Presence of p25alpha-Domain in Seed Plants (Spermatophyta): Microbial/Animal Contaminations and/or Orthologs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Database Homology Search
2.2. Phylogenetic Analysis
3. Results
3.1. Database Homology Search for the p25alpha-Domain in Streptophyta
3.2. Search for Further Contaminations
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steinegger, M.; Salzberg, S.L. Terminating contamination: Large-scale search identifies more than 2,000,000 contaminated entries in GenBank. Genome Biol. 2020, 21, 115. [Google Scholar] [CrossRef] [PubMed]
- Jun, G.; Flickinger, M.; Hetrick, K.N.; Romm, J.M.; Doheny, K.F.; Abecasis, G.R.; Boehnke, M.; Kang, H.M. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012, 91, 839–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orosz, F. Two recently sequenced vertebrate genomes are contaminated with apicomplexan species of the Sarcocystidae family. Int. J. Parasitol. 2015, 45, 871–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurence, M.; Hatzis, C.; Brash, D.E. Common contaminants in nextgeneration sequencing that hinder discovery of low-abundance microbes. PLoS ONE 2014, 9, e97876. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Tan, B.; Zhang, Y.A. Large-scale study into protist-animal interactions based on public genomic data using DNA barcodes. Animals 2023, 13, 2243. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, G.; Pelosi, P. Plant transcriptomes reveal hidden guests. Biochem. Biophys. Res. Commun. 2016, 474, 497–502. [Google Scholar] [CrossRef]
- Reiter, T.; Brown, C.T. Microbial contamination in the genome of the domesticated olive. bioRxiv 2018, 499541. [Google Scholar] [CrossRef]
- Saffar, A.; Matin, M.M. Tracing foreign sequences in plant transcriptomes and genomes using OCT4, a POU domain protein. Mol. Genet. Genomics 2021, 296, 677–688. [Google Scholar] [CrossRef]
- Martín-Blázquez, R.; Bakkali, M.; Ruiz-Estévez, M.; Garrido-Ramos, M.A. Comparison between the gametophyte and the sporophyte transcriptomes of the endangered fern Vandenboschia speciosa. Genes 2023, 14, 166. [Google Scholar] [CrossRef]
- Orosz, F. A new protein superfamily: TPPP-like proteins. PLoS ONE 2012, 7, e49276. [Google Scholar] [CrossRef] [Green Version]
- Orosz, F.; Ovádi, J. TPPP orthologs are ciliary proteins. FEBS Lett. 2008, 582, 3757–3764. [Google Scholar] [CrossRef] [Green Version]
- Tammana, D.; Tammana, T.V.S. Chlamydomonas FAP265 is a tubulin polymerization promoting protein, essential for flagellar reassembly and hatching of daughter cells from the sporangium. PLoS ONE 2017, 12, e0185108. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, D.; Meng, Z.; Zhou, J.; Min, Z.; Deng, S.; Shen, J.; Liu, M. Pyp25α is required for male gametocyte exflagellation. Pathog. Dis. 2022, 80, ftac043. [Google Scholar] [CrossRef] [PubMed]
- Orosz, F. On the TPPP-like proteins of flagellated Fungi. Fung. Biol. 2021, 125, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Orosz, F. Truncated TPPP—An Endopterygota-specific protein. Heliyon 2021, 7, e07135. [Google Scholar] [CrossRef]
- Orosz, F. Wider than thought phylogenetic occurrence of apicortin, a characteristic protein of apicomplexan parasites. J. Mol. Evol. 2016, 82, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Orosz, F. p25alpha domain-containing proteins of apicomplexans and related taxa. Microorganisms 2023, 11, 1528. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixture models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theriault, G.; Michael, P.; Nkongolo, K. Comprehensive transcriptome analysis of responseto nickel stress in white birch (Betula papyrifera). PLoS ONE 2016, 11, e0153762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orosz, F. Tubulin Polymerization Promoting Proteins (TPPPs) of Aphelidiomycota: Correlation between the incidence of p25alpha domain and the eukaryotic flagellum. J. Fungi 2023, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, R.; Okamura, Y.; Haeger, W.; Vogel, H.; Kunert, G.; Pauchet, Y. Metabolic novelty originating from horizontal gene transfer is essential for leaf beetle survival. Proc. Natl. Acad. Sci. USA 2022, 119, e2205857119. [Google Scholar] [CrossRef]
- Orosz, F. Apicortin, a unique protein, with a putative cytoskeletal role, shared only by apicomplexan parasites and the placozoan Trichoplax adhaerens. Infect. Genet. Evol. 2009, 9, 1275–1286. [Google Scholar] [CrossRef]
- Orosz, F. Apicortin, a constituent of apicomplexan conoid/apical complex and its tentative role in pathogen—Host interaction. Trop. Med. Infect. Dis. 2021, 6, 118. [Google Scholar] [CrossRef]
- Ogura, Y. History of discovery of spermatozoids in Ginkgo biloba and Cycas revoluta. Phytomorphology 1967, 17, 109–114. [Google Scholar]
- Borner, J.; Burmester, T. Parasite infection of public databases: A data mining approach to identify apicomplexan contaminations in animal genome and transcriptome assemblies. BMC Genom. 2017, 18, 100. [Google Scholar] [CrossRef] [Green Version]
- Orosz, F. On the benefit of publishing uncurated genome assembly data. J. Bacteriol. Parasitol. 2017, 8, 4. [Google Scholar] [CrossRef]
- Lopes, R.J.; Mérida, A.M.; Carneiro, M. Unleashing the potential of public genomic resources to find parasite genetic data. Trends Parasitol. 2017, 33, 750–753. [Google Scholar] [CrossRef]

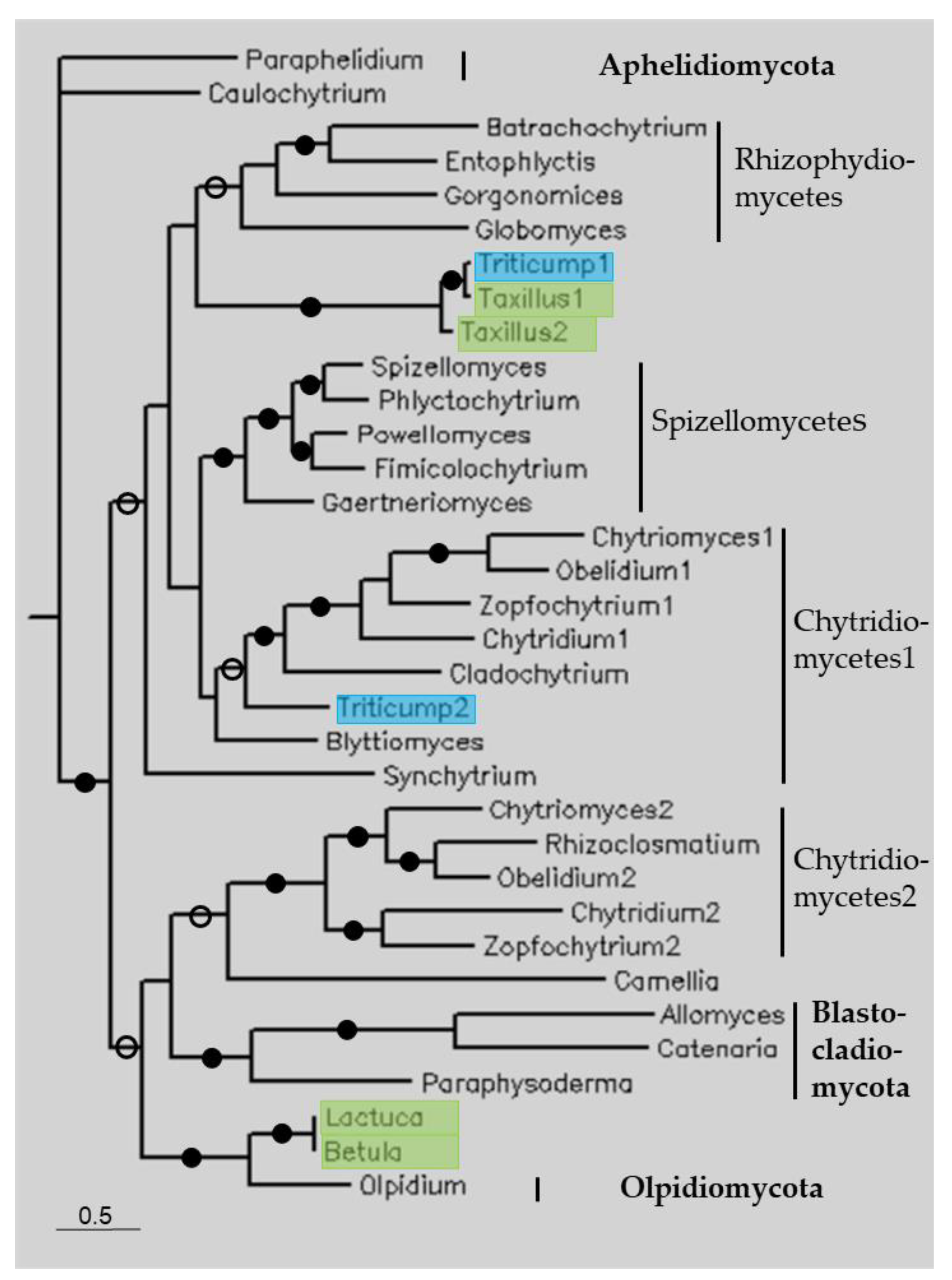



| Plant Species | Order | Accession No 1 | Species | Accession No | Cover % | Identity % |
|---|---|---|---|---|---|---|
| Fungal-type | ||||||
| Lactuca serriola | Asterales 2 | JO041594 | Spizellomyces punctatus 3 | XP_016604112 | 92 | 42.81 |
| Taxillus chinensis 1 | Santalales | GHNL01117630 | S. punctatus | XP_016604112 | 61 | 40.51 |
| T. chinensis 2 | Santalales | GHNL01117629 | S. punctatus | XP_016604112 | 89 | 38.01 |
| Betula papyrifera | Fagales | GEIC01019178 4 GEIC01019177 4 | Quaeritorhiza haematococci | KAJ3085108 | 76 82 | 49.74 |
| Triticum polonicum 1 | Poales | GEDP01099476 | S. punctatus | XP_016604112 | 60 | 40.51 |
| T. polonicum 2 | Poales | GEDP01150747 | Powellomyces hirtus | TPX57673 | 85 | 61.73 |
| Long | ||||||
| Humulus lupulus | Rosales | GAAW01037957 | Tetranychus urticae | XP_015786377 | 65 | 100 |
| Myosoton aquaticum | Caryophyllales | GGTY01056430 | Frankliniella occidentalis | XP_026285276 | 60 | 100 |
| Jasminum sambac | Lamiales | GHOY01138054 | Contarinia nasturtii | XP_031639744 | 51 | 92.02 |
| Cosmos caudatus | Asterales | GJBF01051822 | Adineta steineri | CAF1404786 | 91 | 79.43 |
| Zostera noltei | Alismatales | HACV01012836 | Helobdella robusta | XP_009008741 | 46 | 51.23 |
| Elodea nuttallii | Alismatales | GBEN01147374 | Bulinus truncatus | KAH9498372 | 71 | 88.24 |
| Pinus lambertiana | Pinales | GEUZ01024616 | Adineta vaga | UJR13967 | 99 | 82.22 |
| Oryza sativa | Poales | CT849204 * | Brachionus calyciflorus | CAF0835781 | 99 | 76.09 |
| Hordeum vulgare | Poales | BM815954 * | Adineta vaga | UJR13967 | 100 | 80.00 |
| Alnus glutinosa | Fagales | FQ350563 * | B. calyciflorus | CAF0835781 | 100 | 75.62 |
| Sesamum indicum | Lamiales | JK067166 *,5 JK062224 *,5 | Lucilia cuprina | XP_023301338 | 100 | 84.75 |
| Short | ||||||
| Panax ginseng 1 | Apiales | GDQW01019137 | Tetrahymena thermophila | XP_001023601 7 | 98 | 70.27 |
| P ginseng 2 | Apiales | GDQW01005616 | Trypanosoma brucei | XP_011772860 | 95 | 62.50 |
| B. papyrifera | Fagales | GEIC01017558 | Bodo saltans | CUE71550 | 90 | 73.57 |
| Nicotiana. tabacum | Solanales | AM817762 *,6 AM824543 *,6 | Coccomyxa sp. | BDA43246 | 100 | 47.24 |
| Colobanthus quitensis | Caryophyllales | GCIB01125581 | T. thermophila | XP_001023601 | 98 | 68.03 |
| Persicaria minor | Caryophyllales | GALN01112310 | T. thermopila | XP_001023599 | 79 | 58.62 |
| Chromolaena odorata | Asterales | GACH01135300 | Paramecium sonneborni | CAD8055868 | 100 | 60.34 |
| O. sativa | Poales | CT850609 * | T. thermophila | XP_001023601 | 92 | 58.11 |
| Triticum aestivum | Poales | CD868723 * | T. thermophila | XP_001023599 | 96 | 61.22 |
| Truncated | ||||||
| Cenostigma pyramidale | Fabales | GIYP01283228 | Anastrepha ludens | XP_053956472 | 100 | 98.29 |
| Apicortin | ||||||
| Camellia sinensis | Ericales | GFMV01019718 | Vitrella brassicaformis | CEM06711 | 44 | 41.83 |
| N. tabacum | Solanales | AM844195 * | Jimgerdemannia flammicorona | RUS30044 | 73 | 50.67 |
| Silene dioica | Caryophyllales | GFCH01066796 | J. flammicorona | RUS30044 | 100 | 48.57 |
| Salicornia europaea | Caryophyllales | GAMH01042109 | Rosella allomycis | EPZ32946 | 85 | 51.00 |
| T. polonicum | Poales | GEDP01156285 | Trichoplax adhaerens | XP_002111209 | 65 | 47.24 |
| Ginkgo biloba | Ginkgoales | GHLL01465948 | T. adhaerens | XP_002111209 | 78 | 54.49 |
| Pinus flexilis | Pinales | GHWB01415589 | J. flammicorona | RUS30044 | 91 | 48.78 |
| Accession Number | Best Hit | ||||
|---|---|---|---|---|---|
| Accession Number | Species | Phylum | Query Cover, % | Identity, % | |
| GEIC01019180 | KAH7131324 | Dactylonectria macrodidyma | Ascomycota | 72 | 86.84 |
| KFY33973 | Pseudogymnoascus sp. | Ascomycota | 73 | 61.84 | |
| GEIC01019179 | XP_015895121 | Ziziphus jujuba | Streptophyta | 95 | 78.67 |
| GEIC01019178 | KAJ3085108 | Quaeritorhiza haematococci | Chytridiomycota | 76 | 49.74 |
| KAJ3185965 | Gaertneriomyces sp. | Chytridiomycota | 99 | 42.13 | |
| GEIC01019177 | KAJ3085108 | Quaeritorhiza haematococci | Chytridiomycota | 82 | 49.74 |
| KAJ3031848 | Rhizophlyctis rosea | Chytridiomycota | 91 | 42.17 | |
| GEIC01019176 | XP_018131936 | Pseudogymnoascus verrucosus | Ascomycota | 99 | 100 |
| GEIC01019175 | KAF1315998 | Globisporangium splendens | Oomycota | 99 | 90.43 |
| GEIC01019174 | KIJ40333 | Sphaerobolus stellatus | Basidiomycota | 65 | 39.13 |
| GEIC01019173 | PNP46136 | Trichoderma gamsii | Ascomycota | 99 | 97.53 |
| GEIC01019172 | KAH7141718 | Dactylonectria macrodidyma | Ascomycota | 98 | 99.34 |
| GEIC01019171 | KAH7121618 | Dactylonectria macrodidyma | Ascomycota | 98 | 94.90 |
| GEIC01019170 | TFK80833 | Polyporus arcularius | Basidiomycota | 40 | 61.83 |
| XP_008038329 | Trametes versicolor | Basidiomycota | 47 | 54.25 | |
| GEIC01017560 | KAE8022277 | Carpinus fangiana | Streptophyta | 100 | 91.74 |
| GEIC01017559 | CUE71550 | Bodo saltans | Euglenozoa | 86 | 75.56 |
| EPY25997 | Angomonas deanei | Euglenozoa | 94 | 63.27 | |
| GEIC01017558 | CUE71550 | Bodo saltans | Euglenozoa | 90 | 73.57 |
| GEIC01017557 | ELT89137 | Capitella teleta | Annelida | 87 | 53.04 |
| GEIC01017556 | KAH7144037 | Dactylonectria macrodidyma | Ascomycota | 99 | 98.57 |
| GEIC01017555 | XP_022516040 | Fonsecaea monophora | Ascomycota | 22 | 100 |
| GEIC01017554 | XP_022516040 | Fonsecaea monophora | Ascomycota | 27 | 100 |
| GEIC01017553 | - | - | - | - | - |
| GEIC01017552 | - | - | - | - | - |
| GEIC01017551 | TYZ60970 | Pythium brassicae | Oomycota | 99 | 74.36 |
| GEIC01017550 | TYZ60970 | Pythium brassicae | Oomycota | 81 | 75.76 |
| KAG7389301 | Phytophthora pseudosyringae | Oomycota | 83 | 62.69 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orosz, F. Presence of p25alpha-Domain in Seed Plants (Spermatophyta): Microbial/Animal Contaminations and/or Orthologs. Life 2023, 13, 1664. https://doi.org/10.3390/life13081664
Orosz F. Presence of p25alpha-Domain in Seed Plants (Spermatophyta): Microbial/Animal Contaminations and/or Orthologs. Life. 2023; 13(8):1664. https://doi.org/10.3390/life13081664
Chicago/Turabian StyleOrosz, Ferenc. 2023. "Presence of p25alpha-Domain in Seed Plants (Spermatophyta): Microbial/Animal Contaminations and/or Orthologs" Life 13, no. 8: 1664. https://doi.org/10.3390/life13081664
APA StyleOrosz, F. (2023). Presence of p25alpha-Domain in Seed Plants (Spermatophyta): Microbial/Animal Contaminations and/or Orthologs. Life, 13(8), 1664. https://doi.org/10.3390/life13081664






