Biogas Residues Improved Microbial Diversity and Disease Suppression Function under Extent Indigenous Soil Microbial Biomass
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Soil and Plant Sampling
2.3. Treatment of Oat Seed
2.4. Soil Physicochemical Properties and Crop Yield
2.5. DNA Extraction
2.6. Amplicon Generation and Illumina MiSeq Sequencing
2.7. Sequencing Data Processing
2.8. Statistical Analyses
3. Results
3.1. Oat Seedling Production and Soil Physicochemical Properties
3.2. Alpha Diversity and βeta Diversity of Bacteria and Fungi
3.2.1. Alpha Diversity
3.2.2. Beta Diversity
3.3. Soil Bacterial and Fungal Community
3.3.1. ISMB Level Changes the Soil Bacterial and Fungal Community
3.3.2. Fertilizer Type Changes the Soil Bacterial and Fungal Community
3.3.3. Soil Habitat Changes the Soil Bacterial and Fungal Community
3.4. Co-Occurrence Pattern for Soil Bacteria and Fungi Community
3.5. Variations of Fungal Functional Potential Pathogen Groups
4. Discussion
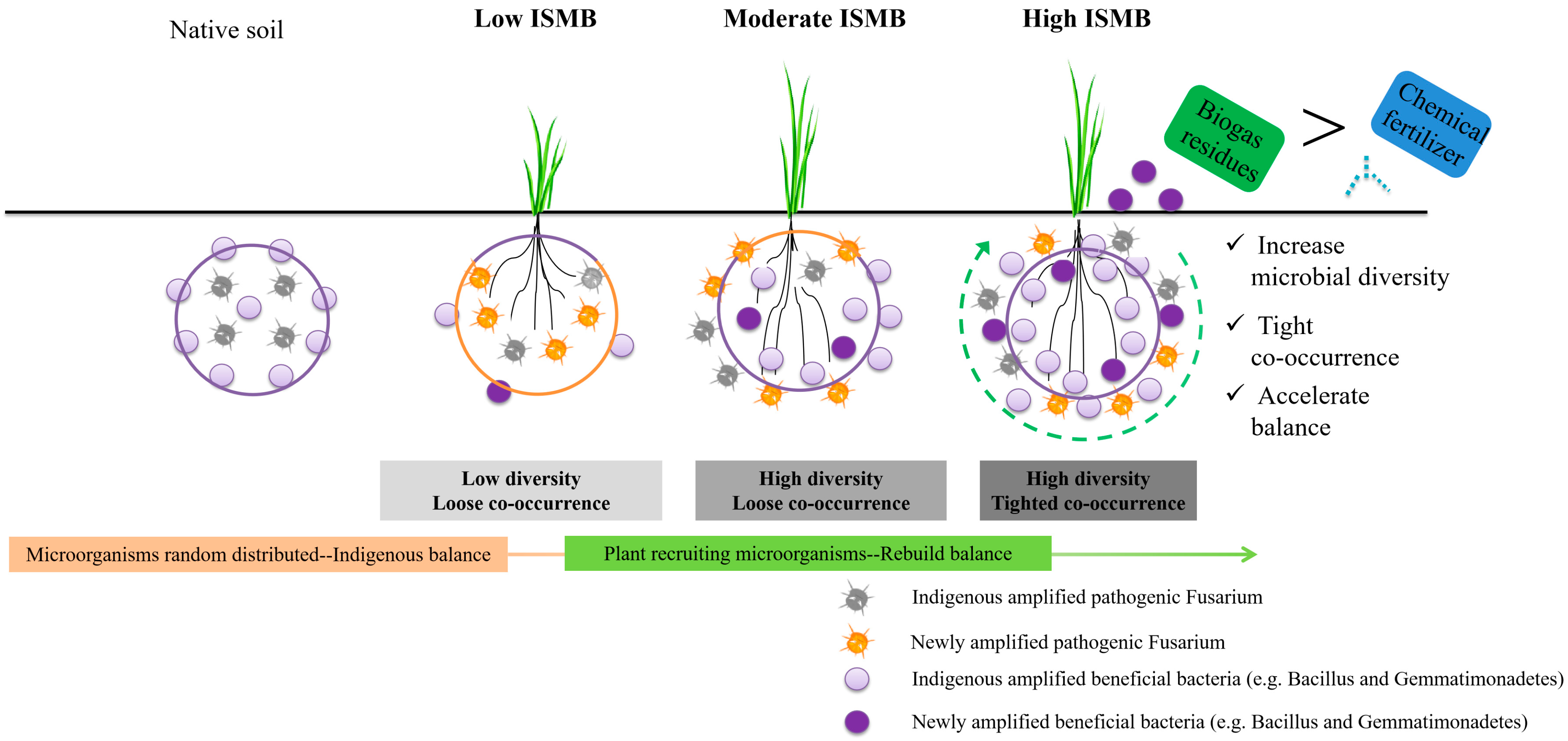
4.1. Microbial Community Formation and Co-Occurrence Pattern under Different Levels of Soil ISMB
4.2. Interactions of Plant and Soil Microorganisms under Different Levels of Soil ISMB
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bardgett, R.D.; Van Der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.Y.; Zhu, Y.G. Editorial: China soil microbiome thematic issue. FEMS Microbiol. Ecol. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Wall, D.H.; Nielsen, U.N.; Six, J. Soil biodiversity and human health. Nature 2015, 528, 69. [Google Scholar] [CrossRef]
- Tóth, G.; Hermann, T.; Silva, M.R.; Montanarella, L. Monitoring soil for sustainable development and land degradation neutrality. Environ. Monit. Assess. 2018, 190, 57. [Google Scholar] [CrossRef]
- Pascual, J.A.; Garcia, C.; Hernandez, T.; Moreno, J.L.; Ros, M. Soil microbial activity as a biomarker of degradation and remediation processes. Soil Biol. Biochem. 2000, 32, 1883. [Google Scholar] [CrossRef]
- Singh, J.S.; Gupta, V.K. Soil microbial biomass: A key soil driver in management of ecosystem functioning. Sci. Total Environ. 2018, 634, 497–500. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; XU, Y.C.; Shen, Q.R.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef]
- Xun, W.; Li, W.; Xiong, W.; Ren, Y.; Liu, Y.P.; Miao, Y.Z.; Xu, Z.H.; Zhang, N.; Shen, Q.R.; Zhang, R.F. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 2019, 10, 3833. [Google Scholar] [CrossRef]
- Tarafdar, J.C. Role of Soil Biology on Soil Health for Sustainable Agricultural Production. In Structure and Functions of Pedosphere; Springer: Berlin/Heidelberg, Germany, 2022; pp. 67–81. [Google Scholar]
- Li, Y.J.; Hu, Y.H.; Yan, C.Q.; Xiong, J.B.; Qiu, Q.F. pH and salinity are the dominant limiting factors for the application of mariculture sludge to paddy soil. Appl. Soil Ecol. 2022, 175, 104463. [Google Scholar] [CrossRef]
- Li, F.X.; Guo, Y.Z.; Wang, Z.J.; Wu, Y.X. Influence of different phytoremediation on soil microbial diversity and community composition in saline-alkaline land. Int. J. Phytoremediat. 2022, 24, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Hicks, N.; Liu, X.; Gregory, R.; Kenny, J.; Lucaci, A.; Lenzi, A.; Paterson, D.M.; Duncan, K.R. Temperature driven changes in benthic bacterial diversity influences biogeochemical cycling in coastal sediments. Front. Microbiol. 2018, 9, 1730. [Google Scholar] [CrossRef]
- Richardson, K. A safe operating space for humanity. Nature 2009, 461, 472–475. [Google Scholar]
- Stavi, I.; Bel, G.; Zaady, E. Soil functions and ecosystem services in conventional, conservation, and integrated agricultural systems. A review. Agron. Sustain. Dev. 2016, 36, 32. [Google Scholar] [CrossRef]
- Alves, T.D.S.; Campos, L.L.; Neto, N.E.; Matsuoka, M.; Loureiro, M.F. Biomass and soil microbial activity under native vegetation and different soil managements. Acta Sci. Agron. 2011, 33, 341–347. [Google Scholar]
- Wagg, C.; Bender, S.F.; Widmer, F.; Heijden, M.G.A.V.D. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef]
- Savari, M.; Gharechaee, H. Utilizing the theory of planned behavior to predict iranian farmers’intention for safe use of chemical fertilizers. J. Clean. Prod. 2020, 263, 121512. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, Z.; Huang, Z.L.; Dong, M.; Yu, X.L.; Ning, P. A new strategy for co-composting dairy manure with rice straw: Addition of different inocula at three stages of composting. Waste Manag. 2015, 40, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Ruan, Y.Z.; Wang, B.B.; Zhang, J.; Waseem, R.; Huang, Q.W.; Shen, Q.R. Plant growth-promoting rhizobacteria strain bacillus amyloliquefaciens njn-6-enriched bio-organic fertilizer suppressed fusarium wilt and promoted the growth of banana plants. J. Agron. Food Chem. 2013, 61, 3774–3780. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.Q.; Qiao, Y.H.; Wu, W.L.; Smith, P.; Scott, S. Environmental impacts and production performances of organic agriculture in china: A monetary valuation. J. Environ. Manag. 2017, 188, 49–57. [Google Scholar] [CrossRef]
- Seufert, V.; Ramankutty, N.; Foley, J.A. Comparing the yields of organic and conventional agriculture. Nature 2012, 485, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, R.K.; Bhardwaj, D.; Tuteja, N. Biofertilizers: A Sustainable Eco-Friendly Agricultural Approach to Crop Improvement. In Plant Acclimation to Environmental Stress; Springer: New York, NY, USA, 2013; pp. 403–432. [Google Scholar]
- Yao, Y.L.; Zhang, M.; Tian, Y.H.; Zhao, M.; Zeng, K.; Zhang, B.W.; Zhao, M.; Yin, B. Azolla biofertilizer for improving low nitrogen use efficiency in an intensive rice cropping system. Field Crops Res. 2018, 216, 158–164. [Google Scholar] [CrossRef]
- Tsachidou, B.; Scheuren, M.; Gennen, J.; Debbaut, V.; Toussaint, B.; Hissler, C. Biogas residues in substitution for chemical fertilizers: A comparative study on a grassland in the Walloon Region. Sci. Total Environ. 2019, 666, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Sieling, K.; Herrmann, A.; Wienforth, B.; Taube, F.; Ohl, S.; Hartung, E.; Kage, H. Biogas cropping systems: Short term response of yield performance and N use efficiency to biogas residue application. Eur. J. Agron. 2013, 47, 44–54. [Google Scholar] [CrossRef]
- Zhu, N.M.; Luo, T.; Guo, X.J.; Zhang, H.; Deng, Y. Nutrition potential of biogas residues as organic fertilizer regarding the speciation and leachability of inorganic metal elements. Environ. Technol. 2015, 36, 992–1000. [Google Scholar] [CrossRef]
- Kaur, R. Governance of Sustainable Agriculture Schemes in India with Special Reference to Biogas Residues Project and Analysis. In Proceedings of the Second International Conference on Recent Advances in Bioenergy Research, Rome, Italy, 8–10 August 2016; Springer: Singapore, 2018; pp. 147–156. [Google Scholar]
- Abubaker, J.; Cederlund, H.; Arthurson, V.; Pell, M. Bacterial community structure and microbial activity in different soils amended with biogas residues and cattle slurry. Appl. Soil Ecol. 2013, 72, 171–180. [Google Scholar] [CrossRef]
- Karimi, B.; Sadet-Bourgeteau, S.; Cannavacciuolo, M.; Chauvin, C.; Flamin, C.; Haumont, A.; Jean-Baptiste, V.; Reibel, A.; Vrignaud, G.; Ranjard, L. Impact of biogas digestates on soil microbiota in agriculture: A review. Environ. Chem. Lett. 2022, 20, 1–24. [Google Scholar] [CrossRef]
- Zhang, M.; Tian, Y.; Zhao, M.; Yin, B.; Zhu, Z.L. The assessment of nitrate leaching in a rice–wheat rotation system using an improved agronomic practice aimed to increase rice crop yields. Agron. Ecosyst. Environ. 2017, 241, 100–109. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, G.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Zhao, Z.B.; He, J.Z.; Geisen, S.; Han, L.L.; Wang, J.T.; Shen, J.P.; Wei, W.X.; Fang, X.T.; Li, P.P.; Zhang, L.M. Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 2019, 7, 33. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. March, Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Shen, H.; Yan, W.; Yang, X.; He, X.H.; Wang, X.; Zhang, Y.T.; Wang, B.; Xia, Q.Y. Co-occurrence network analyses of rhizosphere soil microbial plfas and metabolites over continuous cropping seasons in tobacco. Plant Soil 2020, 452, 119–135. [Google Scholar] [CrossRef]
- Ling, F.Q.; Hwang, C.C.; LeChevallier, M.W.; Andersen, G.L.; Liu, V.T. Core-satellite populations and seasonality of water meter biofilms in a metropolitan drinking water distribution system. ISME J. 2015, 10, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Mason-Jones, K.; Robinson, S.L.; Veen, G.F.; Manzoni, S.; Putten, W.H.V.D. Microbial storage and its implications for soil ecology. ISME J. 2022, 16, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Ikotun, T.; Adekunle, F. Inhibition of growth of some plant pathogenic fungi by some antagonistic microorganisms isolated from soil. J. Basic Microb. 1990, 30, 95–98. [Google Scholar] [CrossRef]
- Liu, C.M.; Yang, Z.F.; He, P.F.; Munir, S.; Munir, S.; Wu, Y.X.; Ho, H.M.; He, Y.Q. Deciphering the bacterial and fungal communities in clubroot-affected cabbage rhizosphere treated with Bacillus subtilis XF-1. Agr. Ecosyst. Environ. 2018, 256, 12–22. [Google Scholar] [CrossRef]
- Izquierdo, L.F.; González-Almario, A.; Cotes, A.M.; Moreno-Velandia, C.A. Trichoderma virens gl006 and bacillus velezensis bs006: A compatible interaction controlling fusarium wilt of cape gooseberry. Sci. Rep. 2020, 10, 6857. [Google Scholar] [CrossRef] [PubMed]
- Orwin, K.H.; Wardle, D.A. New indices for quantifying the resistance and resilience of soil biota to exogenous disturbances. Soil Biol. Biochem. 2004, 36, 1907–1912. [Google Scholar] [CrossRef]
- Delmont, T.O.; Francioli, D.; Jacquesson, S.; Laoudi, S.; Mathieu, A.; Nesme, J.; Ceccherini, M.T.; Nannipieri, P.; Simonet, P.; Vogel, T.M. Microbial community development and unseen diversity recovery in inoculated sterile soil. Biol. Fert. Soils 2014, 50, 1069–1076. [Google Scholar] [CrossRef]
- Cai, F.; Pang, G.; Li, R.X.; Li, R.; Gu, X.L.; Shen, Q.R.; Chen, W. Bioorganic fertilizer maintains a more stable soil microbiome than chemical fertilizer for monocropping. Biol. Fert. Soils 2017, 53, 861–872. [Google Scholar] [CrossRef]
- Lopez-Lozano, N.E.; Echeverria Molinar, A.; Ortiz Duran, E.A.; Rosales, M.H.; Souza, V. Bacterial Diversity and Interaction Networks of Agave lechuguilla Rhizosphere Differ Significantly From Bulk Soil in the Oligotrophic Basin of Cuatro Cienegas. Front. Plant Sci. 2020, 11, 1028. [Google Scholar] [CrossRef]
- Fan, K.K.; Delgado-Baquerizo, M.; Guo, X.S.; Wang, D.Z.; Wu, Y.Y.; Zhu, M.; Yu, W.; Yao, H.Y.; Zhu, Y.G.; Chu, H.Y. Suppressed n fixation and diazotrophs after four decades of fertilization. Microbiome 2019, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Oates, L.G.; Duncan, D.S.; Sanford, G.R.; Liang, C.; Jackson, R.D. Bioenergy cropping systems that incorporate native grasses stimulate growth of plant-associated soil microbes in the absence of nitrogen fertilization. Agron. Ecosyst. Environ. 2016, 233, 396–403. [Google Scholar] [CrossRef]
- Liang, X.; Huadong, R.; Sheng, L.; Leng, X.H.; Yao, X.H. Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front. Microbiol. 2017, 8, 2377. [Google Scholar]
- Guo, M.X.; Gong, Z.Q.; Miao, R.H.; Su, D.; Li, X.J.; Jia, C.Y.; Zhuang, J. The influence of root exudates of maize and soybean on polycyclic aromatic hydrocarbons degradation and soil bacterial community structure. Ecol. Eng. 2017, 99, 22–30. [Google Scholar] [CrossRef]
- Parkinson, L. Investigating Soilborne Nectriaceous Fungi Impacting Avocado Tree Establishment in Australia. Ph.D. Thesis, University of Queensland, St. Lucia, Australia, 2017. [Google Scholar]
- Nikitin, D.A.; Ivanova, E.A.; Semenov, M.V.; Zhelezova, A.D.; Ksenofontova, N.A.; Tkhakakhova, A.K.; Kholodov, V.A. Diversity, Ecological Characteristics and Identification of Some Problematic Phytopathogenic Fusarium in Soil: A Review. Diversity 2023, 15, 49. [Google Scholar] [CrossRef]
- Wang, Q.; Song, R.; Fan, S.; Coleman, J.J.; Xu, X.M.; Hu, X.P. Diversity of Fusarium community assembly shapes mycotoxin accumulation of diseased wheat heads. Mol. Ecol. 2022. [Google Scholar] [CrossRef]
- Barahona, E.; Navazo, A.; Martínez-Granero, F.; Zea-Bonilla, T.; Pérez-Jiménez, R.M.; Martín, M.; Rivilla, R. Pseudomonas fluorescens f113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microb. 2011, 77, 5412. [Google Scholar] [CrossRef]
- Fu, L.; Penton, C.R.; Ruan, Y.Z.; Shen, Z.Z.; Xue, C.; Li, R.; Shen, Q.R. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana fusarium wilt disease. Soil Biol. Biochem. 2017, 104, 39–48. [Google Scholar] [CrossRef]

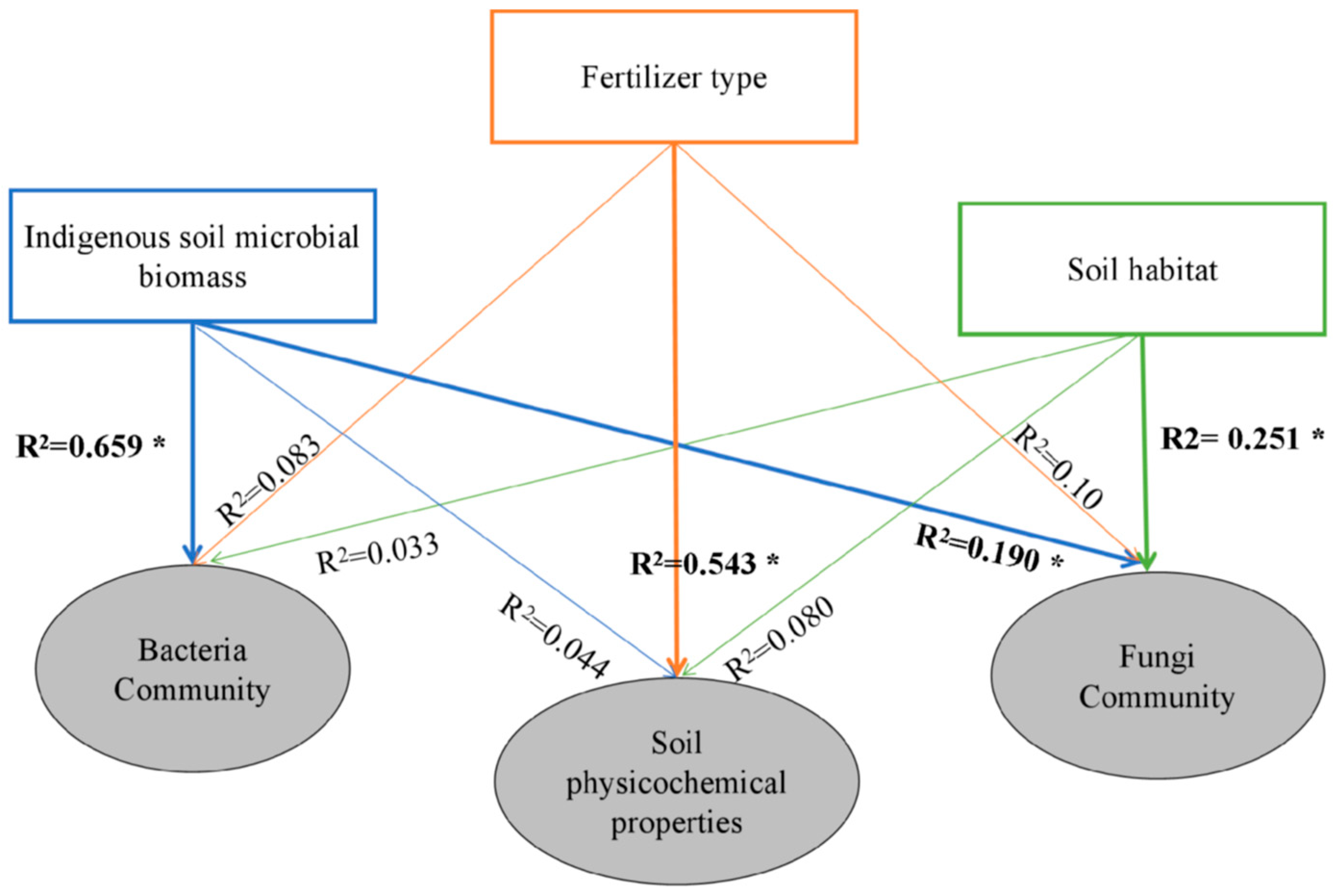
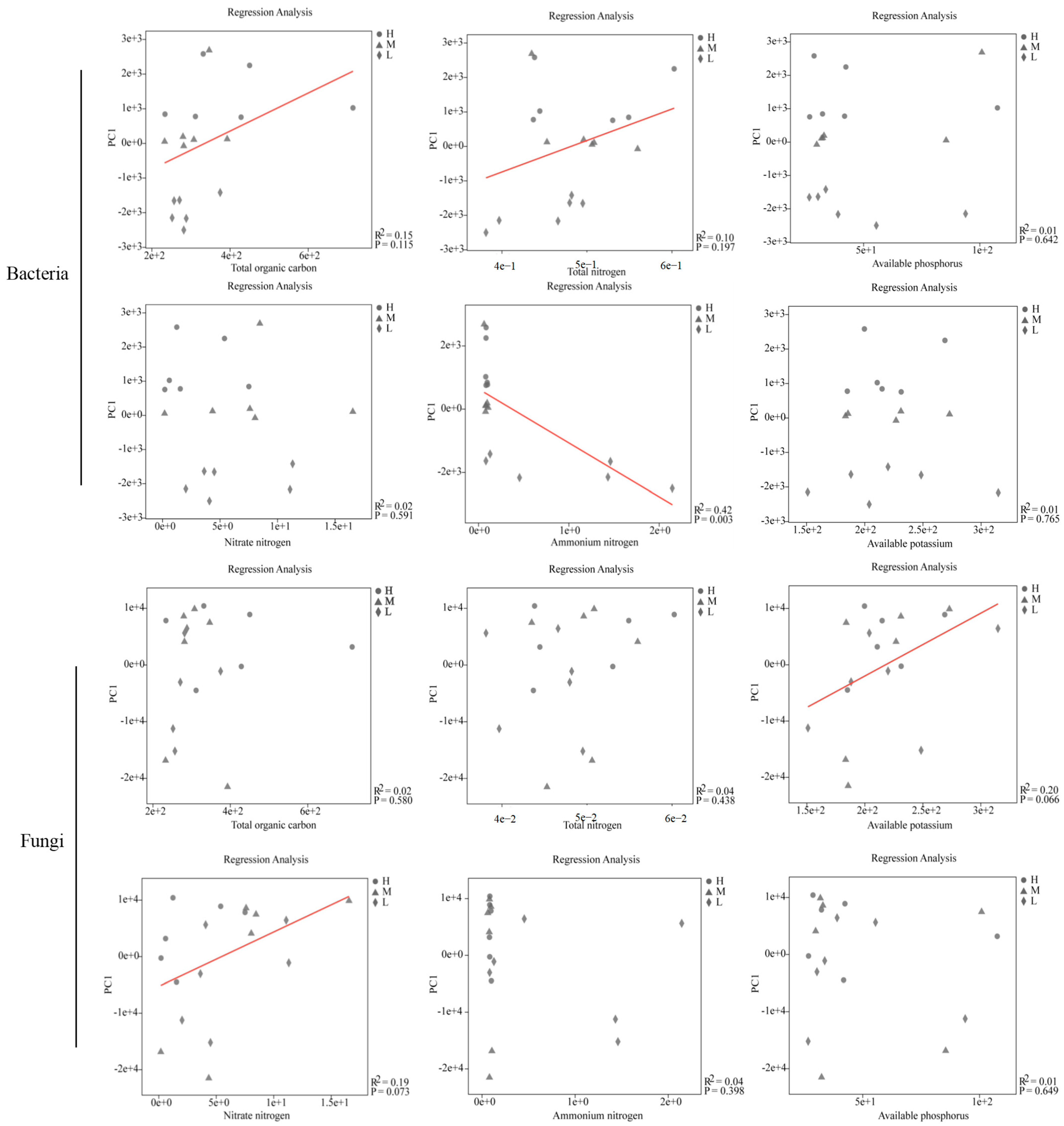
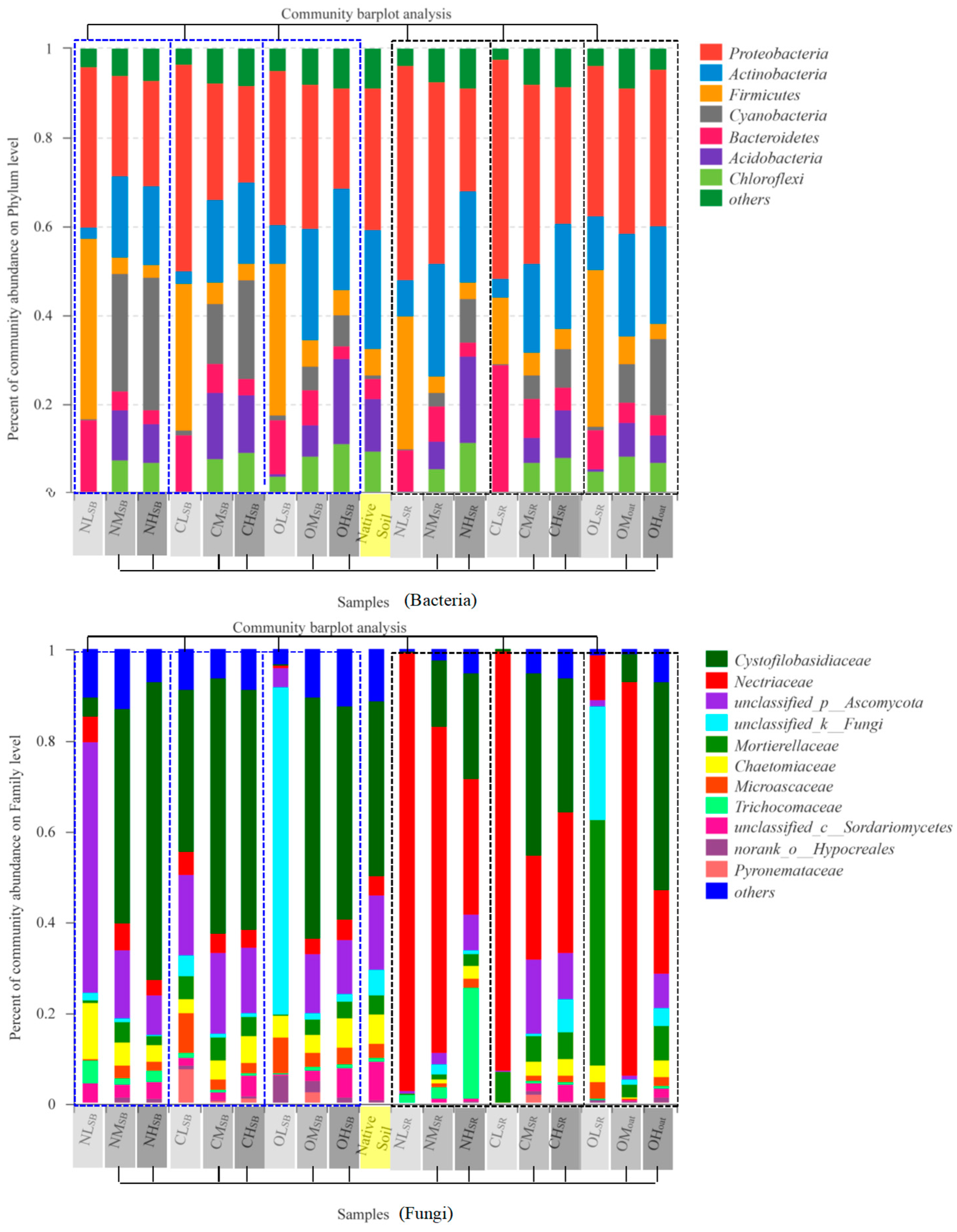
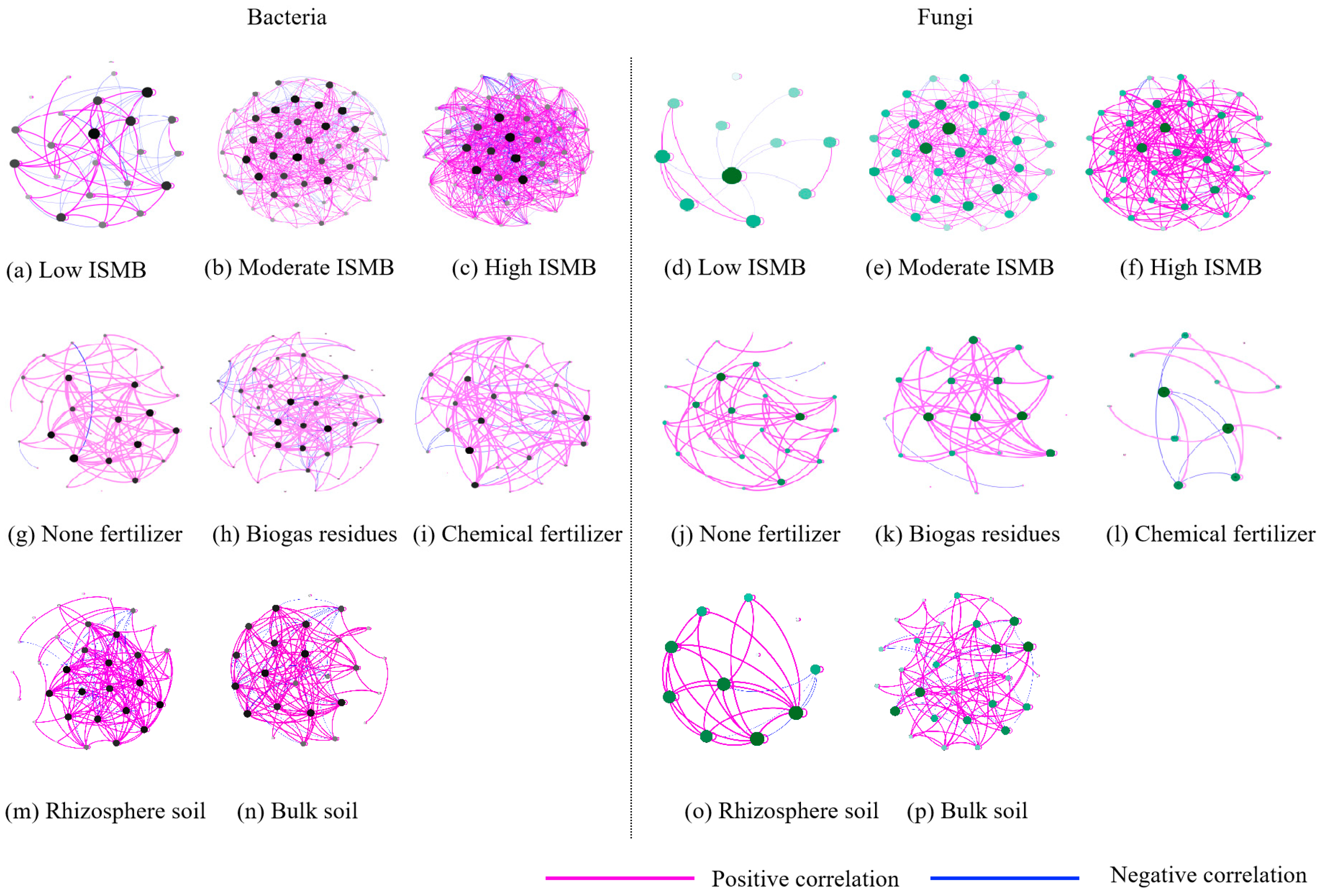
| ISMB | Fertilizer Type | Soil Habitat | Fertilizer Type × ISMB | Fertilizer Type × Soil Habitat | ISMB × Soil Habitat | Fertilizer Type × ISMB × Soil Habitat | ||
|---|---|---|---|---|---|---|---|---|
| Total nitrogen | F | 19.02 | 58.05 | 0.77 | 6.43 | 10.32 | 13.91 | 2.12 |
| P | 0.00 | 0.00 | 0.39 | 0.00 | 0.00 | 0.00 | 0.10 | |
| Ammoniumnitrogen | F | 7891.65 | 2494.42 | 22.01 | 2494.78 | 16.34 | 621.95 | 680.60 |
| P | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Nitrate nitrogen | F | 6111.05 | 11,753.84 | 22,388.83 | 2753.62 | 115.86 | 2420.89 | 435.78 |
| P | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Organic carbon | F | 380.85 | 126.21 | 0.00 | 118.45 | 257.84 | 181.66 | 178.39 |
| P | 0.00 | 0.00 | 0.98 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Available phosphorus | F | 6740.72 | 197,601.14 | 3.11 | 137,813.05 | 66,588.33 | 74,504.35 | 33,461.68 |
| P | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Available Potassium | F | 1.08 | 553.12 | 236.64 | 17.83 | 9.19 | 23.97 | 10.59 |
| P | 0.36 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Dry matter | F | 12.97 | 9.26 | 0.20 | ||||
| P | 0.00 | 0.00 | 0.94 | |||||
| Stem length | F | 12.30 | 5.79 | 0.42 | ||||
| P | 0.00 | 0.01 | 0.80 |
| Alpha Diversity | Treatment | F | P | |
|---|---|---|---|---|
| Bacteria | Chao | Indigenous soil microbial biomass | 95.48 | 0.00 |
| Fertilizer type | 0.16 | 0.85 | ||
| Soil habitat | 0.00 | 0.98 | ||
| Shannon | Indigenous soil microbial biomas | 38.67 | 0.00 | |
| Fertilizer type | 0.36 | 0.70 | ||
| Soil habitat | 0.09 | 0.77 | ||
| Fungi | Chao | Indigenous soil microbial biomas | 16.54 | 0.00 |
| Fertilizer type | 0.05 | 0.95 | ||
| Soil habitat | 1.79 | 0.20 | ||
| Shannon | Indigenous soil microbial biomas | 3.79 | 0.05 | |
| Fertilizer type | 0.17 | 0.85 | ||
| Soil habitat | 3.59 | 0.08 |
| (a) | ||||||||
| ISMB Level | Fertilizer Type | Soil Habitat | ||||||
| Low | Moderate | High | N | O | C | SR | SB | |
| Clustering coefficient | 0.5 | 0.43 | 0.7 | 0.71 | 0.72 | 0.69 | 0.75 | 0.73 |
| Network density | 0.32 | 0.36 | 0.6 | 0.29 | 0.19 | 0.24 | 0.37 | 0.51 |
| Number of nodes | 23 | 46 | 44 | 28 | 47 | 33 | 32 | 25 |
| Number of edges | 82 | 371 | 566 | 108 | 207 | 124 | 185 | 152 |
| Average path length | 2.09 | 1.75 | 1.448 | 2.83 | 2.79 | 2.51 | 1.97 | 1.81 |
| Positive correlation coefficient (%) | 64.63 | 67.65 | 68.37 | 98.15 | 77.78 | 88.71 | 84.32 | 78.29 |
| Negative correlation coefficient (%) | 35.37 | 32.35 | 31.63 | 1.85 | 22.22 | 11.29 | 15.68 | 21.71 |
| (b) | ||||||||
| ISMB Level | Fertilizer Type | Soil Habitat | ||||||
| Low | Moderate | High | N | O | C | SR | SB | |
| Clustering coefficient | 0.21 | 0.32 | 0.76 | 0.73 | 0.76 | 0.62 | 0.65 | 0.66 |
| Network density | 0.44 | 0.35 | 0.65 | 0.24 | 0.36 | 0.37 | 0.67 | 0.28 |
| Number of nodes | 11 | 35 | 41 | 25 | 19 | 13 | 11 | 29 |
| Number of edges | 24 | 208 | 535 | 73 | 62 | 29 | 37 | 114 |
| Average path length | 1.82 | 1.76 | 1.4 | 2.82 | 1.53 | 1.65 | 1.28 | 2.36 |
| Positive correlation coefficient (%) | 66.67 | 89.42 | 61.87 | 98.63 | 98.39 | 75.86 | 89.19 | 84.21 |
| Negative correlation coefficient (%) | 33.33 | 10.58 | 38.13 | 1.37 | 1.61 | 24.14 | 10.81 | 15.79 |
| Fusarium | Unclassified_f__Nectriaceae | ||||
|---|---|---|---|---|---|
| Correlation coefficient | P | Correlation coefficient | P | ||
| No fertilizer | Adhaeribacter | 0.72 | 0.11 | −0.36 | 0.49 |
| Bacillus | 0.54 | 0.27 | −0.01 | 0.98 | |
| Bdellovibrio | 0.68 | 0.14 | −0.27 | 0.61 | |
| Devosia | 0.63 | 0.18 | −0.13 | 0.80 | |
| Ensifer | 0.19 | 0.71 | −0.17 | 0.74 | |
| Flavisolibacter | 0.42 | 0.41 | −0.19 | 0.72 | |
| Massilia | 0.79 | 0.06 | −0.26 | 0.63 | |
| norank_f__Chitinophagaceae | 0.45 | 0.37 | −0.31 | 0.55 | |
| Paenibacillus | 0.58 | 0.23 | 0.39 | 0.45 | |
| Paucimonas | 0.66 | 0.15 | −0.28 | 0.60 | |
| Rhizobium | 0.90 | 0.02 | −0.16 | 0.76 | |
| Chemical fertilizer | Adhaeribacter | 0.06 | 0.92 | 0.23 | 0.66 |
| Bacillus | −0.13 | 0.81 | 0.10 | 0.85 | |
| Brevundimonas | 0.56 | 0.25 | 0.17 | 0.75 | |
| Chthoniobacter | 0.14 | 0.80 | 0.52 | 0.29 | |
| Devosia | 0.55 | 0.26 | 0.85 | 0.03 | |
| Ensifer | −0.16 | 0.76 | 0.78 | 0.07 | |
| Massilia | 0.21 | 0.69 | 0.90 | 0.01 | |
| Paenibacillus | 0.37 | 0.47 | 0.62 | 0.19 | |
| Paenisporosarcina | 0.59 | 0.22 | 0.19 | 0.72 | |
| Pedobacter | 0.37 | 0.47 | 0.52 | 0.29 | |
| Rhizobium | 0.16 | 0.76 | 0.86 | 0.03 | |
| unclassified_f__Oxalobacteraceae | −0.08 | 0.89 | 0.66 | 0.15 | |
| Biogas residues | Bacillus | −0.37 | 0.47 | −0.54 | 0.27 |
| Devosia | −0.11 | 0.84 | 0.17 | 0.75 | |
| Ensifer | −0.08 | 0.88 | 0.32 | 0.54 | |
| Massilia | −0.10 | 0.85 | −0.58 | 0.22 | |
| norank_c__Gemmatimonadetes | −0.31 | 0.55 | −0.66 | 0.15 | |
| norank_f__Cytophagaceae | −0.41 | 0.42 | −0.55 | 0.26 | |
| norank_o__Acidimicrobiales | −0.02 | 0.98 | −0.59 | 0.22 | |
| norank_o__AKYG1722 | −0.12 | 0.82 | −0.45 | 0.37 | |
| Paenibacillus | −0.45 | 0.38 | −0.64 | 0.17 | |
| Paenisporosarcina | −0.45 | 0.38 | −0.56 | 0.25 | |
| Rhizobium | −0.01 | 0.99 | 0.15 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Hu, K.; Yu, J.; Khan, M.T.A.; Cai, Y.; Zhao, X.; Zheng, Z.; Hu, Y.; Cui, Z.; Wang, X. Biogas Residues Improved Microbial Diversity and Disease Suppression Function under Extent Indigenous Soil Microbial Biomass. Life 2023, 13, 774. https://doi.org/10.3390/life13030774
Zhao Y, Hu K, Yu J, Khan MTA, Cai Y, Zhao X, Zheng Z, Hu Y, Cui Z, Wang X. Biogas Residues Improved Microbial Diversity and Disease Suppression Function under Extent Indigenous Soil Microbial Biomass. Life. 2023; 13(3):774. https://doi.org/10.3390/life13030774
Chicago/Turabian StyleZhao, Yubin, Kai Hu, Jiadong Yu, Md. Tariful Alam Khan, Yafan Cai, Xiaoling Zhao, Zehui Zheng, Yuegao Hu, Zongjun Cui, and Xiaofen Wang. 2023. "Biogas Residues Improved Microbial Diversity and Disease Suppression Function under Extent Indigenous Soil Microbial Biomass" Life 13, no. 3: 774. https://doi.org/10.3390/life13030774
APA StyleZhao, Y., Hu, K., Yu, J., Khan, M. T. A., Cai, Y., Zhao, X., Zheng, Z., Hu, Y., Cui, Z., & Wang, X. (2023). Biogas Residues Improved Microbial Diversity and Disease Suppression Function under Extent Indigenous Soil Microbial Biomass. Life, 13(3), 774. https://doi.org/10.3390/life13030774











