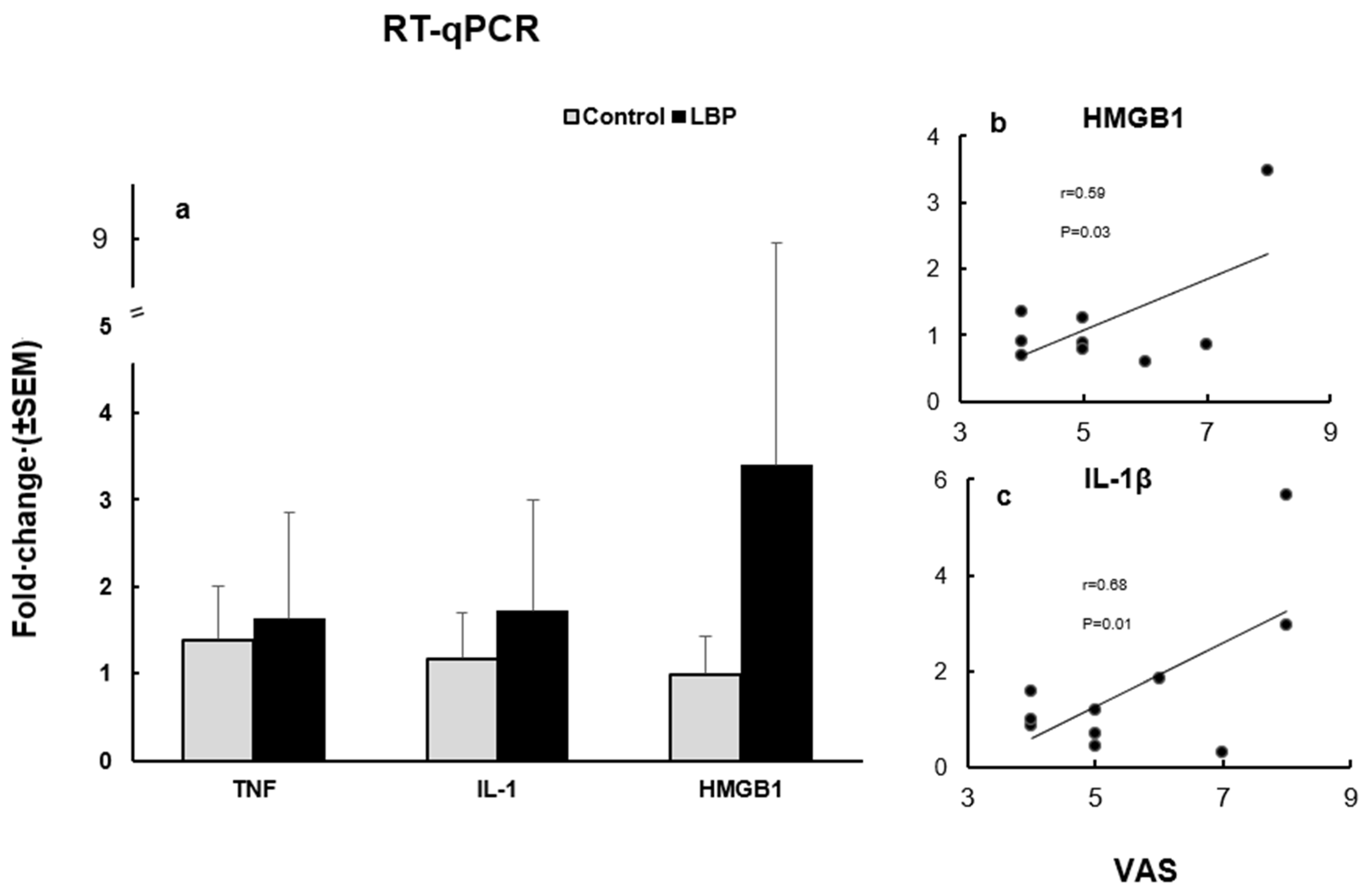
4.3. RT-qPCR
5. Discussion
6. Limitations
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maher, C.; Underwood, M.; Buchbinder, R. Non-specific low back pain. Lancet 2017, 389, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Hartvigsen, J.; Hancock, M.J.; Kongsted, A.; Louw, Q.; Ferreira, M.L.; Genevay, S.; Hoy, D.; Karppinen, J.; Pransky, G.; Sieper, J.; et al. What low back pain is and why we need to pay attention. Lancet 2018, 391, 2356–2367. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.N.; Jacobsen, H.E.; Khan, J.; Filippi, C.G.; Levine, M.; Lehman, R.A.; Riew, K.D.; Lenke, L.G.; Chahine, N.O. Inflammatory biomarkers of low back pain and disc degeneration: A review. Ann. N. Y. Acad. Sci. 2017, 1410, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.Z.; Wang, Y.; Cicuttini, F.M.; Hughes, H.J.; Chou, L.; Urquhart, D.M.; Ong, P.X.; Hussain, S.M. Association between inflammatory biomarkers and non-specific low back pain. Systematic review. Clin. J. Pain 2020, 36, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Teodorczyk-Injeyan, J.A.; Triano, J.J.; Injeyan, H.S. Nonspecific low back pain. Inflammatory profiles of patients with acute and chronic pain. Clin. J. Pain 2019, 35, 818–825. [Google Scholar] [CrossRef] [PubMed]
- Teodorczyk-Injeyan, J.A.; McGregor, M.; Triano, J.J.; Injeyan, S.H. Elevated production of nociceptive CC chemokines and sE-selectin in patients with low back pain and the effects of spinal manipulation. Clin. J. Pain 2018, 34, 68–75. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Liu, Z.-Z.; Duan, D.-P. Inflammation in low back pain may be detected from the peripheral blood: Suggestions for biomarker. Biosci. Rep. 2016, 36, e00361. [Google Scholar] [CrossRef]
- Langlais, D.; Fodil, N.; Gros, P.; Nakayama, T.; Hirahara, K.; Onodera, A.; Endo, Y.; Hosokawa, H.; Shinoda, K.; Tumes, D.J.; et al. Genetics of Infectious and Inflammatory Diseases: Overlapping Discoveries from Association and Exome-Sequencing Studies. Annu. Rev. Immunol. 2017, 35, 1–30. [Google Scholar] [CrossRef]
- Capoor, M.N.; Ruzicka, F.; Machackova, T.; Jancalek, R.; Smrcka, M.; Schmitz, J.E.; Hermanova, M.; Sana, J.; Michu, E.; Baird, J.C.; et al. Prevalence of Propionibacterium acnes in intervertebral discs of patients un-dergoing lumbar microdiscectomy: A prospective cross-sectional study. PLoS ONE 2016, 11, e0161676. [Google Scholar] [CrossRef]
- Gilligan, C.J.; Cohen, S.P.; Fischetti, V.A.; Hirsch, J.A.; Czaplewski, L.G. Chronic low back pain, bacterial infection and treatment with antibiotics. Spine J. 2021, 21, 903–914. [Google Scholar] [CrossRef]
- Chen, G.Y.; Nuñez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 2010, 10, 826–837. [Google Scholar] [CrossRef]
- Tsung, A.; Tohme, S.; Billiar, T.R. High-mobility group box-1 in sterile inflammation. J. Intern. Med. 2014, 276, 425–443. [Google Scholar] [CrossRef]
- Gardella, S.; Andrei, C.; Ferrera, D.; Lotti, L.V.; Torrisi, M.R.; Bianchi, M.E.; Rubartelli, A. The nuclear protein HMGB1 is secreted by monocytes via non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002, 3, 990–1001. [Google Scholar] [CrossRef]
- Andersson, U.; Wang, H.; Palmblad, K.; Aveberger, A.C.; Bloom, O.; Erlandsson-Harris, H.; Janson, A.; Kokkola, R.; Zhang, M.; Yang, H.; et al. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine syn-thesis in human monocytes. J. Exp. Med. 2000, 192, 565–570. [Google Scholar] [CrossRef]
- Venereau, E.; Casalgrandi, M.; Schiraldi, M.; Antoine, D.J.; Cattaneo, A.; De Marchis, F.; Liu, J.; Antonelli, A.; Preti, A.; Raeli, L.; et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proin-flammatory cytokine release. J. Exp. Med. 2012, 209, 1519–1528. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Czura, C.J.; Tracey, K.J. The cytokine activity of HMGB1. J. Leukoc. Biol. 2005, 78, 1–8. [Google Scholar] [CrossRef]
- Yamada, S.; Maruyama, I. HMGB1, a novel inflammatory cytokine. Clin. Chim. Acta 2007, 375, 36–42. [Google Scholar] [CrossRef]
- Gruber, H.E.; Hoelscher, G.L.; Bethea, S.; Ingram, J.; Cox, M.; Hanley, E.N., Jr. High-mobility box 1 gene, a potent proinflammatory mediator, is upregulated in more degenerated human discs in vivo and its receptor upregulated by TNF-alpha exposure in vitro. Exp. Mol. Pathol. 2015, 98, 427–430. [Google Scholar] [CrossRef]
- Shah, B.S.; Burt, K.G.; Jacobsen, T.; Fernandes, T.D.; Alipui, D.O.; Weber, K.T.; Levine, M.; Chavan, S.S.; Yang, H.; Tracey, K.J.; et al. High mobility group box-1 induces pro-inflammatory signalling in human nucleus pulposus cells via Toll-like receptor 4-dependent pathway. J. Orthop. Res. 2019, 37, 220–231. [Google Scholar] [CrossRef]
- Burke, J.G.; Watson RW, G.; McCormack DR, W.G.; Dowling, F.E.; Walsh, M.G.; Fitzpatrick, J.M. Intervertebral discs which cause low back pain secrete high levels of proin-flammatory cytokines. J. Bone Joint Surg. 2002, 84, 196–201. [Google Scholar] [CrossRef]
- Gaillard, C.; Borde, C.; Gozlan, J.; Maréchal, V.; Strauss, F. A high -sensitivity method for detection and measurement of HMGB1 protein concentration by high affinity-biding to DNA hemicatenanes. PLoS ONE 2008, 3, e2855. [Google Scholar] [CrossRef] [PubMed]
- Bonaldi, T.; Talamo, F.; Scaffidi, P.; Ferrera, D.; Porto, A.; Bachi, A.; Rubartelli, A.; Agresti, A.; Bianchi, M.E. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003, 22, 5551–5560. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Chen, G.; Li, J.; Ochani, M.; Rendon-Mitchell, B.; Qiang, X.; Susarla, S.; Ulloa, L.; Yang, H.; Fan, S.; Goyert, S.M.; et al. Bacterial endotoxin stimulates macrophages to release HMGB1 partly through CD14- and TNF-dependent mechanisms. J. Leukoc. Biol. 2004, 76, 994–1001. [Google Scholar] [CrossRef]
- Mϋller, S.; Ronfani, L.; Bianchi, M.E. Regulated expression and localization of HMGB1, a chromatin protein with a cytokine function. J. Inter. Med. 2004, 255, 332–343. [Google Scholar] [CrossRef]
- El Gazzar, M. HMGB1 modulated inflammatory responses in LPS-activated macrophages. Inflamm. Res. 2007, 56, 162–167. [Google Scholar] [CrossRef]
- Amin, R.A.; Islam, A.B. Genomic analysis and differential expression of HMG and S100 family in human arthritis; upregulated expression of chemokines, IL-8 and nitric oxide by HMGB1. DNA Cell Biol. 2014, 33, 550–565. [Google Scholar] [CrossRef]
- Geng, Y.; Munirathinam, G.; Palani, S.; Ross, J.E.; Wang, B.; Chen, A.; Zheng, G. HMGB1-neutralizing IgM antibody is a normal component of blood plasma. J. Immunol. 2020, 15, 407–413. [Google Scholar] [CrossRef]
- Hreggvidsdottir, H.S.; Östberg, T.; Wähämaa, H.; Schierbeck, H.; Aveberger, A.-C.; Klevenvall, L.; Palmblad, K.; Ottosson, L.; Andersson, U.; Harris, H.E. The alarmin HMGB1 acts with synergy with endogenous and exogenous danger signals to promote inflammation. J. Leuc. Biol. 2009, 86, 655–662. [Google Scholar] [CrossRef]
- Treutiger, C.J.; Mullins, G.E.; Johansson, A.-S.M.; Rouhiainen, A.; Rauvala, H.; Erlandsson-Harris, H.; Andersson, U.; Yang, H.; Tracey, K.J.; Palmblad, J.E.W. High mobility group 1 B-box mediates activation of human endothelium. J. Intern. Med. 2003, 254, 375–385. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Ju, Z.; Ragab, A.A.; Lundbäck, P.; Long, W.; Valdés-Ferrer, S.I.; He, M.; Pribis, J.P.; Li, J.; et al. MD-2 is required for disulfide HMGB1---Dependent TLR4 signaling. J. Exp. Med. 2015, 212, 5–14. [Google Scholar] [CrossRef]
- Youn, J.H.; Oh, Y.J.; Kim, E.S.; Choi, J.E.; Shin, J.S. High mobility group box 1 protein biding to lipopolysaccharide facilitates transfer of lipopoly-saccharide to CD14 and enhances lipopolysaccharide-mediated TNFα production in human monocytes. J. Immunol. 2008, 180, 5067–5074. [Google Scholar] [CrossRef]
- Sha, Y.; Zmijewski, J.; Xu, Z.; Abraham, E. HMGB1 Develops Enhanced Proinflammatory Activity by Binding to Cytokines. J. Immunol. 2008, 180, 2531–2537. [Google Scholar] [CrossRef]
- Bianchi, M. HMGB1 likes company. J. Leuc. Biol. 2009, 86, 573–576. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Andersson, U. Targeting Inflammation Driven by HMGB1. Front. Immunol. 2020, 11, 484. [Google Scholar] [CrossRef]
- Kratofil, R.M.; Kubes, P.; Deniset, J.F. Monocyte Conversion During Inflammation and Injury. Arter. Thromb. Vasc. Biol. 2017, 37, 35–42. [Google Scholar] [CrossRef]
- Spahn, J.H.; Kreisel, D. Monocytes in Sterile Inflammation: Recruitment and Functional Consequences. Arch. Immunol. Ther. Exp. 2014, 62, 187–194. [Google Scholar] [CrossRef]
- Rothoerl, R.; Woertgen, C.; Holzschuh, M.; Brehme, K.; Rüschoff, J.; Brawanski, A. Macrophage tissue infiltration, clinical symptoms, and signs in patients with lumbar disc herniation. A clinicopathological study on 179 patients. Acta Neurochir. 1998, 140, 1245–1248. [Google Scholar] [CrossRef]
- Klyne, D.M.; Barbe, M.F.; Hodges, P.W. Systemic inflammatory profiles and their relationships with demographic, behavioural and clinical features in acute low back pain. Brain Behav. Immun. 2017, 60, 84–92. [Google Scholar] [CrossRef]
- Morris, P.; Ali, K.; Merritt, M.; Pelletier, J.; Macedo, L.G. A systematic review of the role of inflammatory biomarkers in acute, subacute and chronic non-specific low back pain. BMC Musculoskelet. Disord. 2020, 21, 142. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zeng, Q.; Silverman, H.A.; Gunasekaran, M.; George, S.J.; Devarajan, A.; Addorisio, M.E.; Li, J.; Tsaava, T.; Shah, V.; et al. HMGB1 released from nociceptors mediates inflammation. Proc. Natl. Acad. Sci. USA 2021, 118, e2102034118. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Ozaki, M.; Kobayashi, Y.; Kiguchi, N.; Kishioka, S. HMGB1 as a Potential Therapeutic Target for Neuropathic Pain. J. Pharmacol. Sci. 2013, 123, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Agalave, N.M.; Svensson, C.I. Extracellular High-Mobility Group Box 1 Protein (HMGB1) as a Mediator of Persistent Pain. Mol. Med. 2014, 20, 569–578. [Google Scholar] [CrossRef]
- Das, N.; Dewan, V.; Grace, P.M.; Gunn, R.J.; Tamura, R.; Tzarum, N.; Watkins, L.R.; Wilsons, I.A.; Yin, H. HMGB1 activated pro-inflammatory signaling via TLR5 leading to allodynia. Cell Rep. 2016, 17, 1128–1140. [Google Scholar] [CrossRef]
- VanPatten, S.; Al-Abed, Y. High Mobility Group Box-1 (HMGb1): Current Wisdom and Advancement as a Potential Drug Target. J. Med. Chem. 2018, 61, 5093–5107. [Google Scholar] [CrossRef]
- Andersson, U.; Tracey, K.J. HMGB1 Is a Therapeutic Target for Sterile Inflammation and Infection. Annu. Rev. Immunol. 2011, 29, 139–162. [Google Scholar] [CrossRef]
- Oliveira, C.B.; Maher, C.G.; Pinto, R.Z.; Traeger, A.C.; Lin, C.-W.C.; Chenot, J.-F.; van Tulder, M.; Koes, B.W. Clinical practice guidelines for the management of non-specific low back pain in primary care: An updated overview. Eur. Spine J. 2018, 27, 2791–2803. [Google Scholar] [CrossRef]
| Mean ± SD | ||
|---|---|---|
| Characteristics | LBP N = 10 | Asymptomatic controls N = 12 |
| Age (range) | 35.2 ± 13 (22–58) | 27.6 ± 3.2 (20–34) |
| Sex: male/female | 8/2 | 6/6 |
| BMI (range) | 27.1 ± 3.4 (23–26) | 25.5 ± 4 (20–34) |
| VAS (range) | 5.6 ± 1.6 (4–8) | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teodorczyk-Injeyan, J.A.; Khella, H.; Injeyan, H.S. Clinical Biomarker of Sterile Inflammation, HMGB1, in Patients with Chronic Non-Specific Low Back Pain: A Pilot Cross-Sectional Study. Life 2023, 13, 468. https://doi.org/10.3390/life13020468
Teodorczyk-Injeyan JA, Khella H, Injeyan HS. Clinical Biomarker of Sterile Inflammation, HMGB1, in Patients with Chronic Non-Specific Low Back Pain: A Pilot Cross-Sectional Study. Life. 2023; 13(2):468. https://doi.org/10.3390/life13020468
Chicago/Turabian StyleTeodorczyk-Injeyan, Julita A., Heba Khella, and H. Stephen Injeyan. 2023. "Clinical Biomarker of Sterile Inflammation, HMGB1, in Patients with Chronic Non-Specific Low Back Pain: A Pilot Cross-Sectional Study" Life 13, no. 2: 468. https://doi.org/10.3390/life13020468
APA StyleTeodorczyk-Injeyan, J. A., Khella, H., & Injeyan, H. S. (2023). Clinical Biomarker of Sterile Inflammation, HMGB1, in Patients with Chronic Non-Specific Low Back Pain: A Pilot Cross-Sectional Study. Life, 13(2), 468. https://doi.org/10.3390/life13020468







