BézierSeg: Parametric Shape Representation for Fast Object Segmentation in Medical Images
Abstract
1. Introduction
- We propose to use parametric curves for shape encoding and reframe the pixel-wise classification problem into a point coordinate autoregression problem, thus providing convenience for many practical applications in clinical scenarios, e.g., manual refinement of prediction and data transmission.
- We propose BézierSeg, an end-to-end solution that can directly output the control points of the Bézier curves that encompass the detected object. We also devise a Bézier Differentiable Shape Decoder (BDSD) that further improves the segmentation performance.
- We validate our model on three medical image datasets. Experimental results show that BézierSeg can reach comparable results to the mainstream pixel-based solution while achieving 98 frames per second on a single Tesla V100 GPU for object segmentation.
2. Related Works
2.1. Pixel-Wise Segmentation
2.2. Contour-Based Segmentation
3. Proposed Approach
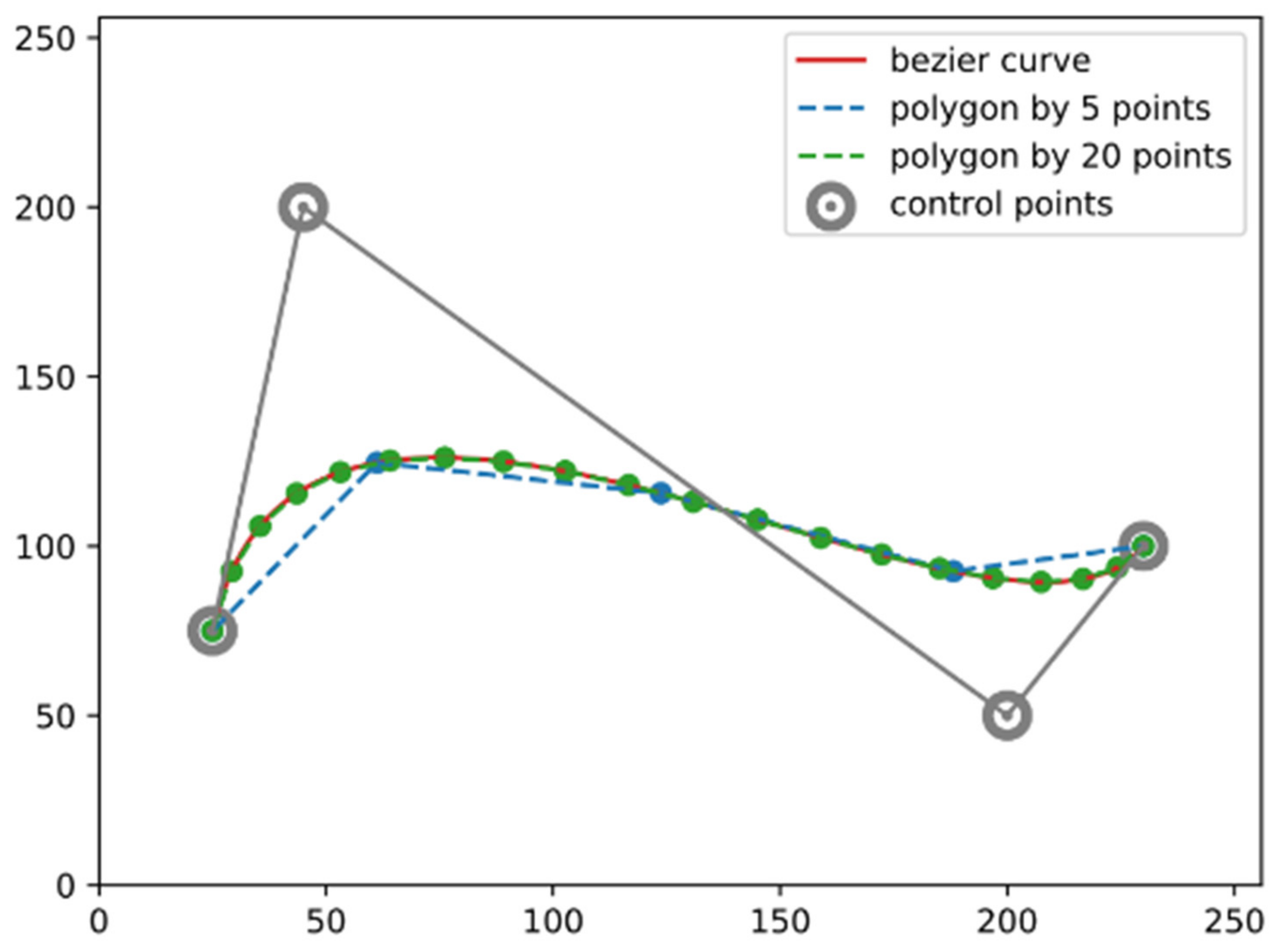
3.1. Parametric Representation
- (1)
- Create the control polygon of the Bézier curve by connecting the consecutive control points.
- (2)
- Insert intermediate points to each line segment, with the ratio .
- (3)
- Treat the intermediate points as the new control points, and repeat step (1) and (2) until you end up with a single point.
- (4)
- As varies from 0 to 1, the trajectory of that single point forms the curve.
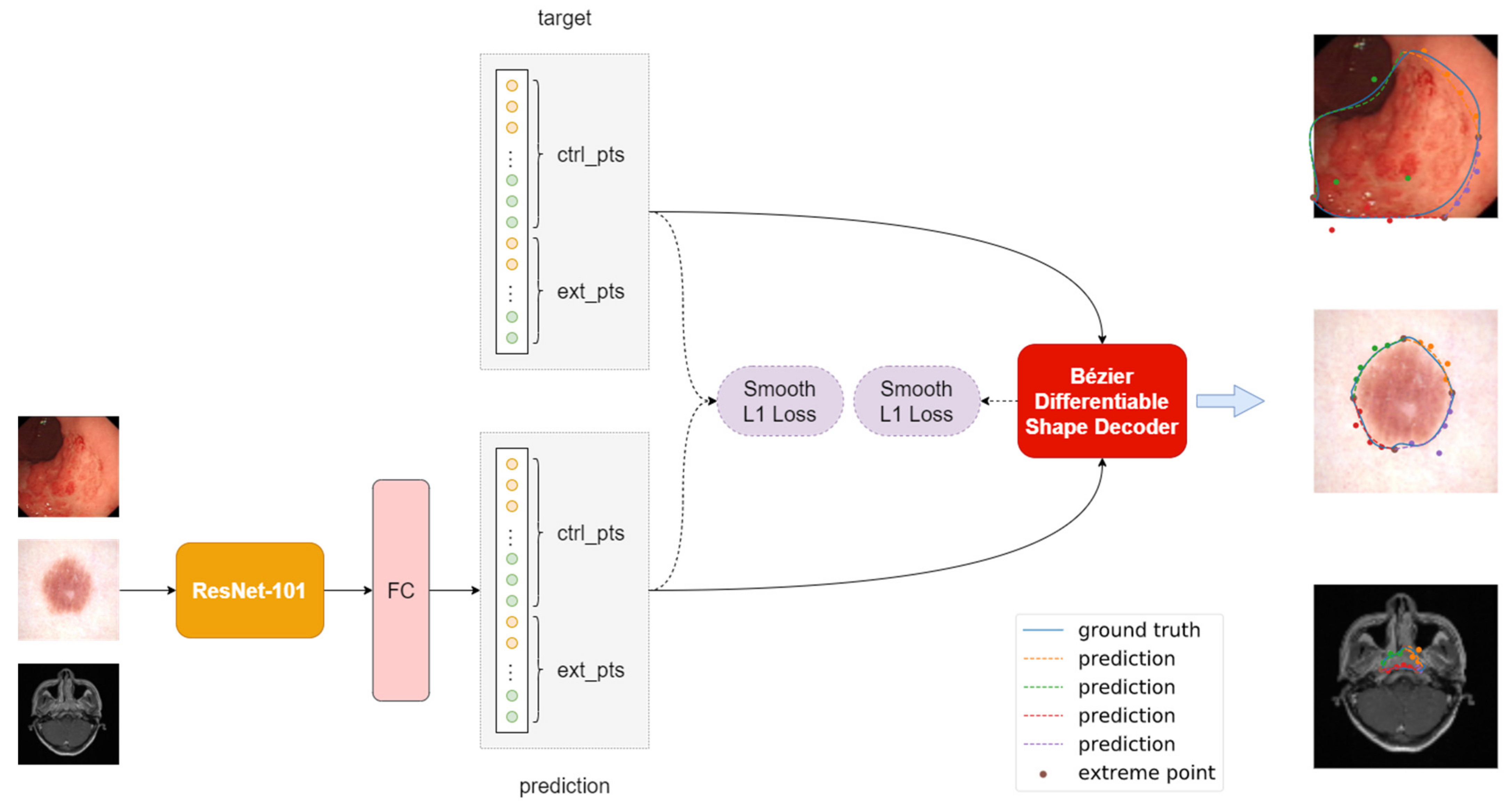
3.2. Model Architecture
3.3. Ground Truth Label Generation
3.4. Bézier Differentiable Shape Decoder
3.5. Loss
4. Experiments
4.1. Datasets and Evaluation Metric
4.2. Implementation Details
5. Results and Discussion
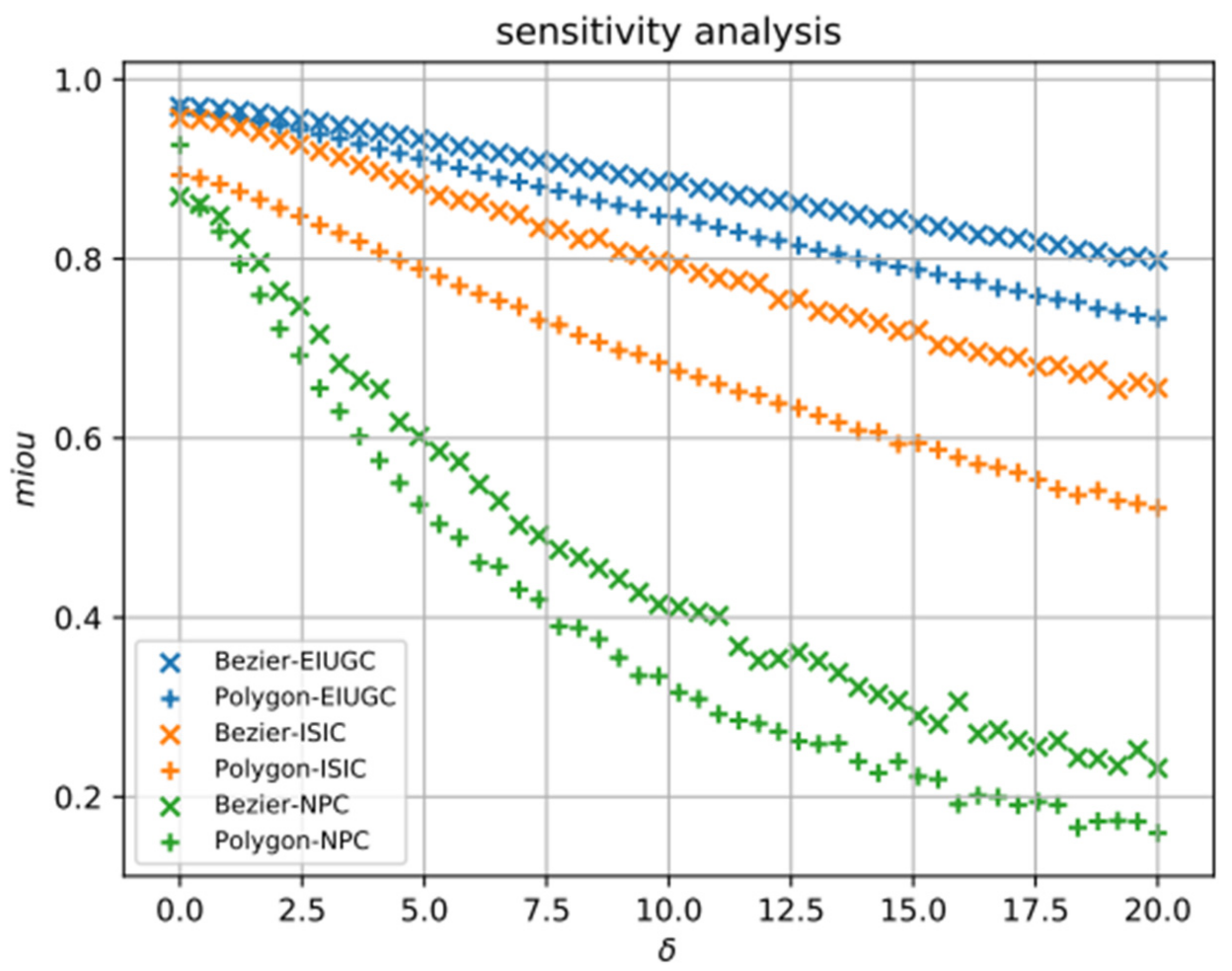
5.1. Quantitative Evaluation
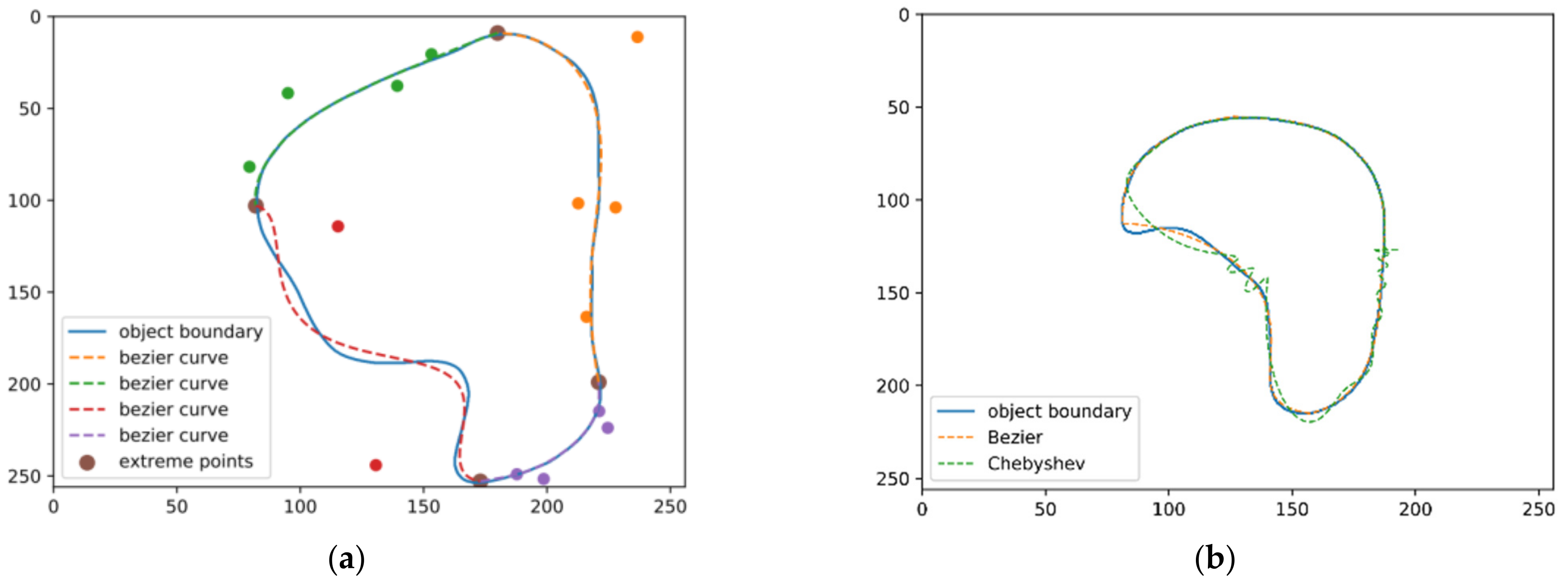
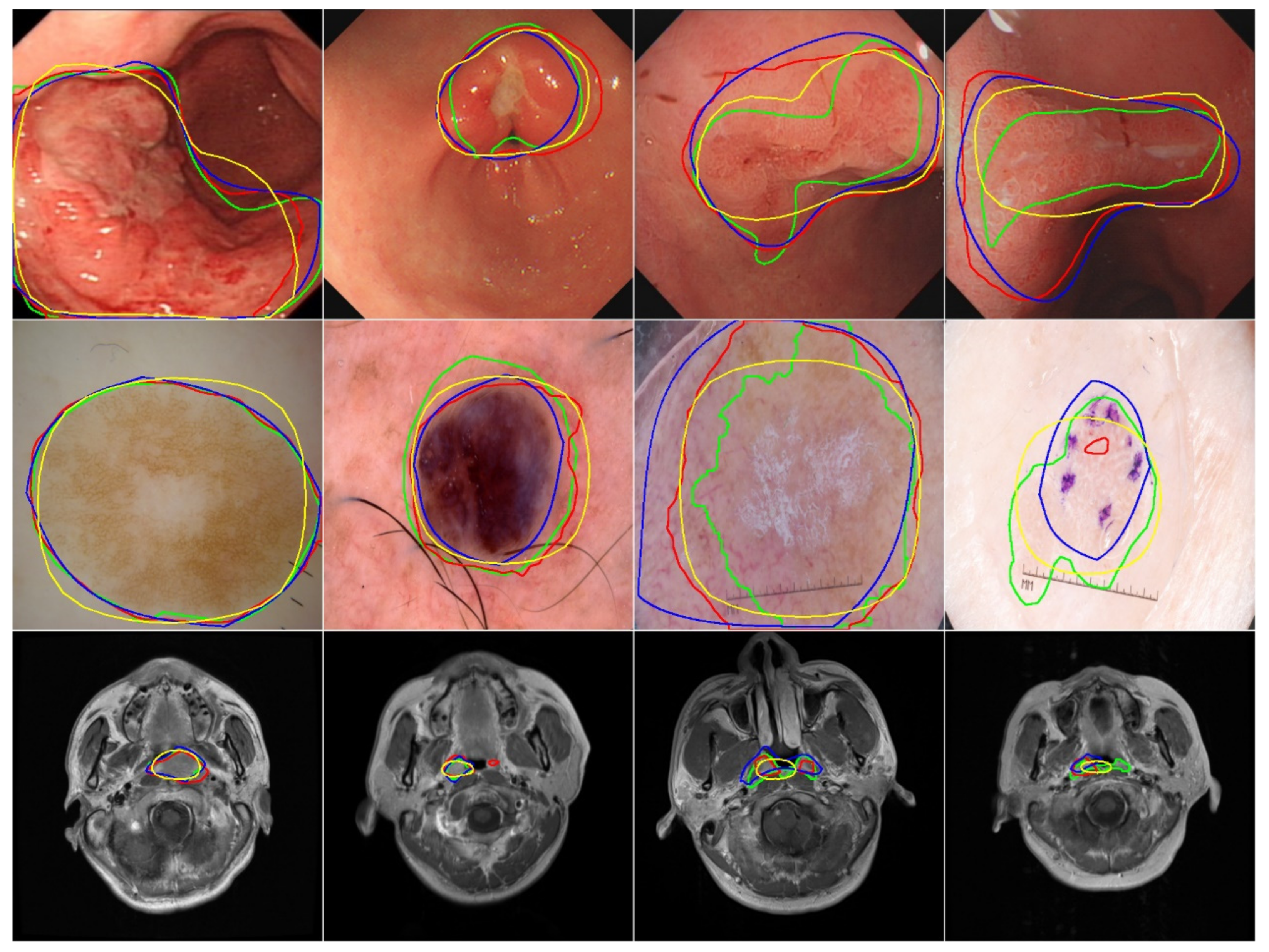
5.2. Qualitative Evaluation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grand Challenge. Available online: https://grand-challenge.org/challenges (accessed on 18 May 2020).
- Wang, X.; Han, S.; Chen, Y.; Gao, D.; Vasconcelos, N. Volumetric Attention for 3D Medical Image Segmentation and Detection. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Springer International Publishing: Cham, Switzerland, 2019; Part VI 22; pp. 175–184. [Google Scholar]
- Wang, S.; Singh, V.K.; Cheah, E.; Wang, X.; Li, Q.; Chou, S.H.; Lehman, C.D.; Kumar, V.; Samir, A.E. Stacked Dilated Convolutions and Asymmetric Architecture for U-Net-based Medical Image Segmentation. Comput. Biol. Med. 2022, 148, 105891. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Merhof, D. MixNet: Multi-modality Mix Network for Brain Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers; Springer International Publishing: Cham, Switzerland, 2018; Part I 4; pp. 367–377. [Google Scholar]
- Heker, M.; Greenspan, H. Joint Liver Lesion Segmentation and Classification via Transfer Learning. arXiv 2020, arXiv:2004.12352. [Google Scholar]
- Ke, L.; Deng, Y.; Xia, W.; Qiang, M.; Chen, X.; Liu, K.; Jing, B.; He, C.; Xie, C.; Guo, X.; et al. Development of a Self-constrained 3D DenseNet Model in Automatic Detection and Segmentation of Nasopharyngeal Carcinoma Using Magnetic Resonance Images. Oral Oncol. 2020, 110, 104862. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Springer International Publishing: Cham, Switzerland, 2015; Part III 18; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A Deep Convolutional Encoder-decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools Prof. Program. 2000, 25, 120–123. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Yu, C.; Wang, J.; Peng, C.; Gao, C.; Yu, G.; Sang, N. Bisenet: Bilateral Segmentation Network for Real-time Semantic Segmentation. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 325–341. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Wang, K.; Liew, J.H.; Zou, Y.; Zhou, D.; Feng, J. Panet: Few-shot Image Semantic Segmentation with Prototype Alignment. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9197–9206. [Google Scholar]
- Bai, M.; Urtasun, R. Deep Watershed Transform for Instance Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5221–5229. [Google Scholar]
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y.J. Yolact: Real-time Instance Segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9157–9166. [Google Scholar]
- Xie, E.; Sun, P.; Song, X.; Wang, W.; Liu, X.; Liang, D.; Shen, C.; Luo, P. Polarmask: Single Shot Instance Segmentation with Polar Representation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12193–12202. [Google Scholar]
- Xu, W.; Wang, H.; Qi, F.; Lu, C. Explicit Shape Encoding for Real-time Instance Segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 5168–5177. [Google Scholar]
- Ling, H.; Gao, J.; Kar, A.; Chen, W.; Fidler, S. Fast Interactive Object Annotation with Curve-gcn. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 5257–5266. [Google Scholar]
- Peng, S.; Jiang, W.; Pi, H.; Li, X.; Bao, H.; Zhou, X. Deep Snake for Real-time Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 8533–8542. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Riaz HU, M.; Benbarka, N.; Zell, A. FourierNet: Compact Mask Representation for Instance Segmentation Using Differentiable Shape Decoders. In Proceedings of the 2020 25th International Conference on Pattern Recognition, Milan, Italy, 10–15 January 2021; pp. 7833–7840. [Google Scholar]
- Huttenlocher, D.P.; Klanderman, G.A.; Rucklidge, W.J. Comparing Images Using the Hausdorff Distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 850–863. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the Predicted and Observed Secondary Structure of T4 Phage Lysozyme. Biochim. Biophys. Acta (BBA)-Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft Coco: Common Objects in Context. In Computer Vision–ECCV 2014, Proceedings of the 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Springer International Publishing: Cham, Switzerland, 2014; Part V 13; pp. 740–755. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. Imagenet: A Large-scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Perez, L.; Wang, J. The Effectiveness of Data Augmentation in Image Classification Using Deep Learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Tasci, E.; Uluturk, C.; Ugur, A. A Voting-based Ensemble Deep Learning Method Focusing on Image Augmentation and Preprocessing Variations for Tuberculosis Detection. Neural Comput. Appl. 2021, 33, 15541–15555. [Google Scholar] [CrossRef] [PubMed]

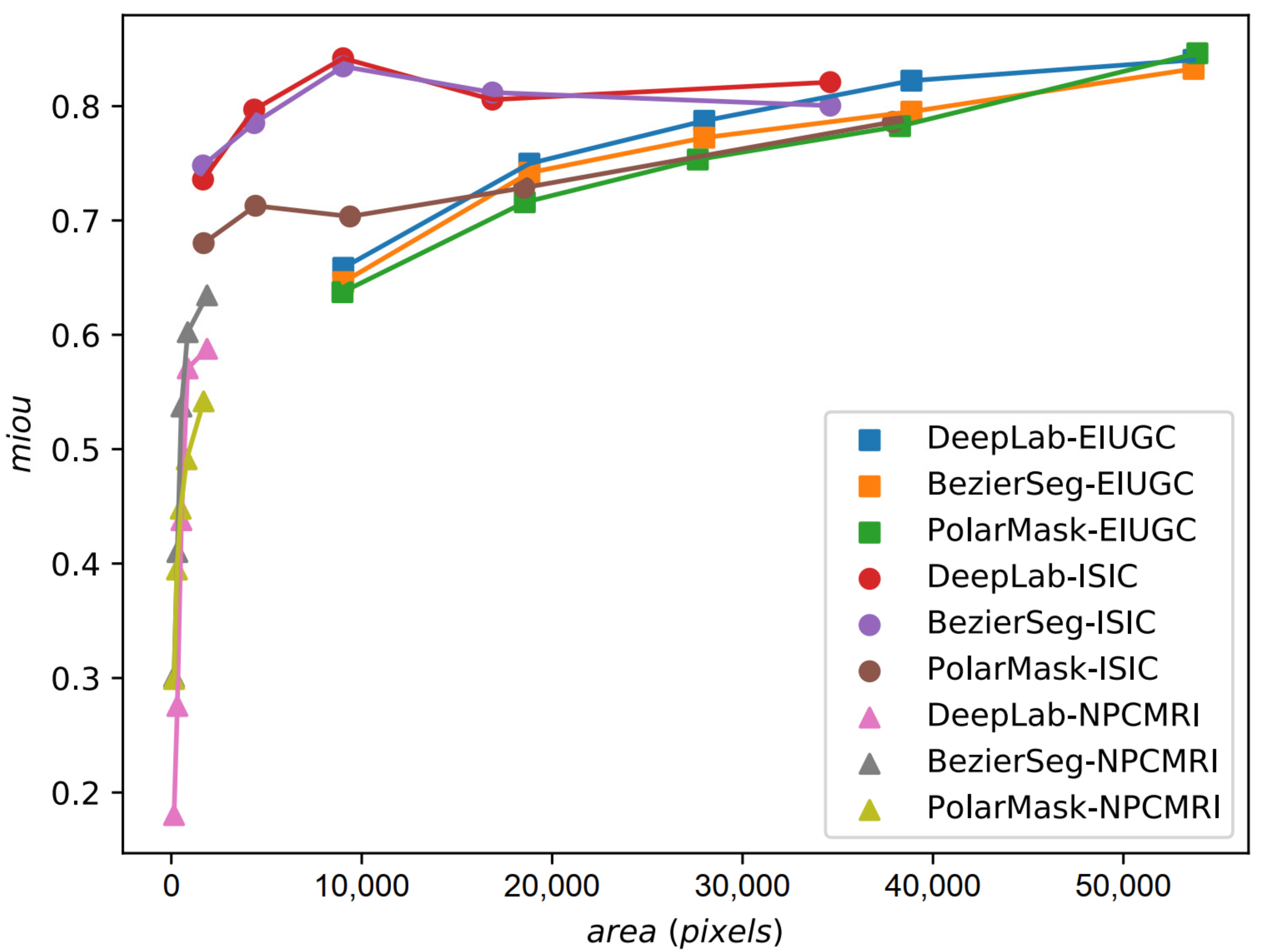
| Degree of Bézier Curve (n) | 3 | 5 | 7 | 9 |
| MIOU | 0.753 | 0.755 | 0.749 | 0.736 |
| Dataset | Trainset | Valset | Testset | MIOU | SIOU |
|---|---|---|---|---|---|
| EIUGC | 30,762 | 3845 | 3846 | 0.970 | 0.019 |
| NPCMRI | 1869 | 234 | 234 | 0.869 | 0.056 |
| ISIC | 2060 | 258 | 258 | 0.957 | 0.021 |
| Dataset | Model | BDSD | Curve MIOU | Mask MIOU | Hausdorff | Mcc | Auc | fp | fn |
|---|---|---|---|---|---|---|---|---|---|
| EIUGC | DeepLab v3+ | - | - | 0.772 | 14.330 | 0.781 | 0.891 | 0.104 | 0.115 |
| PolarMask | - | - | 0.747 | 16.263 | 0.753 | 0.878 | 0.129 | 0.116 | |
| BézierSeg 50 | ◦ | 0.750 | 0.745 | 15.473 | 0.752 | 0.876 | 0.118 | 0.129 | |
| BézierSeg 50 | ✓ | 0.750 | 0.745 | 15.573 | 0.751 | 0.876 | 0.126 | 0.122 | |
| BézierSeg 101 | ◦ | 0.761 | 0.757 | 14.898 | 0.763 | 0.882 | 0.113 | 0.124 | |
| BézierSeg 101 | ✓ | 0.762 | 0.758 | 14.818 | 0.763 | 0.882 | 0.119 | 0.117 | |
| ISIC | DeepLab v3+ | - | - | 0.801 | 8.139 | 0.877 | 0.939 | 0.028 | 0.093 |
| PolarMask | - | - | 0.723 | 8.707 | 0.812 | 0.903 | 0.039 | 0.156 | |
| BézierSeg 50 | ◦ | 0.796 | 0.791 | 7.339 | 0.869 | 0.934 | 0.029 | 0.104 | |
| BézierSeg 50 | ✓ | 0.799 | 0.793 | 7.349 | 0.872 | 0.936 | 0.029 | 0.099 | |
| BézierSeg 101 | ◦ | 0.797 | 0.792 | 7.414 | 0.866 | 0.934 | 0.031 | 0.102 | |
| BézierSeg 101 | ✓ | 0.801 | 0.796 | 7.406 | 0.870 | 0.936 | 0.030 | 0.097 | |
| NPCMRI | DeepLab v3+ | - | - | 0.413 | 9.553 | 0.671 | 0.820 | 0.003 | 0.357 |
| PolarMask | - | - | 0.437 | 5.348 | 0.650 | 0.799 | 0.003 | 0.398 | |
| BézierSeg 50 | ◦ | 0.482 | 0.467 | 5.285 | 0.685 | 0.840 | 0.003 | 0.317 | |
| BézierSeg 50 | ✓ | 0.504 | 0.488 | 4.909 | 0.707 | 0.855 | 0.003 | 0.287 | |
| BézierSeg 101 | ◦ | 0.500 | 0.478 | 5.258 | 0.696 | 0.856 | 0.004 | 0.284 | |
| BézierSeg 101 | ✓ | 0.520 | 0.499 | 4.886 | 0.717 | 0.866 | 0.003 | 0.265 |
| Model | Post-Processing | FPS |
|---|---|---|
| DeepLab v3+ | ◦ | 48.4 |
| ✓ | 45.6 | |
| BézierSeg | ◦ | 103.8 |
| ✓ | 97.8 | |
| PolarMask | ◦ | 103.3 |
| ✓ | 101.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Deng, Y.; Li, B.; Li, Z.; Chen, H.; Jing, B.; Li, C. BézierSeg: Parametric Shape Representation for Fast Object Segmentation in Medical Images. Life 2023, 13, 743. https://doi.org/10.3390/life13030743
Chen H, Deng Y, Li B, Li Z, Chen H, Jing B, Li C. BézierSeg: Parametric Shape Representation for Fast Object Segmentation in Medical Images. Life. 2023; 13(3):743. https://doi.org/10.3390/life13030743
Chicago/Turabian StyleChen, Haichou, Yishu Deng, Bin Li, Zeqin Li, Haohua Chen, Bingzhong Jing, and Chaofeng Li. 2023. "BézierSeg: Parametric Shape Representation for Fast Object Segmentation in Medical Images" Life 13, no. 3: 743. https://doi.org/10.3390/life13030743
APA StyleChen, H., Deng, Y., Li, B., Li, Z., Chen, H., Jing, B., & Li, C. (2023). BézierSeg: Parametric Shape Representation for Fast Object Segmentation in Medical Images. Life, 13(3), 743. https://doi.org/10.3390/life13030743






