Narrowing the Relationship between Human CCR5 Gene Polymorphisms and Chagas Disease: Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategies
2.2. Term Selection, DeCS Analysis and String Construction
2.3. Data Base
2.4. Inclusion and Exclusion Criteria
2.5. Selection of Publications and Quality Analysis
2.6. Data Extraction
2.7. Meta-Analysis
2.8. SNP2TFBS
2.9. Patient Classification
2.10. Statistical Analysis
3. Results
3.1. Characterization of the Studies
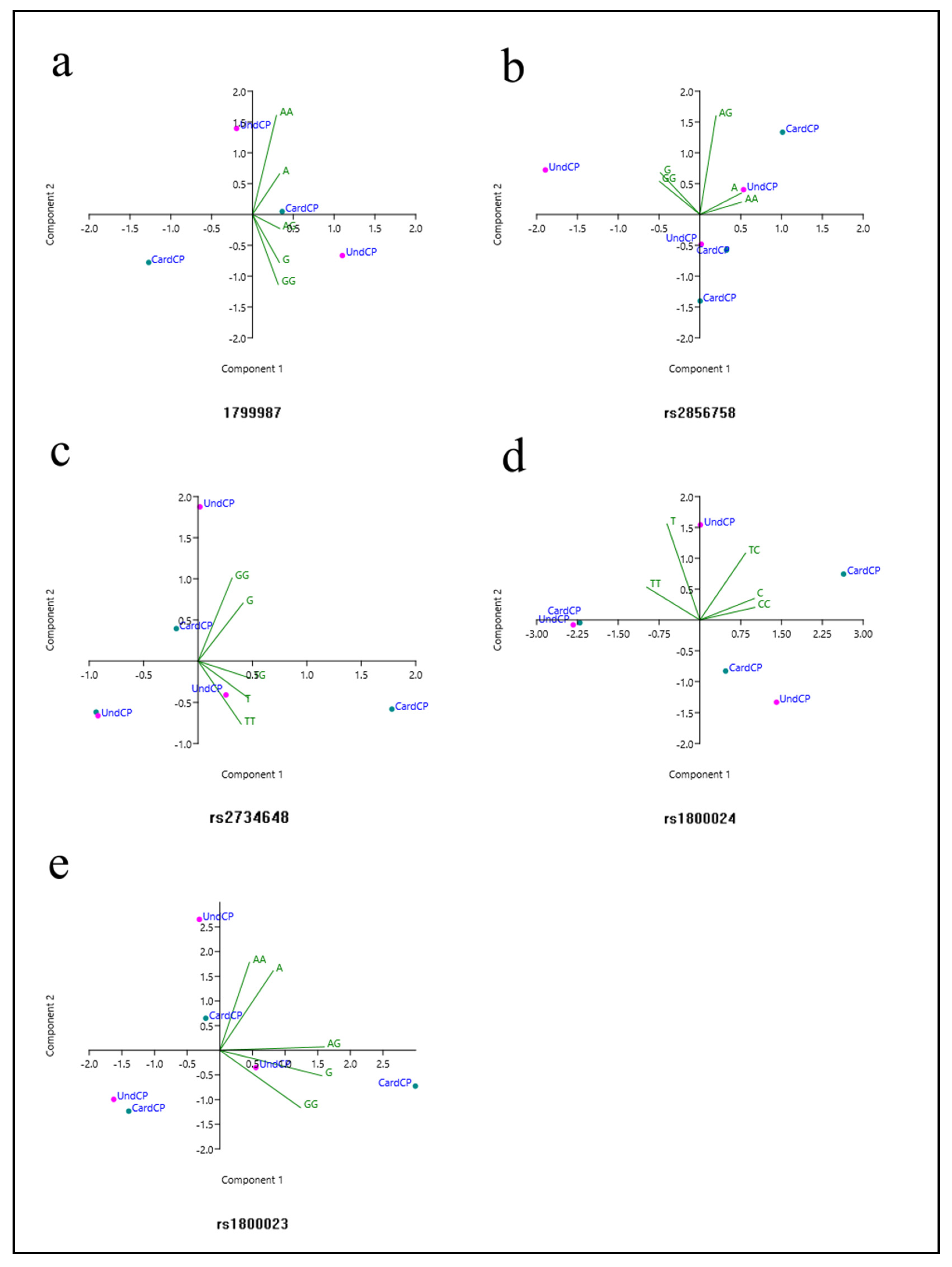
3.2. Meta-Analysis and PCA
3.2.1. rs1799987
3.2.2. rs333
3.2.3. rs2856758
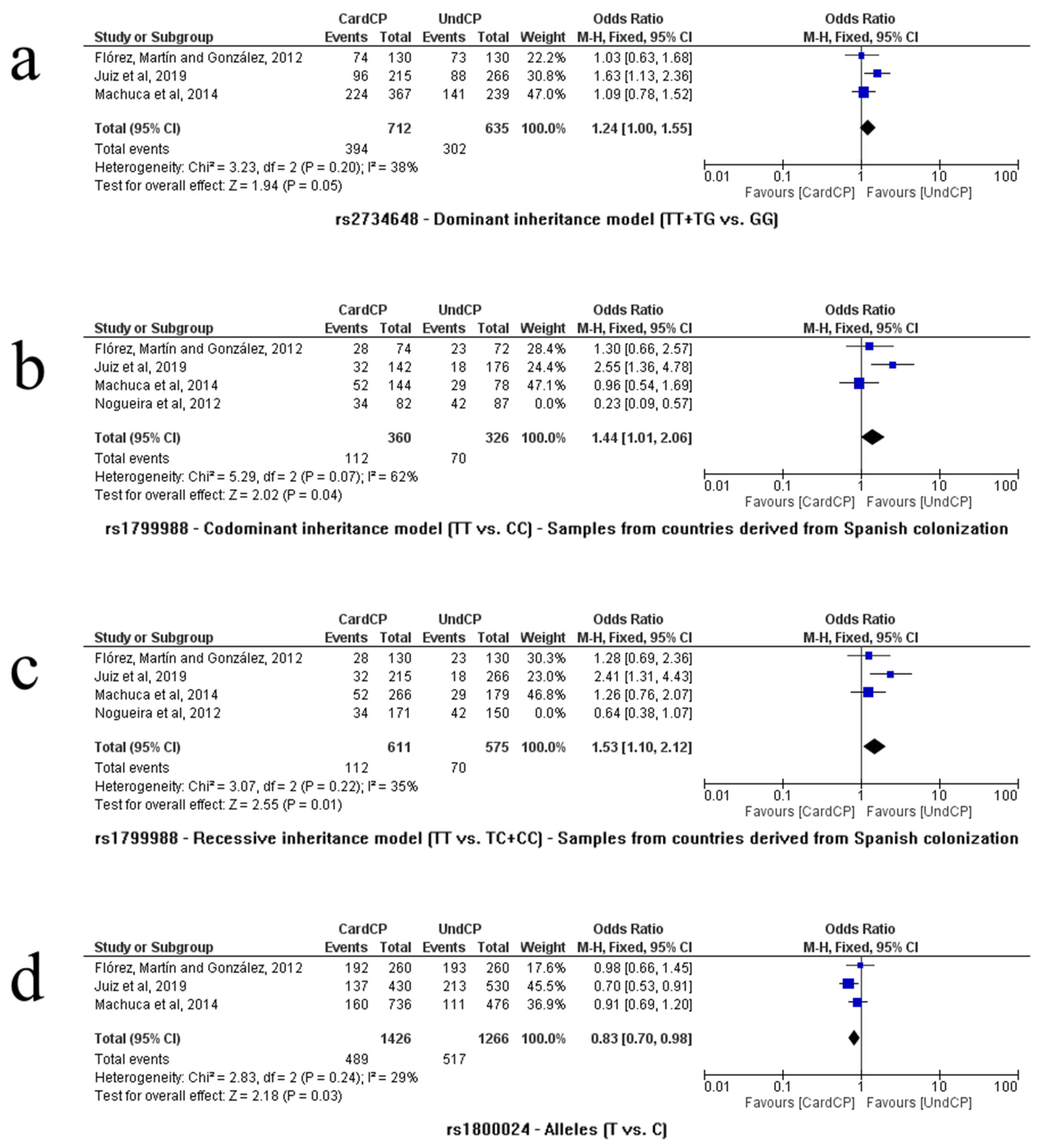
3.2.4. rs2734648
3.2.5. rs1799988
3.2.6. rs41469351
3.2.7. rs1800024
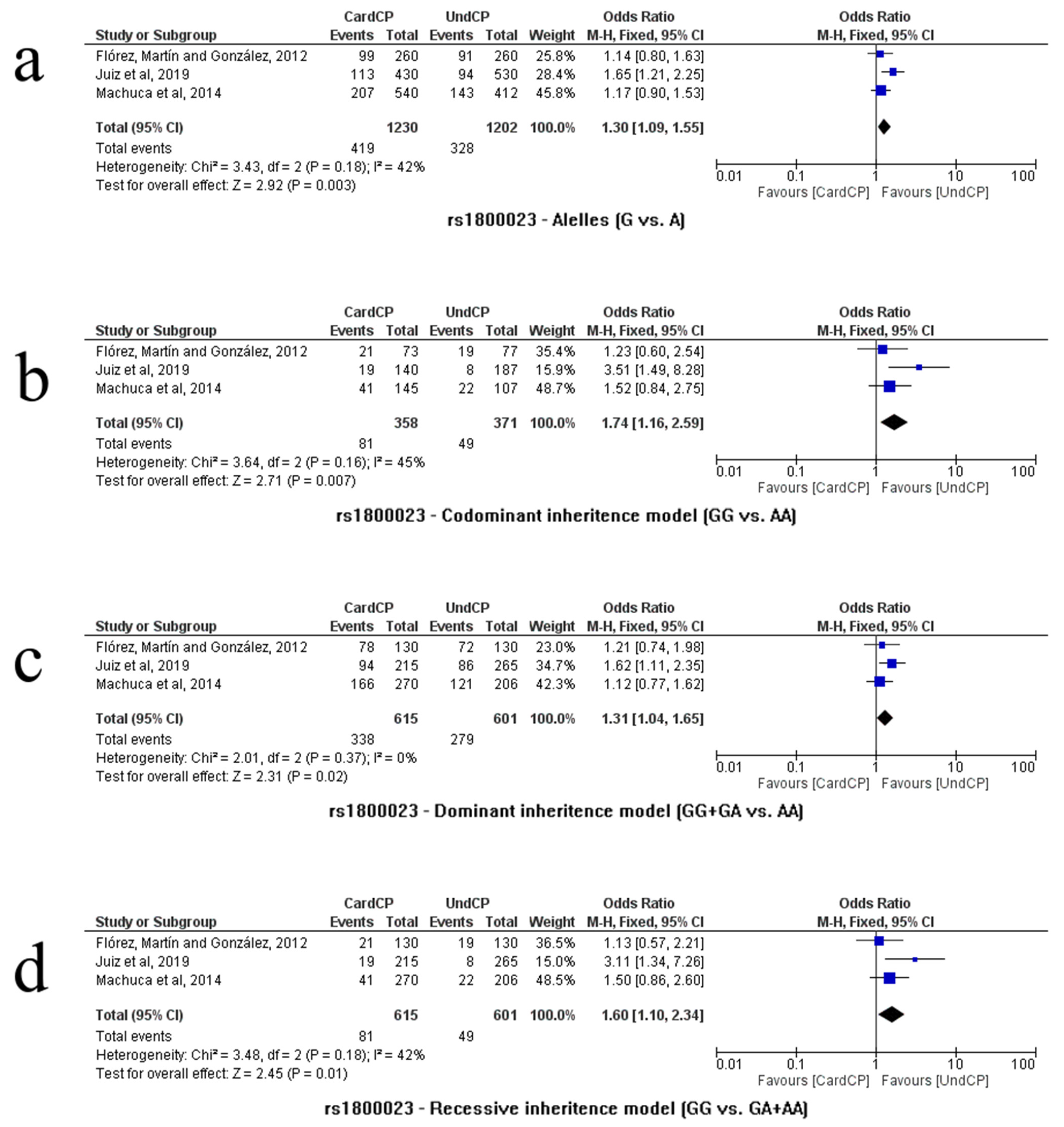
3.2.8. rs1800023
3.3. Publication Biases and Heterogeneity
3.4. SNP2TFBS
3.5. Correlation between HHC Haplotype, Alleles and CD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization—WHO. Chagas Disease (American Trypanosomiasis). Available online: https://www.who.int/health-topics/chagas-disease#tab=tab_1 (accessed on 20 June 2022).
- Pérez-Molina, J.A.; Molina, I. Chagas Disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Suárez, C.; Nolder, D.; García-Mingo, A.; Moore, D.A.J.; Chiodini, P.L. Diagnosis and Clinical Management of Chagas Disease: An Increasing Challenge in Non-Endemic Areas. Res. Rep. Trop. Med. 2022, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Herrera, M.; Strauss, M.; Casares-Marfil, D.; Martín, J.; Network, C.G.C. Genomic Medicine in Chagas Disease. Acta Trop. 2019, 197, 105062. [Google Scholar] [CrossRef] [PubMed]
- Gomes dos Santos, A.; Watanabe, E.H.; Ferreira, D.T.; Oliveira, J.; Nakanishi, É.S.; Oliveira, C.S.; Bocchi, E.; Novaes, C.T.G.; Cruz, F.; Carvalho, N.B. A Specific IL6 Polymorphic Genotype Modulates the Risk of Trypanosoma Cruzi Parasitemia While IL18, IL17A, and IL1B Variant Profiles and HIV Infection Protect against Cardiomyopathy in Chagas Disease. Front. Immunol. 2020, 11, 521409. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, C.; Nunes, J.P.S.; Frade, A.F.; Almeida, R.R.; Pandey, R.P.; Nascimento, M.S.; Kalil, J.; Cunha-Neto, E. Disease Tolerance and Pathogen Resistance Genes May Underlie Trypanosoma Cruzi Persistence and Differential Progression to Chagas Disease Cardiomyopathy. Front. Immunol. 2018, 9, 2791. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Fang, Q.; Liu, S.; Hou, W.; Li, J.; Huang, Y.; Shi, J. Advances of CCR5 Antagonists: From Small Molecules to Macromolecules. Eur. J. Med. Chem. 2020, 208, 112819. [Google Scholar] [CrossRef]
- Batista, A.M.; Alvarado-Arnez, L.E.; Alves, S.M.; Melo, G.; Pereira, I.R.; de Souza Ruivo, L.A.; Da Silva, A.A.; Gibaldi, D.; do E S Protásio da Silva, T.; De Lorena, V.M.B. Genetic Polymorphism at CCL5 Is Associated with Protection in Chagas’ Heart Disease: Antagonistic Participation of CCR1+ and CCR5+ Cells in Chronic Chagasic Cardiomyopathy. Front. Immunol. 2018, 9, 615. [Google Scholar] [CrossRef]
- Calzada, J.E.; Nieto, A.; Beraun, Y.; Martin, J. Chemokine Receptor CCR5 Polymorphisms and Chagas’ Disease Cardiomyopathy. Tissue Antigens 2001, 58, 154–158. [Google Scholar] [CrossRef]
- Flórez, O.; Martín, J.; González, C.I. Genetic Variants in the Chemokines and Chemokine Receptors in Chagas Disease. Hum. Immunol. 2012, 73, 852–858. [Google Scholar] [CrossRef]
- Frade, A.F.; Pissetti, C.W.; Ianni, B.M.; Saba, B.; Lin-Wang, H.T.; Nogueira, L.G.; de Melo Borges, A.; Buck, P.; Dias, F.; Baron, M. Genetic Susceptibility to Chagas Disease Cardiomyopathy: Involvement of Several Genes of the Innate Immunity and Chemokine-Dependent Migration Pathways. BMC Infect. Dis. 2013, 13, 587. [Google Scholar] [CrossRef]
- Lima, A.P.B.; de Oliveira, M.T.; Silva, R.R.; Torres, R.M.; Veloso, V.M.; de Lana, M.; da Silva, G.N. Evaluation of Parasite and Host Genetics in Two Generations of a Family with Chagas Disease. Parasitol. Res. 2018, 117, 3009–3013. [Google Scholar] [CrossRef]
- de Oliveira, A.P.; Bernardo, C.R.; da Silveira Camargo, A.V.; da Ronchi, L.S.; Borim, A.A.; Brandao de Mattos, C.C.; de Campos Junior, E.; Castiglioni, L.; Netinho, J.G.; Cavasini, C.E. Genetic Susceptibility to Cardiac and Digestive Clinical Forms of Chronic Chagas Disease: Involvement of the CCR5 59029 A/G Polymorphism. PLoS ONE 2015, 10, e0141847. [Google Scholar] [CrossRef]
- Brochet, P.; Ianni, B.; Laugier, L.; Frade, A.; Teixeira, P.; Mady, C.; Ferreira, L.; Ferré, Q.; Santos, R.; Kuramoto, A. Epigenetic Regulation of Transcription Factor Binding Motifs Promotes Th1 Response in Chagas Disease Cardiomyopathy. Front. Immunol. 2022, 13, 4536. [Google Scholar] [CrossRef] [PubMed]
- Laugier, L.; Ferreira, L.R.P.; Ferreira, F.M.; Cabantous, S.; Frade, A.F.; Nunes, J.P.; Ribeiro, R.A.; Brochet, P.; Teixeira, P.C.; Santos, R.H.B. MiRNAs May Play a Major Role in the Control of Gene Expression in Key Pathobiological Processes in Chagas Disease Cardiomyopathy. PLoS Negl. Trop. Dis. 2020, 14, e0008889. [Google Scholar] [CrossRef]
- Ferreira, L.R.P.; Ferreira, F.M.; Laugier, L.; Cabantous, S.; Navarro, I.C.; da Silva Cândido, D.; Rigaud, V.C.; Real, J.M.; Pereira, G.V.; Pereira, I.R. Integration of MiRNA and Gene Expression Profiles Suggest a Role for MiRNAs in the Pathobiological Processes of Acute Trypanosoma Cruzi Infection. Sci. Rep. 2017, 7, 17990. [Google Scholar] [CrossRef]
- Santos, C.M.d.C.; Pimenta, C.A.d.M.; Nobre, M.R.C. The PICO Strategy for the Research Question Construction and Evidence Search. Rev. Lat. Am. Enferm. 2007, 15, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization—PAHO. Chagas Disease—PAHO/WHO|Pan American Health Organization. Available online: https://www.paho.org/en/topics/chagas-disease (accessed on 20 June 2022).
- Bern, C. Chagas’ Disease. N. Engl. J. Med. 2015, 373, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring Inconsistency in Meta-Analyses. BMJ Br. Med. J. 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Wijarnpreecha, K.; Werlang, M.; Panjawatanan, P.; Kroner, P.T.; Cheungpasitporn, W.; Lukens, F.J.; Pungpapong, S.; Ungprasert, P. Association between Sarcopenia and Hepatic Encephalopathy: A Systematic Review and Meta-Analysis. Ann. Hepatol. 2020, 19, 245–250. [Google Scholar] [CrossRef]
- Erdfelder, E. Statistical Power Analyses Using G * Power 3.1: Tests for Correlation and Regression Analyses. Behav. Res. Methods 2009, 41, 1149–1160. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A Flexible Statistical Power Analysis Program for the Social, Behavioral, and Biomedical Sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Oliveira, A.P.; Bernardo, C.R.; Camargo, A.V.S.; Villafanha, D.F.; Cavasini, C.E.; de Mattos, C.C.B.; de Godoy, M.F.; Bestetti, R.B.; de Mattos, L.C. CCR5 Chemokine Receptor Gene Variants in Chronic Chagas’ Disease. Int. J. Cardiol. 2014, 176, 520–522. [Google Scholar] [CrossRef]
- Nogueira, L.G.; Santos, R.H.B.; Ianni, B.M.; Fiorelli, A.I.; Mairena, E.C.; Benvenuti, L.A.; Frade, A.; Donadi, E.; Dias, F.; Saba, B. Myocardial Chemokine Expression and Intensity of Myocarditis in Chagas Cardiomyopathy Are Controlled by Polymorphisms in CXCL9 and CXCL10. PLoS Negl. Trop. Dis. 2012, 6, e1867. [Google Scholar] [CrossRef]
- Juiz, N.A.; Estupiñán, E.; Hernández, D.; Garcilazo, A.; Chadi, R.; Morales Sanfurgo, G.; Schijman, A.G.; Longhi, S.A.; González, C.I. Association Study between CCR2-CCR5 Genes Polymorphisms and Chronic Chagas Heart Disease in Wichi and in Admixed Populations from Argentina. PLoS Negl. Trop. Dis. 2019, 13, e0007033. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Mestre, M.T.; Montagnani, S.; Layrisse, Z. Is the CCR5-59029-G/G Genotype a Protective Factor for Cardiomyopathy in Chagas Disease? Hum. Immunol. 2004, 65, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Machuca, M.A.; Suárez, E.U.; Echeverría, L.E.; Martín, J.; González, C.I. SNP/Haplotype Associations of CCR2 and CCR5 Genes with Severity of Chagasic Cardiomyopathy. Hum. Immunol. 2014, 75, 1210–1215. [Google Scholar] [CrossRef]
- Aldinucci, D.; Borghese, C.; Casagrande, N. The CCL5/CCR5 Axis in Cancer Progression. Cancers 2020, 12, 1765. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; Von Stebut, E. Macrophage Inflammatory Protein-1. Int. J. Biochem. Cell Biol. 2004, 36, 1882–1886. [Google Scholar] [CrossRef] [PubMed]
- Donninelli, G.; Studer, V.; Brambilla, L.; Zecca, C.; Peluso, D.; Laroni, A.; Michelis, D.; Mantegazza, R.; Confalonieri, P.; Volpe, E. Immune Soluble Factors in the Cerebrospinal Fluid of Progressive Multiple Sclerosis Patients Segregate into Two Groups. Front. Immunol. 2021, 12, 633167. [Google Scholar] [CrossRef]
- Patterson, B.K.; Seethamraju, H.; Dhody, K.; Corley, M.J.; Kazempour, K.; Lalezari, J.; Pang, A.P.S.; Sugai, C.; Francisco, E.B.; Pise, A. Disruption of the CCL5/RANTES-CCR5 Pathway Restores Immune Homeostasis and Reduces Plasma Viral Load in Critical COVID-19. MedRxiv 2020. [Google Scholar] [CrossRef]
- Zeng, Z.; Lan, T.; Wei, Y.; Wei, X. CCL5/CCR5 Axis in Human Diseases and Related Treatments. Genes Dis. 2021, 9, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.J.; Hayward, J.A.; Huang, C.E.; Huma, Z.; Sanchez, J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int. J. Mol. Sci. 2017, 18, 342. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, P.; Jaimes, J.; Poveda, C.; Ramírez, J.D. A Systematic Review of the Trypanosoma Cruzi Genetic Heterogeneity, Host Immune Response and Genetic Factors as Plausible Drivers of Chronic Chagasic Cardiomyopathy. Parasitology 2019, 146, 269–283. [Google Scholar] [CrossRef]
- Cunha-Neto, E.; Nogueira, L.G.; Teixeira, P.C.; Ramasawmy, R.; Drigo, S.A.; Goldberg, A.C.; Fonseca, S.G.; Bilate, A.M.; Kalil, J. Immunological and Non-Immunological Effects of Cytokines and Chemokines in the Pathogenesis of Chronic Chagas Disease Cardiomyopathy. Mem. Inst. Oswaldo Cruz 2009, 104, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.-A. Human Genetic Susceptibility to Chagas Disease. In American Trypanosomiasis Chagas Disease; Elsevier: Amsterdam, The Netherlands, 2017; pp. 629–652. [Google Scholar]
- de Oliveira, A.P.; Ayo, C.M.; Bestetti, R.B.; de Mattos, C.C.B.; Cavasini, C.E.; de Mattos, L.C. The Role of CCR5 in Chagas Disease-a Systematic Review. Infect. Genet. Evol. 2016, 45, 132–137. [Google Scholar] [CrossRef]
- Rodrigues de Moura, R.; Coelho, A.V.C.; de Queiroz Balbino, V.; Crovella, S.; Brandão, L.A.C. Meta-analysis of Brazilian Genetic Admixture and Comparison with Other Latin America Countries. Am. J. Hum. Biol. 2015, 27, 674–680. [Google Scholar] [CrossRef]
- Du, B.; Gao, W.; Qin, Y.; Zhong, J.; Zhang, Z. Study on the Role of Transcription Factor SPI1 in the Development of Glioma. Chin. Neurosurg. J. 2022, 8, 7. [Google Scholar] [CrossRef]
- Mummidi, S.; Bamshad, M.; Ahuja, S.S.; Gonzalez, E.; Feuillet, P.M.; Begum, K.; Galvis, M.C.; Kostecki, V.; Valente, A.J.; Murthy, K.K. Evolution of Human and Non-Human Primate CC Chemokine Receptor 5 Gene and mRNA: Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcripition factor binding to polymorphic nucleotides. J. Biol. Chem. 2000, 275, 18946–18961. [Google Scholar] [CrossRef]
- Mummidi, S.; Ahuja, S.S.; McDaniel, B.L.; Ahuja, S.K. The Human CC Chemokine Receptor 5 (CCR5) Gene: Multiple Transcripts with 5′-End Heterogeneity, Dual Promoter Usage, and Evidence for Polymorphisms within the Regulatory Regions and Noncoding Exons. J. Biol. Chem. 1997, 272, 30662–30671. [Google Scholar] [CrossRef]
- De Bona, E.; Lidani, K.C.F.; Bavia, L.; Omidian, Z.; Gremski, L.H.; Sandri, T.L.; de Messias Reason, I.J. Autoimmunity in Chronic Chagas Disease: A Road of Multiple Pathways to Cardiomyopathy? Front. Immunol. 2018, 9, 1842. [Google Scholar] [CrossRef]
- D’Ávila, D.A.; Macedo, A.M.; Valadares, H.M.S.; Gontijo, E.D.; de Castro, A.M.; Machado, C.R.; Chiari, E.; Galvão, L.M.C. Probing Population Dynamics of Trypanosoma Cruzi during Progression of the Chronic Phase in Chagasic Patients. J. Clin. Microbiol. 2009, 47, 1718–1725. [Google Scholar] [CrossRef] [PubMed]



| Study Characterization | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| References | Study Country 1 | Study Design | Population/Sample (n) | Group Characterization 2 | STREGA Score | Confirmation Method of T. cruzi Infection | Confirmation Method of Polymorphism | Parasitemia Evaluation | Hardy-Weinberg Equilibrium (HWE) 3 |
| Oliveira et al., 2014 [24] | Brazil | Cross-sectional | Brazilians/n = 168 | CP = 168 (CardCP = 168) | 8—Low quality | ELISA | PCR-RFLP | No | Nd |
| Frade et al., 2013 [11] | Brazil | Cohort | Brazilians/n = 433 | CP = 433 (CardCP = 315 and UndCP = 118) | 13—Medium quality | ELISA, IHA and IIF | Hybridization assay and qPCR | No | Yes 4 |
| Nogueira et al., 2012 [25] | Brazil | Cohort | Brazilians/n = 321 | CP = 22 (CardCP = 171) and UndCP = 150 | 12—Medium quality | ELISA, IHA and IIF | qPCR | No | Yes |
| Juiz et al., 2019 [26] | Argentina | Case-control | Argentinians/n = 480 | CP = 480 (CardCP= 215 and UndCP = 256) | 14—Medium quality | Serologically | qPCR | No | Yes 5 |
| Calzada et al., 2001 [9] | Spain | Case-control | Peruvians/n = 172 | CP = 172 (UndCP = 53; CardCP = 32) and nonCP = 87 | 14—Medium quality | ELISA, IHA and IIF | PCR-RFLP | No | Yes |
| Lima et al., 2018 [12] | Brazil | Observational study—case series | Brazilians/n = 6 | CP = 6 (UndCP = 1 and CardCP = 5) | 10—Medium quality | ELISA, IHA and PCR | PCR and PCR-RFLP | No | Nd |
| Batista et al., 2018 [8] | Brazil | Cross-sectional | Brazilians/n = 406 | CP = 406 (CardCP = 296 and UndCP = 110) | 17—High quality | ELISA and IIF | qPCR | Yes | Nd |
| De Oliveira et al., 2015 [13] | Brazil | Case-control | Brazilians/n = 412 | CP = 240 (CardCP = 121 and DigCP = 98) and nonCP = 172 | 15—Medium quality | ELISA | PCR and PCR-RFLP | No | Yes |
| Flórez; Martín; González, 2012 [10] | Colombia | Case-control | Colombians/n = 260 | CP = 260 (UndCp = 130 and CardCP = 130) | 16—Medium quality | ELISA and IHA | PCR and PCR-RFLP | No | Yes |
| Fernandez-Mestre; Montagnani; Layrisse, 2004 [27] | Venezuela | Cross-sectional | Venezuelans n = 107 | CP = 107 (UndCP = 34 and CardCP = 73) | 11—Medium quality | ELISA, IHA and IIF | PCR-RFLP | No | Nd |
| Machuca et al., 2014 [28] | Colombia | Cohort | Colombians/n = 476 | CP = 476 (UndCP = 206 and CardCP = 270) | 13—Medium quality | ELISA and IHA | qPCR | No | Yes |
| CCR5 Polymorphisms | |||||||||
| Polymorphism | Mutated Nucleotide Localization/Variation Type [and Change]/Consequence 6 | Main Finds of Studies | Quantitative Synthesis by Meta-Analysis (Yes/No) | ||||||
| rs2856758 | 46370170/SNV [A > G]/intron variant, 5 prime UTR variant | Flórez et al., 2012 [10]: G allele increased in asymptomatic group in a comparison with the cardiomyopathy group, indicating the cardiomyopathy risk reduction (p = 0.021, OR = 0.51, 95% CI (0.29–0.91)). Juiz et al., 2019 [26]: not associated. Machuca et al., 2014 [28]: not associated. | Yes | ||||||
| rs2734648 | 46370349/SNV [G > C/G > T]/intron variant | Flórez et al., 2012 [10]: T allele associated with reduced risk of cardiomyopathy progression compared to symptomatic subgroups (group II vs. group III, p = 0.018, OR = 0.44, 95% CI (0.22–0.88)/and group III vs. group IV, p = 0.004, OR = 0.29, 95% CI (0.12–0.68)). Juiz et al., 2019 [26]: not associated. Machuca et al., 2014 [28]: not associated. | Yes | ||||||
| rs1799987 | 46370444/SNV [A > G]/intron variant | Batista et al., 2018 [8]: not associated. Calzada et al., 2001 [9]: G/A increased in asymptomatic group in a comparison with the cardiomyopathy group (p = 0.02, OR = 0.33, 95% CI (0.10–0.94)); G allele increased in asymptomatic group in a comparison with the cardiomyopathy group (p = 0.02, OR = 0.35, 95% CI (0.12–0.96)). Fernández-Mestre et al., 2004 [27]: not associated. Flórez et al., 2012 [10]: not associated. Juiz et al., 2019 [26]: not associated. Lima et al., 2018 [12]: not associated. Oliveira et al., 2015 [13]: A/A frequency was different among patients with digestive and cardiac forms, and health controls (p = 0.036, χ2 = 6.656 (DF = 2)); A/A frequency was different between patients with digestive and cardiac forms (p = 0.013, χ2 = 6.129 (DF = 1)); A/A frequency was different between patients with cardiac form and health controls (p = 0.077, χ2 = 3.128 (DF = 1)); Oliveira et al., 2014 [24]: not associated. Machuca et al., 2014 [28]: not associated. | Yes | ||||||
| rs1799988 | 46370768/SNV [C > T]/intron variant, 5 prime UTR variant | Flórez et al., 2012 [10]: not associated. Juiz et al., 2019 [26]: not associated. Machuca et al., 2014 [28]: not associated. Nogueira et al., 2012 [25]: C/C frequency was increased in CCC severe group than CCC moderate group (p = 0.01, χ2 = 5.55, OR = 2.31, 95% CI (1.14–4.67)); C allele was higher in CCC severe group than CCC moderate group (p = 0.01, χ2 = 6.15, OR = 0.58, 95% CI (0.37–0.89)). | Yes | ||||||
| rs41469351 | 46370771/SNV [C > T]/intron variant, 5 prime UTR variant | Flórez et al., 2012 [10]: not associated. Juiz et al., 2019 [26]: T allele frequency in a sample with Caucasian genetic background was increased in a demonstrated cardiomyopathy group than non-demonstrated cardiomyopathy group (p = 0.028, OR = 4.88, 95% CI (1.03–23.24)). Machuca et al., 2014 [28]: not associated. | Yes | ||||||
| rs1800023 | 46370817/SNV [A > G/A > T]/intron variant, 5 prime UTR variant | Flórez et al., 2012 [10]: not associated. Juiz et al., 2019 [26]: not associated. Machuca et al., 2014 [28]: G allele was more frequent in the group with lower CCC severity compared to symptomatic subgroups (group II vs. group III, p = 0.05, OR = 0.70, 95% CI (0.49–1.00)). | Yes | ||||||
| rs1800024 | 46371068/SNV [C > T]/intron variant | Flórez et al., 2012 [10]: not associated. Juiz et al., 2019 [26]: T allele associated with reduced risk of cardiomyopathy in the group without symptoms of cardiomyopathy (asymptomatic group) when compared to the group with symptoms of cardiomyopathy (cardiomyopathy group) (p = 0.041, OR = 0.69, 95% CI (0.49–0.99)). Machuca et al., 2014 [28]: T allele associated with higher disease severity when compared to symptomatic subgroups (group II vs. group III, p = 0.02, OR = 1.70, 95% CI (1.06–2.71)). | Yes | ||||||
| rs3176763 | 46372790/SNV [G > A/G > T]/intron variant | Frade et al., 2013 [11]: C/C frequency was increased in CCC group than asymptomatic group when considered gender (p = 0.006, OR = 1.79, 95% CI (1.18–2.70)); C/A frequency was increased in CCC group than asymptomatic group when considered left ventricular ejection fraction under 0.4% values (p = 0.005, OR = 1.88, 95% CI (1.20–2.94)). | No | ||||||
| rs333[Δ32] | 46373453–46373487/frameshift variant (Delins, length 35 pb), coding sequence variant, intron variant | Calzada et al., 2001 [9]: not associated. Fernández-Mestre et al., 2004 [27]: not associated. Flórez et al., 2012 [10]: not associated. Oliveira et al., 2015 [13]: not associated. | Yes | ||||||
| rs3087253 | 46377198/SNV [C > T]/intron variant | Frade et al., 2013 [11]: not associated. | No | ||||||
| rs11575815 | 46378679/SNV [A > T]/intron variant | Frade et al., 2013 [11]: A/T and A/A frequecy was increased in the CCC group than asymptomatic group (p = 0.030, OR = 1.41, 95% CI (1.03–1.92)). | No | ||||||
| Reference | Description of CardCP Group Patients |
|---|---|
| Batista et al., 2018 [8] | Group in stage B1 (with structural heart disease, evidenced by ECG or ECHO, but with normal global ventricular function and neither current nor previous signs or symptoms of congestive heart failure) and group in stage C (with ventricular dysfunction and current or previous symptoms of congestive heart failure) |
| Calzada et al., 2001 [9] | Cardiomyopathic group with cardiac symptoms, which were assessed by clinical and electrocardiographic (ECG) characteristics compatible with chagasic cardiomyopathy |
| Fernandez-Mestre, Montagnani and Layrisse, 2004 [27] | Group B (arrhythmia-related symptoms or embolic episodes as a first symptom, and in whom radiological size ranged from normal to severe cardiomegaly and various arrhythmias) and group C (with overt congestive heart failure, most with severe cardiomegaly and various arrhythmias) |
| Flórez, Martín and González, 2012 [10] | Group II (radiology indicative of light heart hypertrophy or minor ECG alterations), group III (moderate heart hypertrophy and considerable ECG alterations, mainly advanced conduction abnormalities) and group IV (severe cardiomegaly and marked ECG alterations, predominantly frequent and/or complex forms of ventricular arrhythmia) |
| Juiz et al., 2019 [26] | CD group (with clinical symptoms and electrocardiography alterations) |
| Lima et al., 2018 [12] | Patients that presented cardiac arrhythmia with stimulus conduction disorder by EKG, compatible with chagasic cardiopathy |
| Machuca et al., 2014 [28] | Symptomatic group (with minor symptoms and alterations) and cardiomyopathic group (with considerable symptoms and alterations) |
| Nogueira et al., 2012 [25] | CCC (moderate group and severe group) |
| Oliveira et al., 2015 [13] | Group of cardiac form (CCHD when presenting with electrocardiographic or echocardiographic abnormalities consistent with the disease) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, J.M.; Santos, B.R.C.d.; Moura, E.L.d.; Santos, A.C.M.d.; Vencioneck Dutra, J.C.; Figueiredo, E.V.M.d.S.; Lima Filho, J.L.d. Narrowing the Relationship between Human CCR5 Gene Polymorphisms and Chagas Disease: Systematic Review and Meta-Analysis. Life 2023, 13, 1677. https://doi.org/10.3390/life13081677
Ferreira JM, Santos BRCd, Moura ELd, Santos ACMd, Vencioneck Dutra JC, Figueiredo EVMdS, Lima Filho JLd. Narrowing the Relationship between Human CCR5 Gene Polymorphisms and Chagas Disease: Systematic Review and Meta-Analysis. Life. 2023; 13(8):1677. https://doi.org/10.3390/life13081677
Chicago/Turabian StyleFerreira, Jean Moisés, Barbara Rayssa Correia dos Santos, Edilson Leite de Moura, Ana Caroline Melo dos Santos, Jean Carlos Vencioneck Dutra, Elaine Virgínia Martins de Sousa Figueiredo, and José Luiz de Lima Filho. 2023. "Narrowing the Relationship between Human CCR5 Gene Polymorphisms and Chagas Disease: Systematic Review and Meta-Analysis" Life 13, no. 8: 1677. https://doi.org/10.3390/life13081677
APA StyleFerreira, J. M., Santos, B. R. C. d., Moura, E. L. d., Santos, A. C. M. d., Vencioneck Dutra, J. C., Figueiredo, E. V. M. d. S., & Lima Filho, J. L. d. (2023). Narrowing the Relationship between Human CCR5 Gene Polymorphisms and Chagas Disease: Systematic Review and Meta-Analysis. Life, 13(8), 1677. https://doi.org/10.3390/life13081677





