Identification of Hub Genes and Biological Mechanisms Associated with Non-Alcoholic Fatty Liver Disease and Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Data Collection
2.2. Differentially Expressed Gene (DEG) Selection
2.3. Functional Classification and Pathway Enrichment of DEGs
2.4. Protein–Protein Interaction (PPI) Establishment and Hub Gene Identification
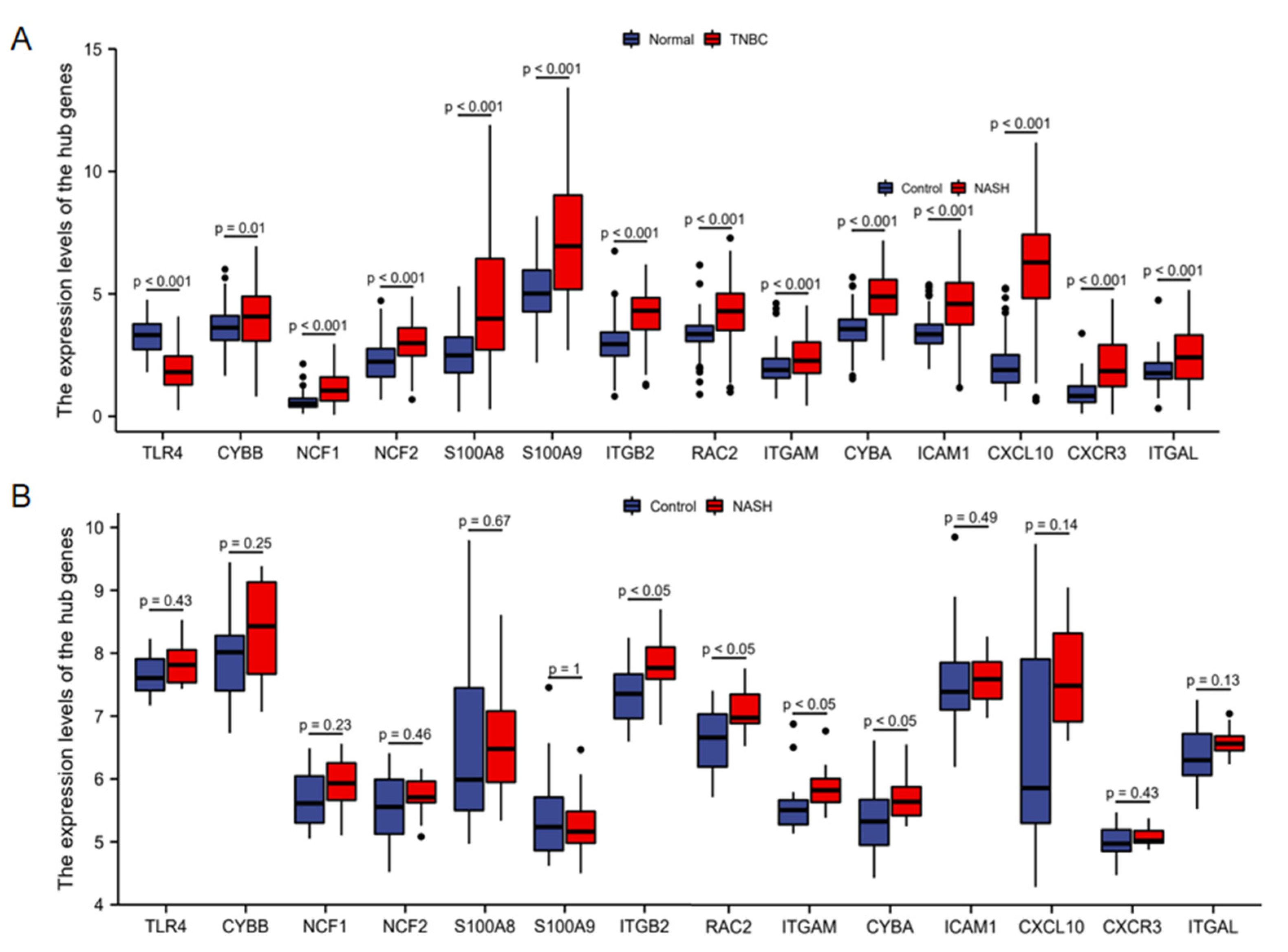
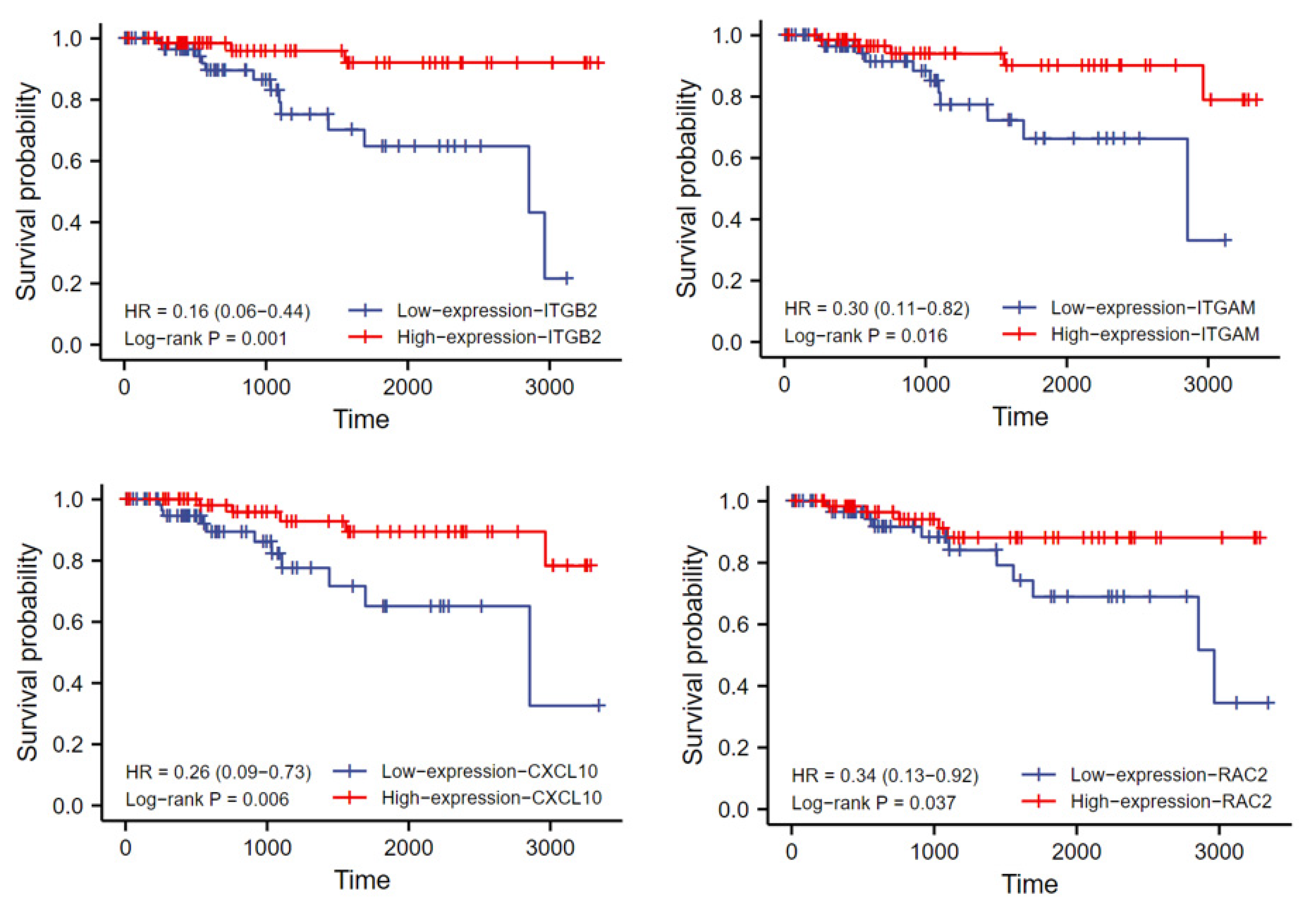
2.5. Hub Gene Expression Validation and Prognostic Analysis
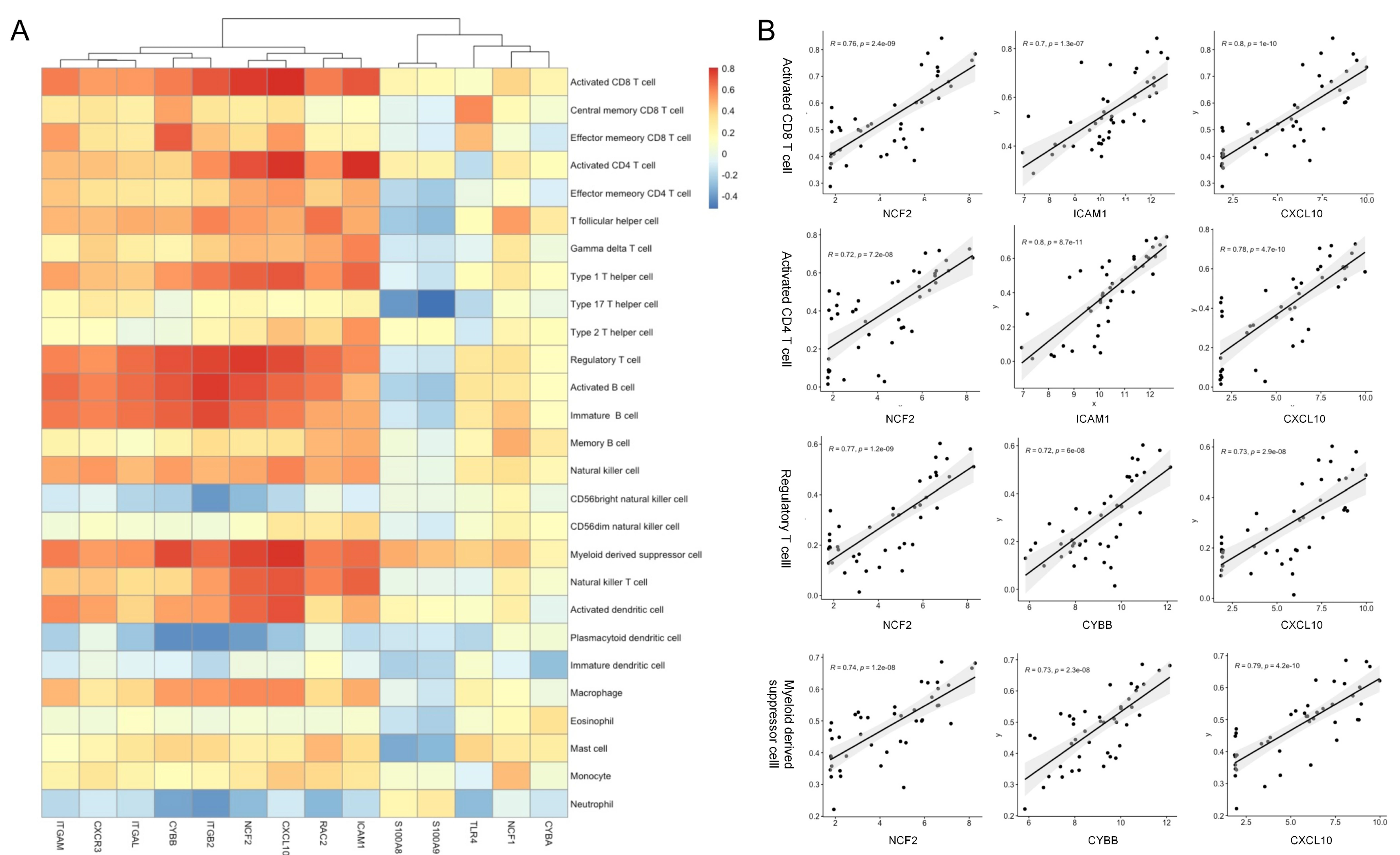
2.6. Immune Infiltration Analysis
3. Results
3.1. DEG Identification in NASH and TNBC
3.2. GO and KEGG Enrichment Pathway Analysis of DEGs
3.3. PPI Network Construction and Hub Gene Identification
3.4. Hub Gene Expression Validation and Prognostic Analysis
3.5. Association between the Hub Genes and Immune Infiltration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Billar, J.A.; Dueck, A.C.; Stucky, C.C.; Gray, R.J.; Wasif, N.; Northfelt, D.W.; McCullough, A.E.; Pockaj, B.A. Triple-negative breast cancers: Unique clinical presentations and outcomes. Ann. Surg. Oncol. 2010, 17 (Suppl. 3), 384–390. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Nofech-Mozes, S.; Trudeau, M.; Kahn, H.K.; Dent, R.; Rawlinson, E.; Sun, P.; Narod, S.A.; Hanna, W.M. Patterns of recurrence in the basal and non-basal subtypes of triple-negative breast cancers. Breast Cancer Res. Treat 2009, 118, 131–137. [Google Scholar] [CrossRef]
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef]
- Cornejo-Moreno, B.A.; Uribe-Escamilla, D.; Salamanca-Gómez, F. Breast cancer genes: Looking for BRACA’s lost brother. Isr. Med. Assoc. J. 2014, 16, 787–792. [Google Scholar] [PubMed]
- Toro, A.L.; Costantino, N.S.; Shriver, C.D.; Ellsworth, D.L.; Ellsworth, R.E. Effect of obesity on molecular characteristics of invasive breast tumors: Gene expression analysis in a large cohort of female patients. BMC Obes. 2016, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.H.; Lundh, M.; Fu, A.; Kriszt, R.; Huang, T.L.; Lynes, M.D.; Leiria, L.O.; Shamsi, F.; Darcy, J.; Greenwood, B.P.; et al. CRISPR-engineered human brown-like adipocytes prevent diet-induced obesity and ameliorate metabolic syndrome in mice. Sci. Transl. Med. 2020, 12, eaaz8664. [Google Scholar] [CrossRef]
- Kazak, L.; Chouchani, E.T.; Lu, G.Z.; Jedrychowski, M.P.; Bare, C.J.; Mina, A.I.; Kumari, M.; Zhang, S.; Vuckovic, I.; Laznik-Bogoslavski, D.; et al. Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab. 2017, 26, 693. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Oyekunle, T.; Salako, O.; Gupta, A.; Alatise, O.; Ogun, G.; Adeniyi, A.; Deveaux, A.; Hall, A.; Ayandipo, O.; et al. Metabolic Syndrome and Risk of Breast Cancer by Molecular Subtype: Analysis of the MEND Study. Clin. Breast Cancer 2022, 22, e463–e472. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology 2012, 142, 1592–1609. [Google Scholar] [CrossRef] [Green Version]
- Lomonaco, R.; Sunny, N.E.; Bril, F.; Cusi, K. Nonalcoholic fatty liver disease: Current issues and novel treatment approaches. Drugs 2013, 73, 1–14. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Ren, Z.; Hu, S.; Gao, Y.; Sun, R.; Lv, S.; Yang, G.; Yu, Z.; Kan, Q. Development and validation of a simple risk model to predict major cancers for patients with nonalcoholic fatty liver disease. Cancer Med. 2020, 9, 1254–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, T.G.; Roelstraete, B.; Sharma, R.; Khalili, H.; Hagström, H.; Ludvigsson, J.F. Cancer Risk in Patients With Biopsy-ConfirMed. Nonalcoholic Fatty Liver Disease: A Population-Based Cohort Study. Hepatology 2021, 74, 2410–2423. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Yan, Y.; Su, L.; Chen, D.; Zhang, C. Development of a risk-stratification scoring system for predicting risk of breast cancer based on non-alcoholic fatty liver disease, non-alcoholic fatty pancreas disease, and uric acid. Open Med. (Wars) 2022, 17, 619–625. [Google Scholar] [CrossRef]
- Nseir, W.; Abu-Rahmeh, Z.; Tsipis, A.; Mograbi, J.; Mahamid, M. Relationship between Non-Alcoholic Fatty Liver Disease and Breast Cancer. Isr. Med. Assoc. J. 2017, 19, 242–245. [Google Scholar]
- Bilici, A.; Ozguroglu, M.; Mihmanli, I.; Turna, H.; Adaletli, I. A case-control study of non-alcoholic fatty liver disease in breast cancer. Med. Oncol. 2007, 24, 367–371. [Google Scholar] [CrossRef]
- Yang, Y.J.; Kim, K.M.; An, J.H.; Lee, D.B.; Shim, J.H.; Lim, Y.S.; Lee, H.C.; Lee, Y.S.; Ahn, J.H.; Jung, K.H.; et al. Clinical significance of fatty liver disease induced by tamoxifen and toremifene in breast cancer patients. Breast 2016, 28, 67–72. [Google Scholar] [CrossRef]
- Taroeno-Hariadi, K.W.; Putra, Y.R.; Choridah, L.; Widodo, I.; Hardianti, M.S.; Aryandono, T. Fatty Liver in Hormone Receptor-Positive Breast Cancer and Its Impact on Patient’s Survival. J. Breast Cancer 2021, 24, 417–427. [Google Scholar] [CrossRef]
- Zheng, Q.; Xu, F.; Nie, M.; Xia, W.; Qin, T.; Qin, G.; An, X.; Xue, C.; Peng, R.; Yuan, Z.; et al. Selective Estrogen Receptor Modulator-Associated Nonalcoholic Fatty Liver Disease Improved Survival in Patients With Breast Cancer: A Retrospective Cohort Analysis. Medicine (Baltim.) 2015, 94, e1718. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, L.; Zhang, X.; Shang, J.; Duan, X. Exploring the molecular mechanisms and shared gene signatuRes. between rheumatoid arthritis and diffuse large B cell lymphoma. Front. Immunol. 2022, 13, 1036239. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zeng, N.; Ge, Y.; Wang, D.; Qin, X.; Zhang, W.; Jiang, F.; Liu, Y. Identification of the Shared Gene SignatuRes. and Biological Mechanism in Type 2 Diabetes and Pancreatic Cancer. Front. Endocrinol. (Lausanne) 2022, 13, 847760. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ma, L.; Ge, Z.; Pan, Y.; Xie, L. Key Genes Associated With Non-Alcoholic Fatty Liver Disease and Polycystic Ovary Syndrome. Front. Mol. Biosci. 2022, 9, 888194. [Google Scholar] [CrossRef]
- Cotter, T.G.; Rinella, M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology 2020, 158, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Wong, G.L.; Tsang, S.W.; Fan, T.; Chu, W.C.; Woo, J.; Chan, A.W.; Choi, P.C.; Chim, A.M.; Lau, J.Y.; et al. High prevalence of colorectal neoplasm in patients with non-alcoholic steatohepatitis. Gut 2011, 60, 829–836. [Google Scholar] [CrossRef]
- Craven, K.E.; Gökmen-Polar, Y.; Badve, S.S. CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer. Sci. Rep. 2021, 11, 4691. [Google Scholar] [CrossRef]
- Finotello, F.; Trajanoski, Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol. Immunother. 2018, 67, 1031–1040. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [Green Version]
- Sanna, C.; Rosso, C.; Marietti, M.; Bugianesi, E. Non-Alcoholic Fatty Liver Disease and Extra-Hepatic Cancers. Int. J. Mol. Sci. 2016, 17, 717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.J.; Chang, H.T.; Lee, C.H. Association between tamoxifen treatment and the development of different stages of nonalcoholic fatty liver disease among breast cancer patients. J. Formos. Med. Assoc. 2016, 115, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Yan, M.; Wang, J.; Xuan, Q.; Dong, T.; He, J.; Zhang, Q. The Relationship Between Tamoxifen-associated Nonalcoholic Fatty Liver Disease and the Prognosis of Patients With Early-stage Breast Cancer. Clin. Breast Cancer 2017, 17, 195–203. [Google Scholar] [CrossRef]
- Wu, W.; Chen, J.; Ye, W.; Li, X.; Zhang, J. Fatty liver decreases the risk of liver metastasis in patients with breast cancer: A two-center cohort study. Breast Cancer Res. Treat 2017, 166, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Chinetti, G.; Fruchart, J.C.; Staels, B. Peroxisome proliferator-activated receptors (PPARs): Nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 2000, 49, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Michalik, L.; Wahli, W. PPARs Mediate Lipid Signaling in Inflammation and Cancer. PPAR Res. 2008, 2008, 134059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zúñiga, J.; Cancino, M.; Medina, F.; Varela, P.; Vargas, R.; Tapia, G.; Videla, L.A.; Fernández, V. N-3 PUFA supplementation triggers PPAR-α activation and PPAR-α/NF-κB interaction: Anti-inflammatory implications in liver ischemia-reperfusion injury. PLoS ONE 2011, 6, e28502. [Google Scholar] [CrossRef] [Green Version]
- Dana, N.; Haghjooy Javanmard, S.; Vaseghi, G. The effect of fenofibrate, a PPARα activator on toll-like receptor-4 signal transduction in melanoma both in vitro and in vivo. Clin. Transl. Oncol. 2020, 22, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, Q.; Zhang, J.; Yang, G.; Shao, Z.; Luo, J.; Fan, M.; Ni, C.; Wu, Z.; Hu, X. Fenofibrate induces apoptosis of triple-negative breast cancer cells via activation of NF-κB pathway. BMC Cancer 2014, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Crosas-Molist, E.; Fabregat, I. Role of NADPH oxidases in the redox biology of liver fibrosis. Redox Biol. 2015, 6, 106–111. [Google Scholar] [CrossRef] [Green Version]
- García-Ruiz, I.; Blanes Ruiz, N.; Rada, P.; Pardo, V.; Ruiz, L.; Blas-García, A.; Valdecantos, M.P.; Grau Sanz, M.; Solís Herruzo, J.A.; Valverde, Á.M. Protein tyrosine phosphatase 1b deficiency protects against hepatic fibrosis by modulating nadph oxidases. Redox Biol. 2019, 26, 101263. [Google Scholar] [CrossRef]
- Hecker, L.; Vittal, R.; Jones, T.; Jagirdar, R.; Luckhardt, T.R.; Horowitz, J.C.; Pennathur, S.; Martinez, F.J.; Thannickal, V.J. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat. Med. 2009, 15, 1077–1081. [Google Scholar] [CrossRef] [Green Version]
- Bondi, C.D.; Manickam, N.; Lee, D.Y.; Block, K.; Gorin, Y.; Abboud, H.E.; Barnes, J.L. NAD(P)H oxidase mediates TGF-beta1-induced activation of kidney myofibroblasts. J. Am. Soc. Nephrol. 2010, 21, 93–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell Mol. Immunol. 2015, 12, 5–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matuz-Mares, D.; Vázquez-Meza, H.; Vilchis-Landeros, M.M. NOX as a Therapeutic Target in Liver Disease. Antioxidants 2022, 11, 2038. [Google Scholar] [CrossRef]
- Ma, Y.; Lee, G.; Heo, S.Y.; Roh, Y.S. Oxidative Stress Is a Key Modulator in the Development of Nonalcoholic Fatty Liver Disease. Antioxidants 2021, 11, 91. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Arrington, M.E.; Temple, B.; Schaefer, A.; Campbell, S.L. The molecular basis for immune dysregulation by the hyperactivated E62K mutant of the GTPase RAC2. J. Biol. Chem. 2020, 295, 12130–12142. [Google Scholar] [CrossRef]
- Watanabe, M.; Terasawa, M.; Miyano, K.; Yanagihara, T.; Uruno, T.; Sanematsu, F.; Nishikimi, A.; Côté, J.F.; Sumimoto, H.; Fukui, Y. DOCK2 and DOCK5 act additively in neutrophils to regulate chemotaxis, superoxide production, and extracellular trap formation. J. Immunol. 2014, 193, 5660–5667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Jun, H.; Yang, C.; Yang, F.; Xu, Y. The Pyroptosis-Related Risk Genes APOBEC3D, TNFRSF14, and RAC2 Were Used to Evaluate Prognosis and as Tumor Suppressor Genes in Breast Cancer. J. Oncol. 2022, 2022, 3625790. [Google Scholar] [CrossRef]
- Abram, C.L.; Lowell, C.A. The ins and outs of leukocyte integrin signaling. Annu. Rev. Immunol. 2009, 27, 339–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas, K.; Baliu-Piqué, M.; Manzano, A.; Saiz-Ladera, C.; García-Barberán, V.; Cimas, F.J.; Pérez-Segura, P.; Pandiella, A.; Győrffy, B.; Ocana, A. In silico transcriptomic mapping of integrins and immune activation in Basal-like and HER2+ breast cancer. Cell Oncol. (Dordr.) 2021, 44, 569–580. [Google Scholar] [CrossRef]
- Rose, D.M.; Liu, S.; Woodside, D.G.; Han, J.; Schlaepfer, D.D.; Ginsberg, M.H. Paxillin binding to the alpha 4 integrin subunit stimulates LFA-1 (integrin alpha L beta 2)-dependent T cell migration by augmenting the activation of focal adhesion kinase/proline-rich tyrosine kinase-2. J. Immunol. 2003, 170, 5912–5918. [Google Scholar] [CrossRef] [Green Version]
- Harjunpää, H.; Llort Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jialal, I.; Adams-Huet, B.; Devaraj, S. Monocyte cell adhesion molecule receptors in nascent metabolic syndrome. Clin. BioChem. 2016, 49, 505–507. [Google Scholar] [CrossRef] [PubMed]
- Winer, S.; Winer, D.A. The adaptive immune system as a fundamental regulator of adipose tissue inflammation and insulin resistance. Immunol. Cell Biol. 2012, 90, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Osborn, O.; Olefsky, J.M. The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 2012, 18, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Zhang, X.; Lau, J.; Yu, J. C-X-C motif chemokine 10 in non-alcoholic steatohepatitis: Role as a pro-inflammatory factor and clinical implication. Expert Rev. Mol. Med. 2016, 18, e16. [Google Scholar] [CrossRef]
- Zhang, X.; Han, J.; Man, K.; Li, X.; Du, J.; Chu, E.S.; Go, M.Y.; Sung, J.J.; Yu, J. CXC chemokine receptor 3 promotes steatohepatitis in mice through mediating inflammatory cytokines, macrophages and autophagy. J. Hepatol. 2016, 64, 160–170. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Sun, A.; Yu, W.; Hong, C.; Liu, Z. CXCL10 mediates breast cancer tamoxifen resistance and promotes estrogen-dependent and independent proliferation. Mol. Cell Endocrinol. 2020, 512, 110866. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, J.; Wang, T.; Zhang, X.; Liu, D. CXCL10/CXCR3 axis promotes the invasion of gastric cancer via PI3K/AKT pathway-dependent MMPs production. BioMed. Pharmacother. 2016, 82, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Zhu, X.; Ueda, R.; Sasaki, K.; Kohanbash, G.; Kastenhuber, E.R.; McDonald, H.A.; Gibson, G.A.; Watkins, S.C.; Muthuswamy, R.; et al. Effective immunotherapy against murine gliomas using type 1 polarizing dendritic cells—significant roles of CXCL10. Cancer Res. 2009, 69, 1587–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Mo, Y.; Jiang, S.; Shang, C.; Feng, Y.; Zeng, X. CXC chemokine ligand-10 promotes the accumulation of monocyte-like myeloid-derived suppressor cells by activating p38 MAPK signaling under tumor conditions. Cancer Sci. 2022, 114, 142–151. [Google Scholar] [CrossRef]
- Mowat, C.; Mosley, S.R.; Namdar, A.; Schiller, D.; Baker, K. Anti-tumor immunity in mismatch repair-deficient colorectal cancers requiRes. type I IFN-driven CCL5 and CXCL10. J. Exp. Med. 2021, 218, e20210108. [Google Scholar] [CrossRef] [PubMed]
- Reschke, R.; Gajewski, T.F. CXCL9 and CXCL10 bring the heat to tumors. Sci. Immunol. 2022, 7, eabq6509. [Google Scholar] [CrossRef]
- Hoch, T.; Schulz, D.; Eling, N.; Gómez, J.M.; Levesque, M.P.; Bodenmiller, B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes featuRes. of the response to immunotherapy. Sci. Immunol. 2022, 7, eabk1692. [Google Scholar] [CrossRef]
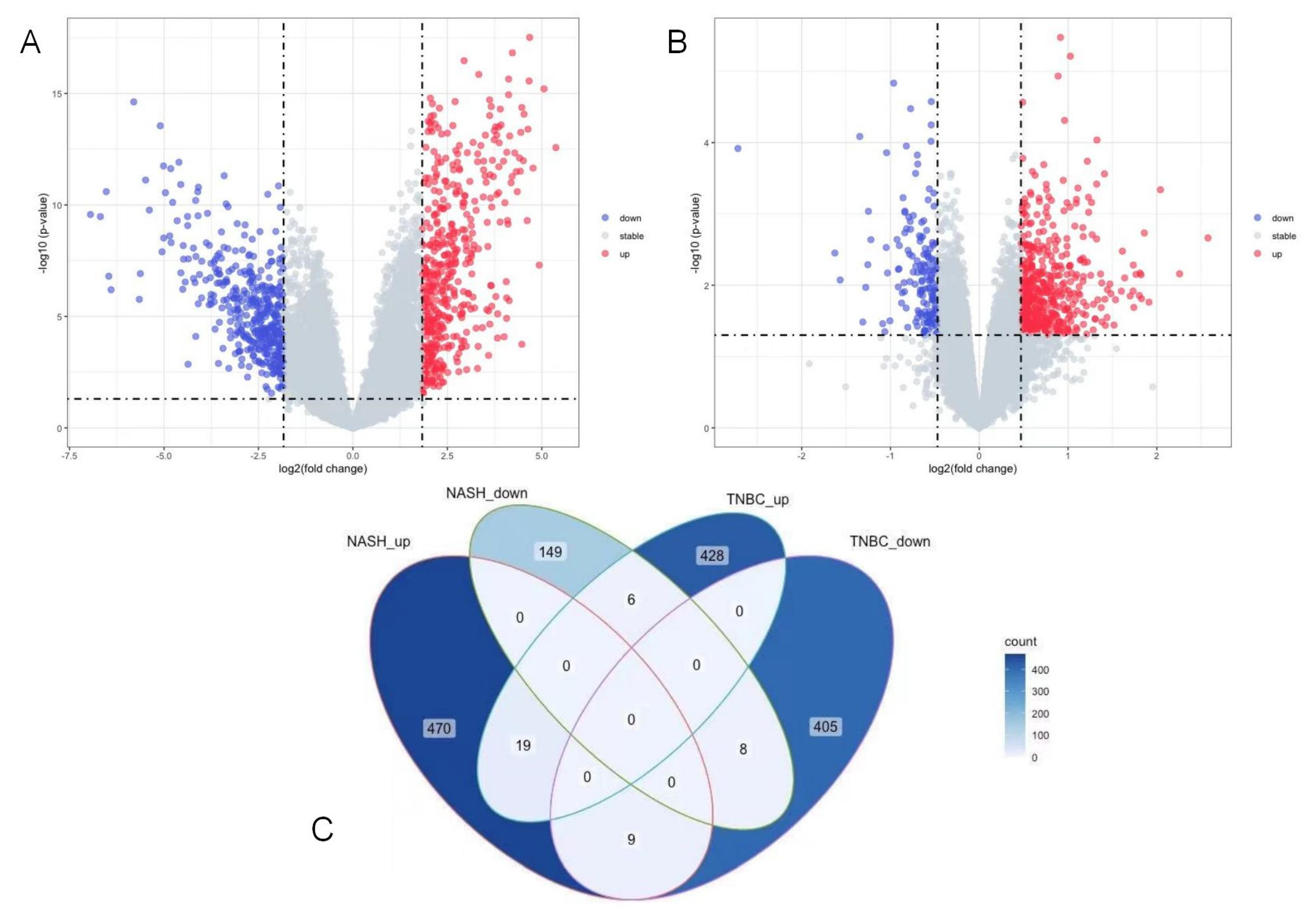
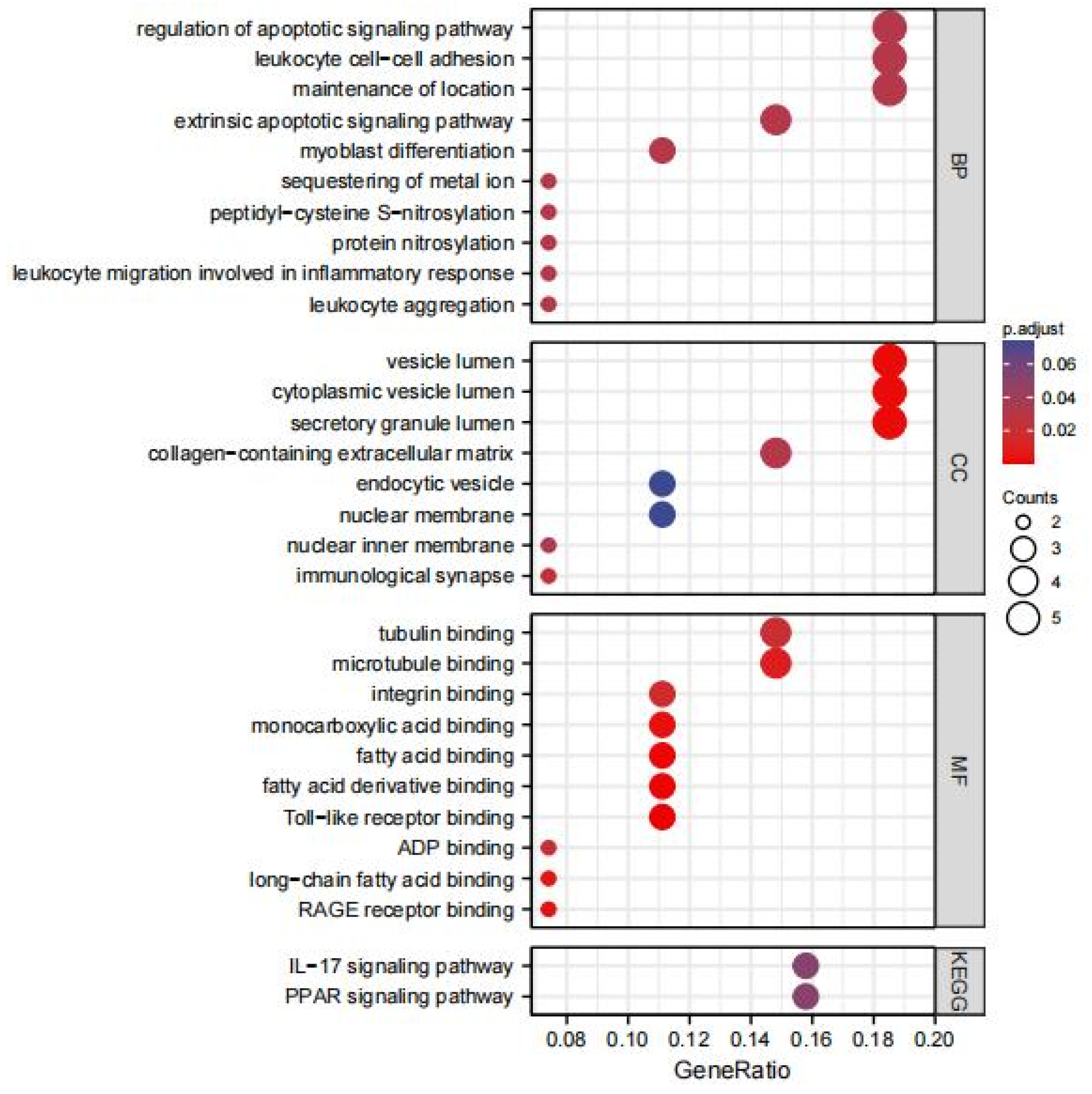
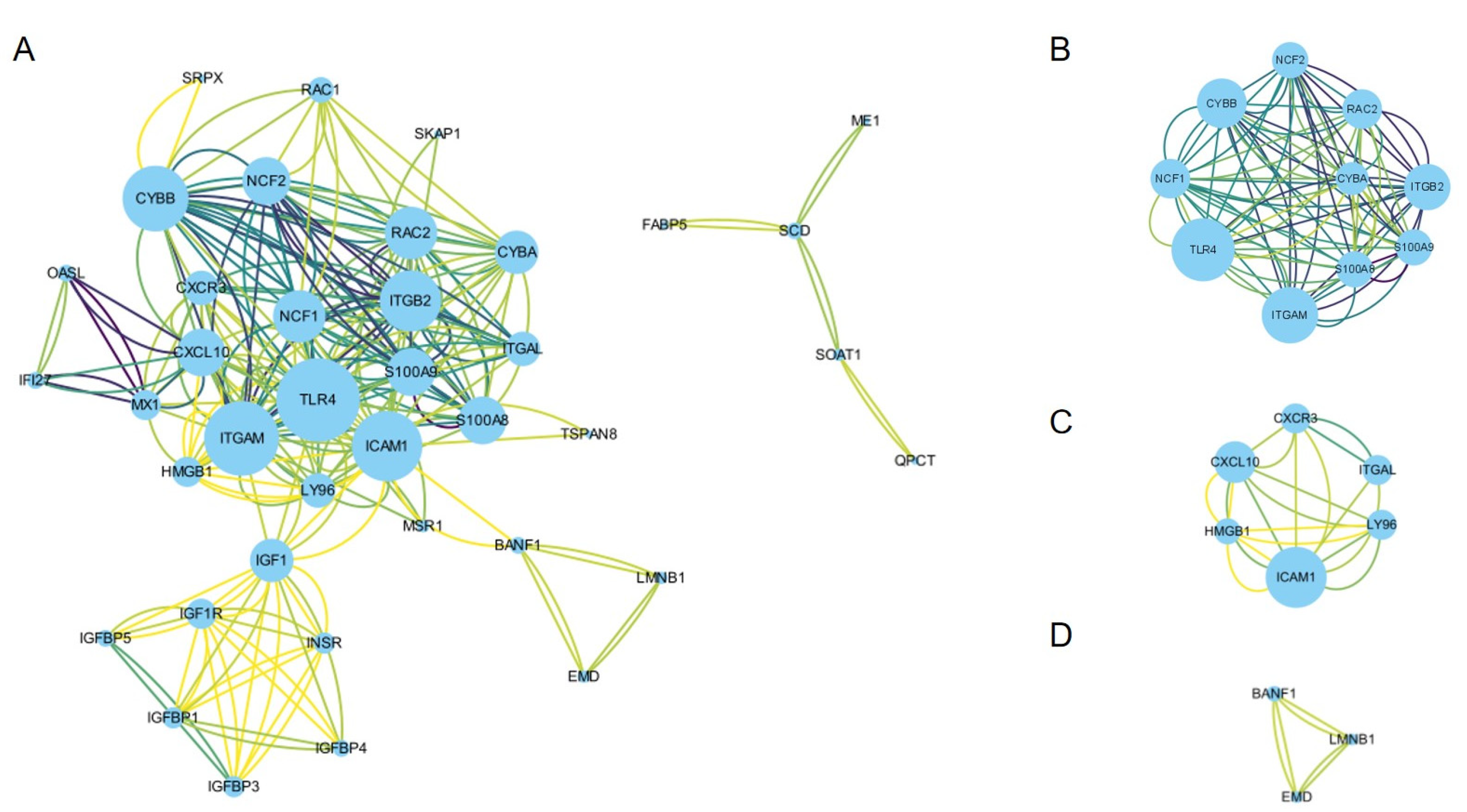
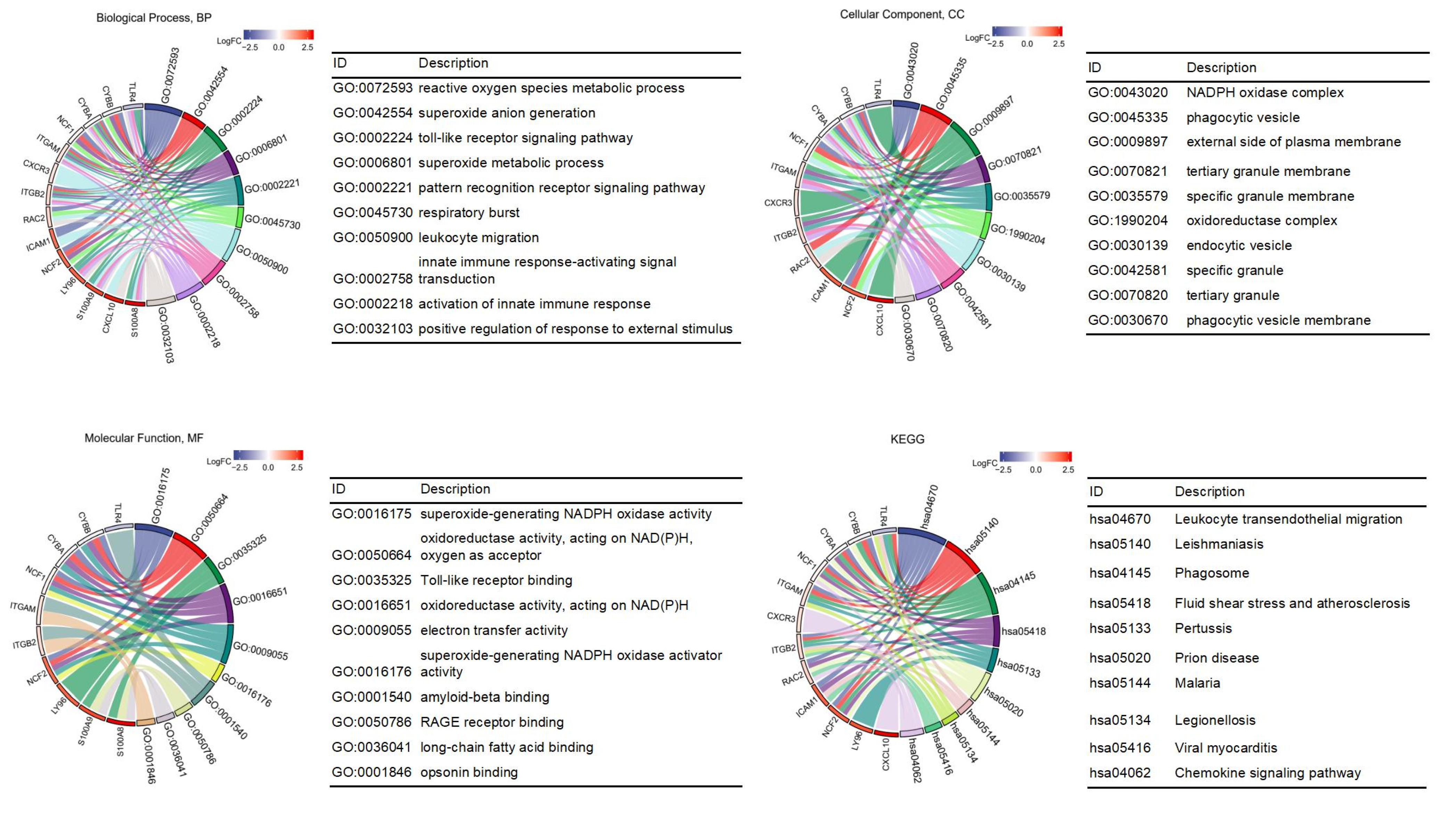
| Series | Country | Status | Platforms | Type of Samples | Numbers |
|---|---|---|---|---|---|
| GSE63067 | Sweden | Public on 7 November 2014 | GPL570 | non-alcoholic steatohepatitis | 9 |
| steatosis | 2 | ||||
| healthy | 7 | ||||
| GSE48452 | Germany | Public on 8 August 2013 | GPL11532 | non-alcoholic steatohepatitis | 18 |
| steatosis | 14 | ||||
| healthy obese | 27 | ||||
| control | 14 | ||||
| GSE38959 | Japan | Public on 21 December 2012 | GPL4133 | triple-negative breast cancer | 30 |
| normal mammary ductal cells | 13 | ||||
| normal human vital organs including heart, lung, liver, and kidney | 4 |
| Factor | Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | ||
|---|---|---|---|---|
| HR (95%CI) | p-Value | HR (95%CI) | p-Value | |
| TLR4 | 0.640 (0.237–1.728) | 0.378 | ||
| CYBB | 0.509 (0.183–1.411) | 0.194 | ||
| NCF1 | 0.431 (0.155–1.200) | 0.107 | ||
| NCF2 | 0.622 (0.228–1.694) | 0.352 | ||
| S100A8 | 0.810 (0.295–2.226) | 0.683 | ||
| S100A9 | 0.523 (0.181–1.507) | 0.230 | ||
| ITGB2 | 0.157 (0.044–0.556) | 0.004 | 0.213 (0.033–1.376) | 0.104 |
| RAC2 | 0.341 (0.118–0.984) | 0.047 | 1.067 (0.282–4.036) | 0.924 |
| ITGAM | 0.282 (0.094–0.842) | 0.023 | 1.392 (0.318–6.100) | 0.661 |
| CYBA | 1.750 (0.632–4.847) | 0.282 | ||
| ICAM1 | 0.676 (0.250–1.828) | 0.440 | ||
| CXCL10 | 0.244 (0.082–0.725) | 0.011 | 0.430 (0.108–1.718) | 0.232 |
| CXCR3 | 0.419 (0.152–1.156) | 0.093 | ||
| ITGAL | 0.413 (0.148–1.151) | 0.091 | ||
| Age | 0.773 (0.249–2.406) | 0.657 | ||
| Race | 2.830 (1.019–7.860) | 0.046 | 2.090 (0.631–6.922) | 0.227 |
| T stage | ||||
| (T2 vs. T1, T3/T4 vs. T1) | 1.717 (0.471–6.255) | 0.194 | ||
| 4.427 (0.858–22.838) | ||||
| N stage | ||||
| (N1/N2/N3 vs. N0) | 5.641 (1.815–17.534) | 0.003 | 4.681 (1.452–15.089) | 0.010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Min, N.; Gong, W.; Chen, Y.; Li, X. Identification of Hub Genes and Biological Mechanisms Associated with Non-Alcoholic Fatty Liver Disease and Triple-Negative Breast Cancer. Life 2023, 13, 998. https://doi.org/10.3390/life13040998
Zhu J, Min N, Gong W, Chen Y, Li X. Identification of Hub Genes and Biological Mechanisms Associated with Non-Alcoholic Fatty Liver Disease and Triple-Negative Breast Cancer. Life. 2023; 13(4):998. https://doi.org/10.3390/life13040998
Chicago/Turabian StyleZhu, Jingjin, Ningning Min, Wenye Gong, Yizhu Chen, and Xiru Li. 2023. "Identification of Hub Genes and Biological Mechanisms Associated with Non-Alcoholic Fatty Liver Disease and Triple-Negative Breast Cancer" Life 13, no. 4: 998. https://doi.org/10.3390/life13040998
APA StyleZhu, J., Min, N., Gong, W., Chen, Y., & Li, X. (2023). Identification of Hub Genes and Biological Mechanisms Associated with Non-Alcoholic Fatty Liver Disease and Triple-Negative Breast Cancer. Life, 13(4), 998. https://doi.org/10.3390/life13040998




