Transcriptome Analysis Reveals Genes Associated with Flooding Tolerance in Mulberry Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. cDNA Library Construction and RNA Sequencing
2.3. Transcriptome Assembly and Data Analysis
2.4. Prediction of Simple Sequence Repeats
2.5. Quantitative Reverse-Transcription PCR (qRT-PCR)
2.6. Statistical Analysis
3. Results
3.1. RNA Sequencing and Assembly
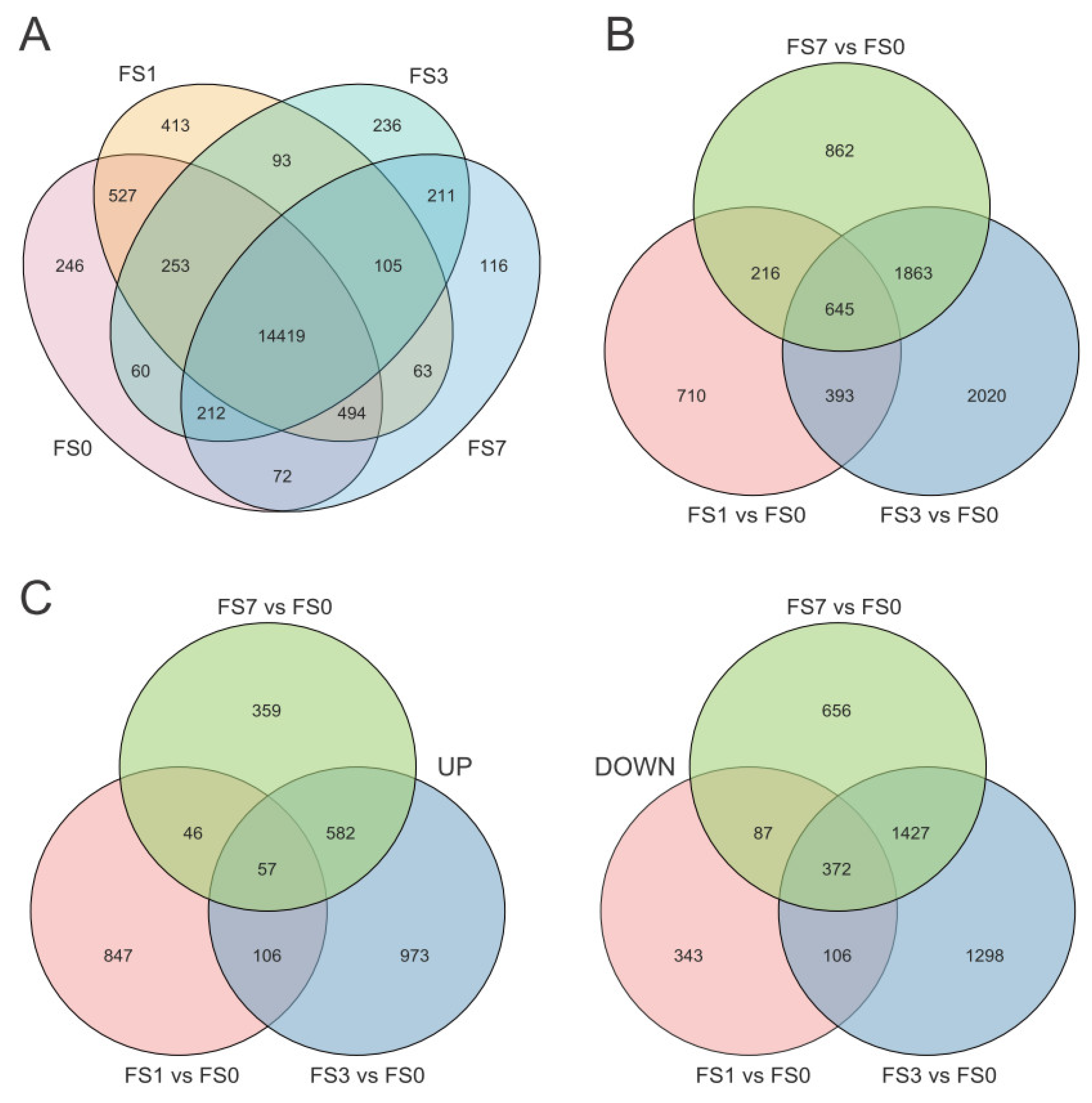
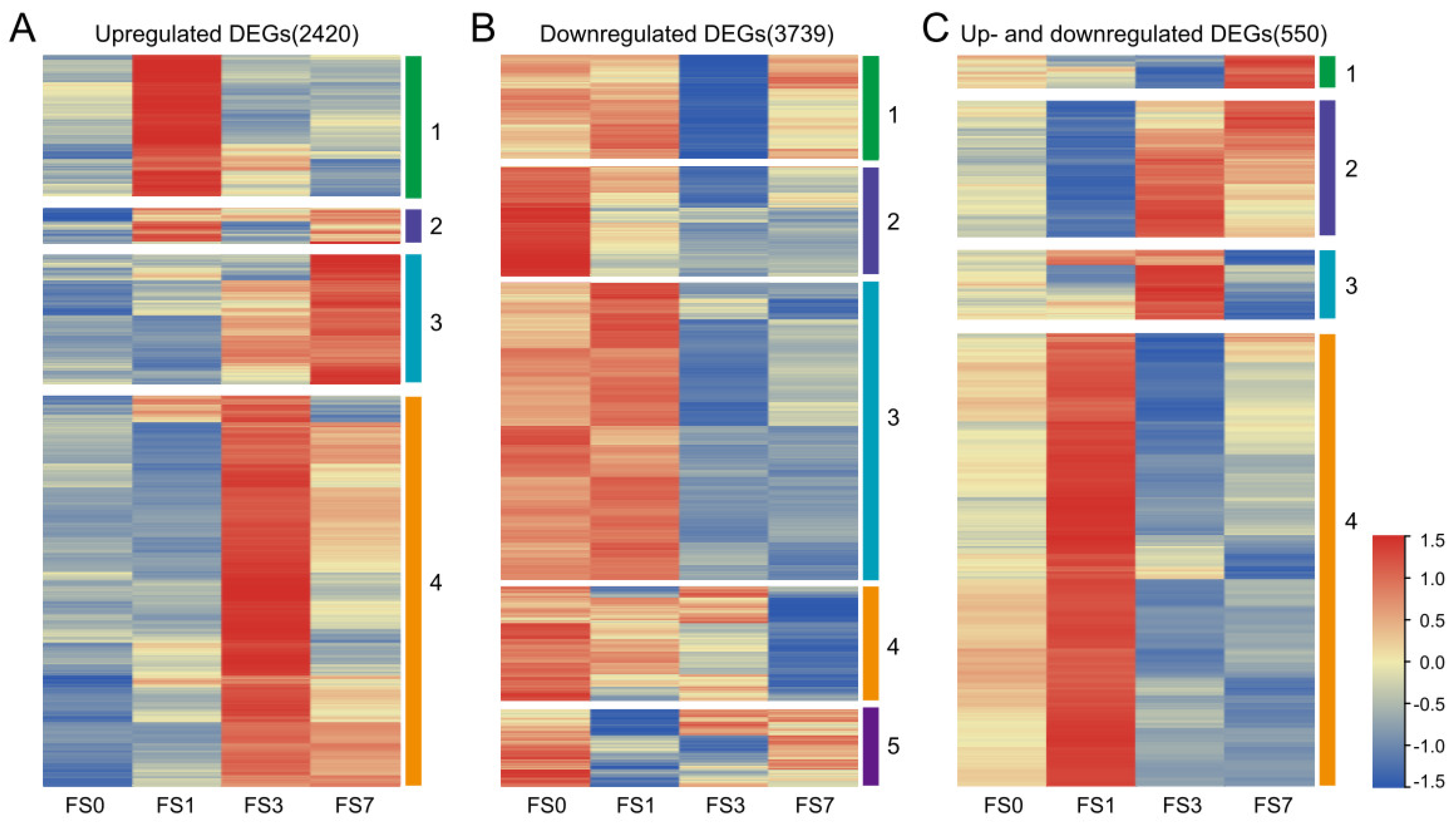
3.2. Identification of DEGs under Flooding Stress
3.3. GO Enrichment Analysis of DEGs
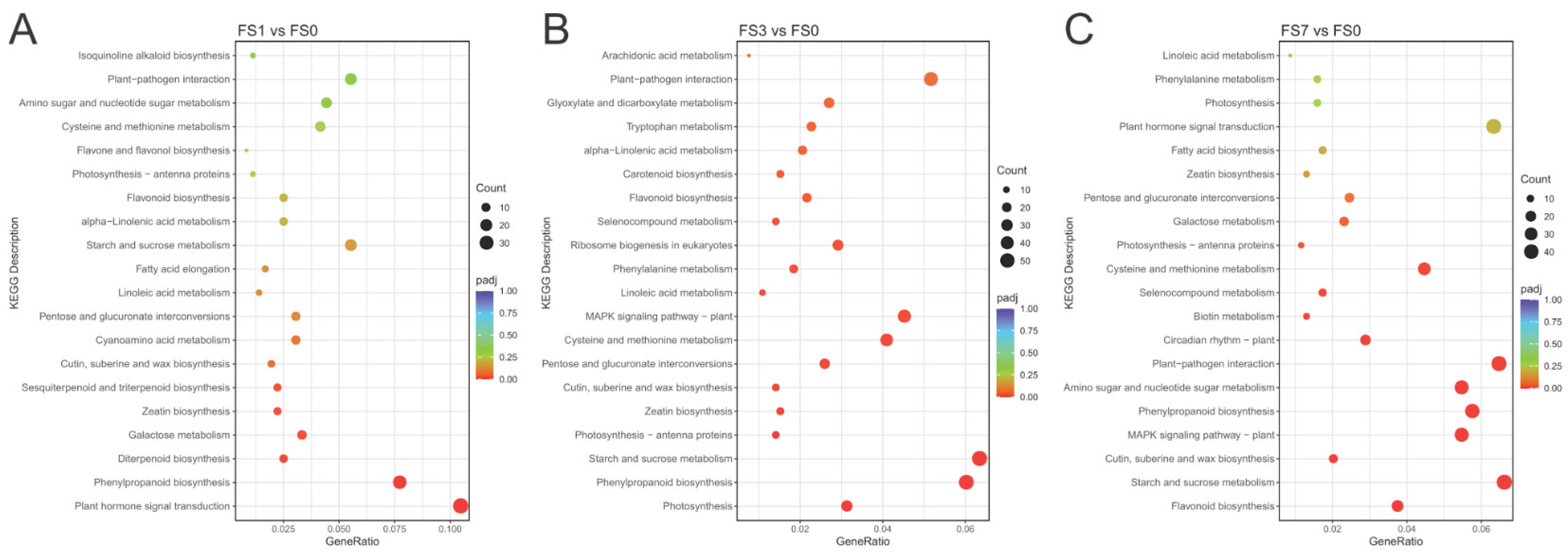
3.4. KEGG Enrichment Analyses of DEGs
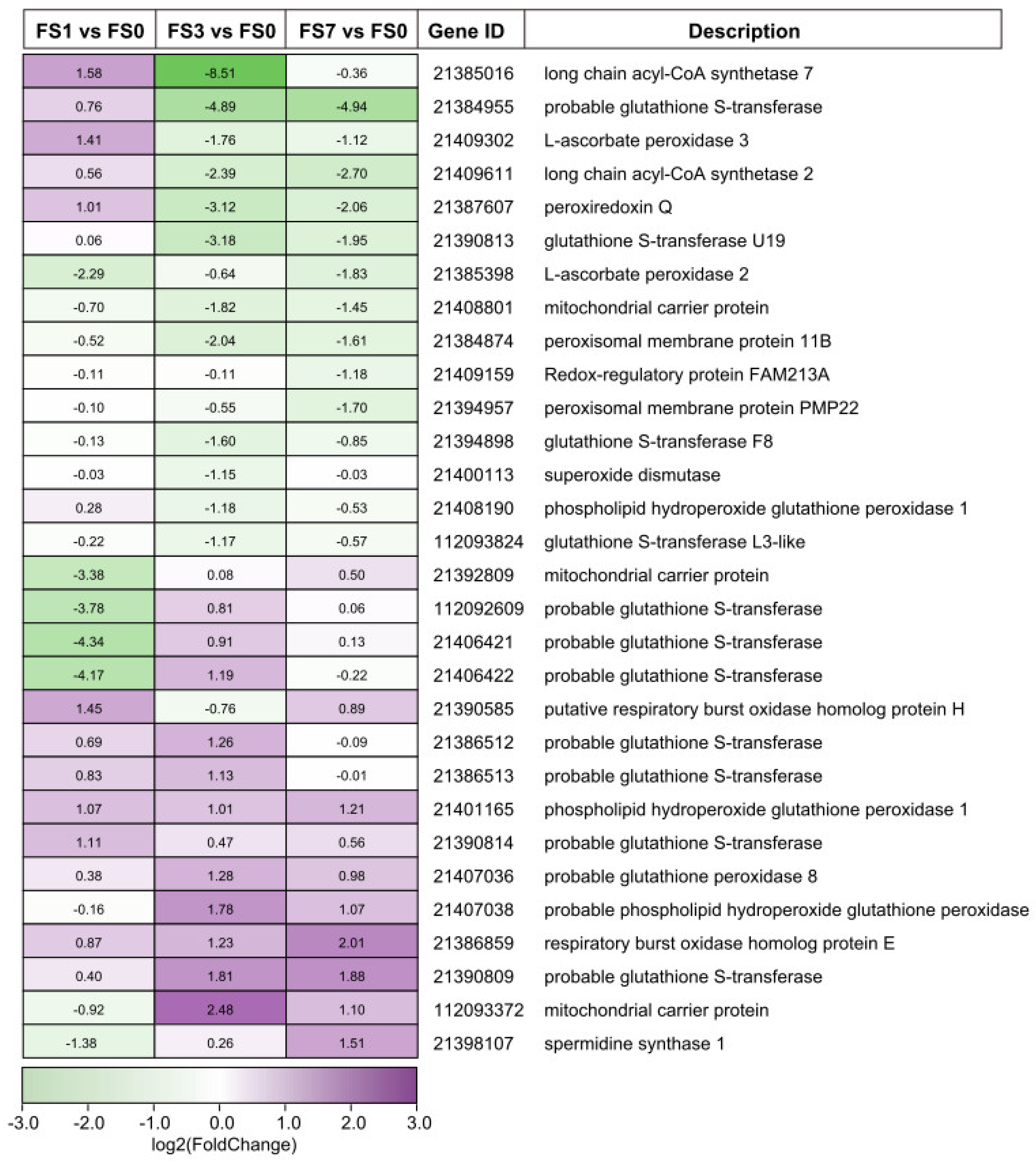
3.5. DEGs Involved in ROS Signaling
3.6. DEGs Involved in MAPK Signaling
3.7. DEGs Involved in Plant Hormone Signaling
3.8. DEGs Related to Energy Metabolism
3.9. Other DEGs Detected in Mulberry Plants after Flooding Stress
3.10. SSR Prediction
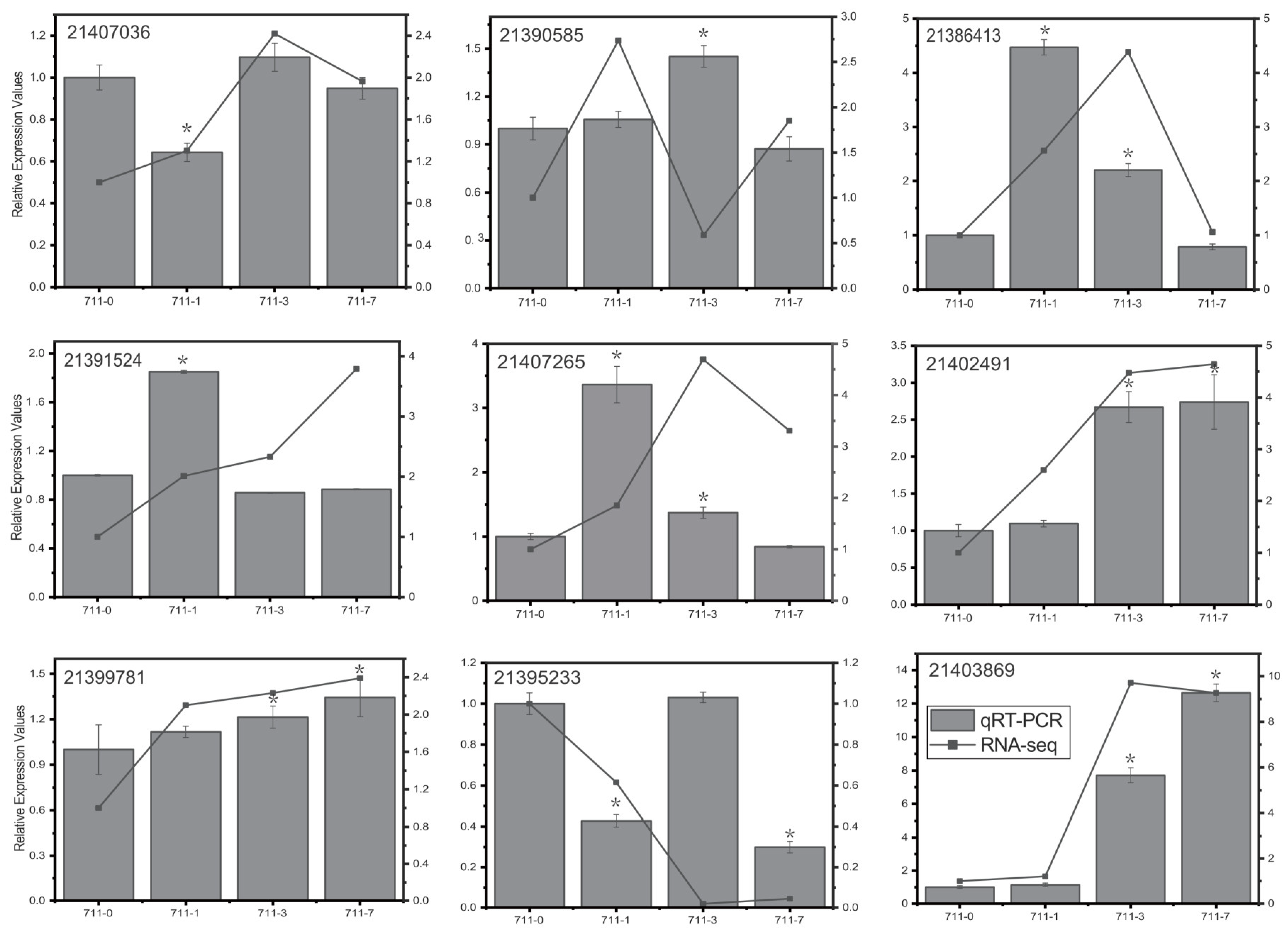
3.11. Validation of RNA-Seq Data by qRT-PCR
4. Discussion
4.1. DEGs Associated with ROS Signaling
4.2. DEGs Associated with Hormones
4.3. DEGs Associated with Energy Metabolism
4.4. Distribution of EST-SSRs in Mulberry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, S.C.; Mustroph, A.; Sasidharan, R.; Vashisht, D.; Pedersen, O.; Oosumi, T.; Voesenek, L.A.; Bailey-Serres, J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011, 190, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Perata, P.; Armstrong, W.; Voesenek, L.A. Plants and flooding stress. New Phytol. 2011, 190, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Colmer, T.; Voesenek, L. Flooding tolerance: Suites of plant traits in variable environments. Funct. Plant Biol. 2009, 36, 665–681. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Voesenek, L. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef]
- Loreti, E.; Yamaguchi, J.; Alpi, A.; Perata, P. Sugar Modulation of α-Amylase Genes under Anoxia. Ann. Bot. 2003, 91, 143–148. [Google Scholar] [CrossRef]
- Fukao, T.; Kennedy, R.A.; Yamasue, Y.; Rumpho, M.E. Genetic and biochemical analysis of anaerobically-induced enzymes during seed germination of Echinochloa crus-galli varieties tolerant and intolerant of anoxia. J. Exp. Bot. 2003, 54, 1421–1429. [Google Scholar] [CrossRef]
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Perata, P. Transcript Profiling of The Anoxic Rice Coleoptile. Plant Physiol. 2007, 144, 218–231. [Google Scholar] [CrossRef]
- Lin, Y.; Li, W.; Zhang, Y.; Xia, C.; Liu, Y.; Wang, C.; Xu, R.; Zhang, L. Identification of genes/proteins related to submergence tolerance by transcriptome and proteome analyses in soybean. Sci. Rep. 2019, 9, 14688. [Google Scholar] [CrossRef]
- Ren, C.G.; Kong, C.C.; Yan, K.; Zhang, H.; Luo, Y.M.; Xie, Z.H. Elucidation of the molecular responses to waterlogging in Sesbania cannabina roots by transcriptome profiling. Sci. Rep. 2017, 7, 9256. [Google Scholar] [CrossRef]
- Xu, X.; Chen, M.; Ji, J.; Xu, Q.; Qi, X.; Chen, X. Comparative RNA-seq based transcriptome profiling of waterlogging response in cucumber hypocotyls reveals novel insights into the de novo adventitious root primordia initiation. BMC Plant Biol. 2017, 17, 129. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, X.; Dai, J.; Luo, Z.; Li, Z.; Lu, H.; Xu, S.; Tang, W.; Zhang, D.; Li, W. Global gene expression in cotton (Gossypium hirsutum L.) leaves to waterlogging stress. PLoS ONE 2017, 12, e0185075. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Khan, A.L.; Al-Harrasi, A.S. Early events in plant abiotic stress signaling: Interplay between calcium, reactive oxygen species and phytohormones. J. Plant Growth Regul. 2018, 37, 1033–1049. [Google Scholar] [CrossRef]
- Steffens, B.; Steffen-Heins, A.; Sauter, M. Reactive oxygen species mediate growth and death in submerged plants. Front. Plant Sci. 2013, 4, 179. [Google Scholar] [CrossRef]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Sharmin, R.A.; Bhuiyan, M.R.; Lv, W.; Yu, Z.; Chang, F.; Kong, J.; Bhat, J.A.; Zhao, T. RNA-Seq based transcriptomic analysis revealed genes associated with seed-flooding tolerance in wild soybean (Glycine soja Sieb. & Zucc.). Environ. Exp. Bot. 2020, 171, 103906. [Google Scholar]
- New, T.; Xie, Z. Impacts of large dams on riparian vegetation: Applying global experience to the case of China’s Three Gorges Dam. Biodivers. Conserv. 2008, 17, 3149–3163. [Google Scholar] [CrossRef]
- Wu, J.; Huang, J.; Han, X.; Gao, X.; He, F.; Jiang, M.; Jiang, Z.; Primack, R.B.; Shen, Z. The three gorges dam: An ecological perspective. Front. Ecol. Environ. 2004, 2, 241–248. [Google Scholar] [CrossRef]
- Fu, B.-J.; Wu, B.-F.; Lü, Y.-H.; Xu, Z.-H.; Cao, J.-H.; Niu, D.; Yang, G.-S.; Zhou, Y.-M. Three Gorges Project: Efforts and challenges for the environment. Prog. Phys. Geogr. 2010, 34, 741–754. [Google Scholar] [CrossRef]
- Shi, R. Ecological environment problems of the Three Gorges Reservoir Area and countermeasures. Procedia Environ. Sci. 2011, 10, 1431–1434. [Google Scholar] [CrossRef]
- Wang, Q.; Yuan, X.; Willison, J.M.; Zhang, Y.; Liu, H. Diversity and above-ground biomass patterns of vascular flora induced by flooding in the drawdown area of China’s Three Gorges Reservoir. PLoS ONE 2014, 9, e100889. [Google Scholar] [CrossRef]
- Yang, F.; Liu, W.-W.; Wang, J.; Liao, L.; Wang, Y. Riparian vegetation’s responses to the new hydrological regimes from the Three Gorges Project: Clues to revegetation in reservoir water-level-fluctuation zone. Acta Ecol. Sin. 2012, 32, 89–98. [Google Scholar] [CrossRef]
- He, X.; Wang, T.; Wu, K.; Wang, P.; Qi, Y.; Arif, M.; Wei, H. Responses of swamp cypress (Taxodium distichum) and Chinese willow (Salix matsudana) roots to periodic submergence in mega-reservoir: Changes in organic acid concentration. Forests 2021, 12, 203. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Y.; Chan, Z. Perspectives on screening winter-flood-tolerant woody species in the riparian protection forests of the Three Gorges Reservoir. PLoS ONE 2014, 9, e108725. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Song, P.; Ma, B.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. Identification of the mulberry genes involved in ethylene biosynthesis and signaling pathways and the expression of MaERF-B2–1 and MaERF-B2–2 in the response to flooding stress. Funct. Integr. Genom. 2014, 14, 767–777. [Google Scholar] [CrossRef]
- Rao, L.; Li, S.; Cui, X. Leaf morphology and chlorophyll fluorescence characteristics of mulberry seedlings under waterlogging stress. Sci. Rep. 2021, 11, 13379. [Google Scholar] [CrossRef]
- Liu, Y.; Willison, J.M. Prospects for cultivating white mulberry (Morus alba) in the drawdown zone of the Three Gorges Reservoir, China. Environ. Sci. Pollut. Res. 2013, 20, 7142–7151. [Google Scholar] [CrossRef]
- Zhang, J.; Ren, R.; Zhu, J.; Song, C.; Liu, J.; Fu, J.; Hu, H.; Wang, J.; Li, H.; Xu, J. Preliminary experimentation on flooding resistance of mulberry trees along the water-fluctuation belt of the Three Gorges Reservoir. Sci. Silvae Sin. 2012, 48, 154–158. [Google Scholar]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor–like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana . Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef]
- Zhang, Q.; Tang, S.; Li, J.; Fan, C.; Xing, L.; Luo, K. Integrative transcriptomic and metabolomic analyses provide insight into the long-term submergence response mechanisms of young Salix variegata stems. Planta 2021, 253, 88. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- Shukla, P.; Reddy, R.A.; Ponnuvel, K.M.; Rohela, G.K.; Shabnam, A.A.; Ghosh, M.; Mishra, R.K. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses. Mol. Biol. Rep. 2019, 46, 1809–1817. [Google Scholar] [CrossRef]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef]
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef]
- Lee, K.-W.; Chen, P.-W.; Lu, C.-A.; Chen, S.; Ho, T.-H.D.; Yu, S.-M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009, 2, ra61. [Google Scholar] [CrossRef]
- Lee, K.W.; Chen, P.W.; Yu, S.M. Metabolic adaptation to sugar/O2 deficiency for anaerobic germination and seedling growth in rice. Plant Cell Environ. 2014, 37, 2234–2244. [Google Scholar]
- Ye, T.; Shi, H.; Wang, Y.; Chan, Z. Contrasting changes caused by drought and submergence stresses in bermudagrass (Cynodon dactylon). Front. Plant Sci. 2015, 6, 951. [Google Scholar] [CrossRef]
- Qi, X.; Li, Q.; Ma, X.; Qian, C.; Wang, H.; Ren, N.; Shen, C.; Huang, S.; Xu, X.; Xu, Q. Waterlogging-induced adventitious root formation in cucumber is regulated by ethylene and auxin through reactive oxygen species signalling. Plant Cell Environ. 2019, 42, 1458–1470. [Google Scholar] [CrossRef]
- Wu, Y.-S.; Yang, C.-Y. Physiological responses and expression profile of NADPH oxidase in rice (Oryza sativa) seedlings under different levels of submergence. Rice 2016, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Ni, X.; Arif, M.; Dong, Z.; Zhang, L.; Tan, X.; Li, J.; Li, C. Transcriptomic analysis of the photosynthetic, respiration, and aerenchyma adaptation strategies in bermudagrass (Cynodon dactylon) under different submergence stress. Int. J. Mol. Sci. 2021, 22, 7905. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, H.-H.; Hua, S.; Lin, K.-H.; Chen, N.; Zhang, Y.; You, Z.; Kuo, Y.-W.; Chen, S.-P. Cloning and overexpression of the ascorbate peroxidase gene from the yam (Dioscorea alata) enhances chilling and flood tolerance in transgenic Arabidopsis . J. Plant Res. 2019, 132, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Liu, F.; Pi, B.; Zhao, T.; Yu, B. Transcriptomic analysis of Glycine soja and G. max seedlings and functional characterization of GsGSTU24 and GsGSTU42 genes under submergence stress. Environ. Exp. Bot. 2020, 171, 103963. [Google Scholar] [CrossRef]
- Wen, X.-P.; Pang, X.-M.; Matsuda, N.; Kita, M.; Inoue, H.; Hao, Y.-J.; Honda, C.; Moriguchi, T. Over-expression of the apple spermidine synthase gene in pear confers multiple abiotic stress tolerance by altering polyamine titers. Transgenic Res. 2008, 17, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, G.; Xiang, W.; Zhu, H.; Zhang, H.; Zhang, J.; Ding, Z.; Liu, J.; Wu, D. Integrated Transcriptomic and Proteomic Analyses Uncover the Regulatory Mechanisms of Myricaria laxiflora under Flooding Stress. Front. Plant Sci. 2022, 13, 924490. [Google Scholar] [CrossRef]
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef]
- Jackson, M.B. Ethylene-promoted elongation: An adaptation to submergence stress. Ann. Bot. 2008, 101, 229–248. [Google Scholar] [CrossRef]
- Yamamoto, F.; Kozlowski, T. Regulation by auxin and ethylene of responses of Acer negundo seedlings to flooding of soil. Environ. Exp. Bot. 1987, 27, 329–340. [Google Scholar] [CrossRef]
- Sasidharan, R.; Voesenek, L.A. Ethylene-mediated acclimations to flooding stress. Plant Physiol. 2015, 169, 3–12. [Google Scholar] [CrossRef]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef]
- Van Veen, H.; Mustroph, A.; Barding, G.A.; Vergeer-van Eijk, M.; Welschen-Evertman, R.A.; Pedersen, O.; Visser, E.J.; Larive, C.K.; Pierik, R.; Bailey-Serres, J. Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell 2013, 25, 4691–4707. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef]
- Loreti, E.; Valeri, M.C.; Novi, G.; Perata, P. Gene regulation and survival under hypoxia requires starch availability and metabolism. Plant Physiol. 2018, 176, 1286–1298. [Google Scholar] [CrossRef]
- Quimio, C.A.; Torrizo, L.B.; Setter, T.L.; Ellis, M.; Grover, A.; Abrigo, E.M.; Oliva, N.P.; Ella, E.S.; Carpena, A.L.; Ito, O. Enhancement of submergence tolerance in transgenic rice overproducing pyruvate decarboxylase. J. Plant Physiol. 2000, 156, 516–521. [Google Scholar] [CrossRef]
- Tougou, M.; Hashiguchi, A.; Yukawa, K.; Nanjo, Y.; Hiraga, S.; Nakamura, T.; Nishizawa, K.; Komatsu, S. Responses to flooding stress in soybean seedlings with the alcohol dehydrogenase transgene. Plant Biotechnol. 2012, 29, 301–305. [Google Scholar] [CrossRef]
- Komatsu, S.; Thibaut, D.; Hiraga, S.; Kato, M.; Chiba, M.; Hashiguchi, A.; Tougou, M.; Shimamura, S.; Yasue, H. Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol. Biol. 2011, 77, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Xuewen, X.; Huihui, W.; Xiaohua, Q.; Qiang, X.; Xuehao, C. Waterlogging-induced increase in fermentation and related gene expression in the root of cucumber (Cucumis sativus L.). Sci. Hortic. 2014, 179, 388–395. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, X.; Yang, G.; Li, Z.; Lu, H.; Kong, X.; Eneji, A.E.; Dong, H. Physiological and molecular adjustment of cotton to waterlogging at peak-flowering in relation to growth and yield. Field Crops Res. 2015, 179, 164–172. [Google Scholar] [CrossRef]
- Zhang, P.; Lyu, D.; Jia, L.; He, J.; Qin, S. Physiological and de novo transcriptome analysis of the fermentation mechanism of Cerasus sachalinensis roots in response to short-term waterlogging. BMC Genom. 2017, 18, 649. [Google Scholar] [CrossRef]
- Huang, X.; Yan, H.-D.; Zhang, X.-Q.; Zhang, J.; Frazier, T.P.; Huang, D.-J.; Lu, L.; Huang, L.-K.; Liu, W.; Peng, Y. De novo transcriptome analysis and molecular marker development of two Hemarthria species. Front. Plant Sci. 2016, 7, 496. [Google Scholar] [CrossRef]
- Huang, C.-J.; Chu, F.-H.; Huang, Y.-S.; Tu, Y.-C.; Hung, Y.-M.; Tseng, Y.-H.; Pu, C.-E.; Hsu, C.T.; Chao, C.-H.; Chou, Y.-S. SSR individual identification system construction and population genetics analysis for Chamaecyparis formosensis. Sci. Rep. 2022, 12, 4126. [Google Scholar] [CrossRef]
- Vatanparast, M.; Shetty, P.; Chopra, R.; Doyle, J.J.; Sathyanarayana, N.; Egan, A.N. Transcriptome sequencing and marker development in winged bean (Psophocarpus tetragonolobus; Leguminosae). Sci. Rep. 2016, 6, 29070. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, S.; Hong, Y.; Song, X. Application of genomic SSR locus polymorphisms on the identification and classification of chrysanthemum cultivars in China. PLoS ONE 2014, 9, e104856. [Google Scholar] [CrossRef]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef]
- Wu, Q.; Zang, F.; Xie, X.; Ma, Y.; Zheng, Y.; Zang, D. Full-length transcriptome sequencing analysis and development of EST-SSR markers for the endangered species Populus wulianensis . Sci. Rep. 2020, 10, 16249. [Google Scholar] [CrossRef]
- Yan, L.-P.; Liu, C.-L.; Wu, D.-J.; Li, L.; Shu, J.; Sun, C.; Xia, Y.; Zhao, L.-J. De novo transcriptome analysis of Fraxinus velutina using Illumina platform and development of EST-SSR markers. Biol. Plant. 2017, 61, 210–218. [Google Scholar] [CrossRef]
- Saeed, B.; Baranwal, V.K.; Khurana, P. Comparative transcriptomics and comprehensive marker resource development in mulberry. BMC Genom. 2016, 17, 98. [Google Scholar] [CrossRef]
- Wang, H.; Tong, W.; Feng, L.; Jiao, Q.; Long, L.; Fang, R.; Zhao, W. De novo transcriptome analysis of mulberry (Morus L.) under drought stress using RNA-Seq technology. Russ. J. Bioorganic Chem. 2014, 40, 423–432. [Google Scholar] [CrossRef]
- Das, D.; Bhattacharyya, S.; Bhattacharyya, M.; Sashankar, P.; Ghosh, A.; Mandal, P. Transcriptome analysis of mulberry (Morus alba L.) leaves to identify differentially expressed genes associated with post-harvest shelf-life elongation. Sci. Rep. 2022, 12, 18195. [Google Scholar] [CrossRef]
- Park, S.; Son, S.; Shin, M.; Fujii, N.; Hoshino, T.; Park, S. Transcriptome-wide mining, characterization, and development of microsatellite markers in Lychnis kiusiana (Caryophyllaceae). BMC Plant Biol. 2019, 19, 14. [Google Scholar] [CrossRef]
- Singh, R.; Mahato, A.K.; Singh, A.; Kumar, R.; Singh, A.K.; Kumar, S.; Marla, S.S.; Kumar, A.; Singh, N.K. TinoTranscriptDB: A Database of Transcripts and Microsatellite Markers of Tinospora cordifolia, an Important Medicinal Plant. Genes 2022, 13, 1433. [Google Scholar] [CrossRef]



| Sample | Raw Bases | Clean Reads | Q30 (%) | GC Content | Mapped Reads | Mapping Rate |
|---|---|---|---|---|---|---|
| Y711_0A | 6.79G | 43,491,174 | 90.90 | 46.73 | 32,773,537 | 75.36% |
| Y711_0B | 6.90G | 44,134,582 | 90.97 | 46.34 | 33,061,793 | 74.91% |
| Y711_0C | 6.87G | 43,715,556 | 90.76 | 46.35 | 32,528,709 | 74.41% |
| Y711_1A | 6.80G | 43,524,524 | 90.84 | 46.57 | 32,618,756 | 74.94% |
| Y711_1B | 6.49G | 41,667,112 | 90.43 | 46.85 | 31,207,395 | 74.90% |
| Y711_1C | 6.80G | 43,708,984 | 90.94 | 46.71 | 32,294,751 | 73.89% |
| Y711_3A | 7.01G | 44,921,118 | 91.10 | 45.97 | 32,755,910 | 72.92% |
| Y711_3B | 6.18G | 39,817,062 | 90.51 | 46.00 | 28,896,894 | 72.57% |
| Y711_3C | 6.85G | 44,029,322 | 90.81 | 45.91 | 32,117,560 | 72.95% |
| Y711_7A | 6.89G | 44,784,490 | 90.62 | 45.86 | 33,199,121 | 74.13% |
| Y711_7B | 6.64G | 43,138,614 | 91.05 | 45.82 | 31,792,726 | 73.70% |
| Y711_7C | 6.90G | 44,404,530 | 91.17 | 45.79 | 32,669,513 | 73.57% |
| TF Family | FS1 | FS3 | FS7 | |||
|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | |
| AP2 | 2 | 14 | 12 | 23 | 6 | 25 |
| bZIP | 3 | 1 | 2 | 8 | 4 | 4 |
| GRAS | 0 | 4 | 4 | 8 | 2 | 9 |
| bHLH | 6 | 2 | 3 | 17 | 3 | 13 |
| Homeobox | 3 | 3 | 6 | 7 | 6 | 7 |
| HSF | 0 | 3 | 3 | 6 | 1 | 4 |
| MYB | 8 | 12 | 10 | 28 | 6 | 23 |
| NAC | 1 | 4 | 7 | 6 | 5 | 11 |
| WRKY | 3 | 8 | 6 | 15 | 1 | 18 |
| MADS | 3 | 1 | 4 | 1 | 1 | 2 |
| YABBY | 0 | 0 | 0 | 2 | 0 | 2 |
| GATA | 0 | 1 | 0 | 6 | 0 | 5 |
| E2F | 0 | 0 | 0 | 3 | 0 | 2 |
| CO-like | 0 | 1 | 2 | 4 | 1 | 4 |
| NFYA | 0 | 2 | 0 | 1 | 0 | 0 |
| B3 | 0 | 3 | 0 | 5 | 0 | 5 |
| Dof | 0 | 0 | 2 | 2 | 3 | 1 |
| C3H | 1 | 1 | 1 | 3 | 0 | 1 |
| TCP | 1 | 0 | 1 | 1 | 1 | 1 |
| ARF | 0 | 0 | 1 | 0 | 1 | 0 |
| ZF-HD | 0 | 2 | 0 | 5 | 0 | 3 |
| Total | 31 | 62 | 64 | 151 | 41 | 140 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Duan, Y.; Yang, J.; Gan, L.; Chen, W.; Yang, J.; Xiao, G.; Guan, L.; Chen, J. Transcriptome Analysis Reveals Genes Associated with Flooding Tolerance in Mulberry Plants. Life 2023, 13, 1087. https://doi.org/10.3390/life13051087
Hu J, Duan Y, Yang J, Gan L, Chen W, Yang J, Xiao G, Guan L, Chen J. Transcriptome Analysis Reveals Genes Associated with Flooding Tolerance in Mulberry Plants. Life. 2023; 13(5):1087. https://doi.org/10.3390/life13051087
Chicago/Turabian StyleHu, Jingtao, Yanyan Duan, Junnian Yang, Liping Gan, Wenjing Chen, Jin Yang, Guosheng Xiao, Lingliang Guan, and Jingsheng Chen. 2023. "Transcriptome Analysis Reveals Genes Associated with Flooding Tolerance in Mulberry Plants" Life 13, no. 5: 1087. https://doi.org/10.3390/life13051087
APA StyleHu, J., Duan, Y., Yang, J., Gan, L., Chen, W., Yang, J., Xiao, G., Guan, L., & Chen, J. (2023). Transcriptome Analysis Reveals Genes Associated with Flooding Tolerance in Mulberry Plants. Life, 13(5), 1087. https://doi.org/10.3390/life13051087







