Structural Phenomena in a Vesicle Membrane Obtained through an Evolution Experiment: A Study Based on MD Simulations
Abstract
:1. Introduction
2. Computational Methods
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kitadai, N.; Maruyama, S. Origins of building blocks of life: A review. Geosci. Front. 2018, 9, 1117–1153. [Google Scholar] [CrossRef]
- Jortner, J. Conditions for the emergence of life on the early Earth: Summary and reflections. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 1877–1891. [Google Scholar] [CrossRef]
- Harrison, T.M. Could the Hadean Eon Have Been Habitable? In Hadean Earth; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Deamer, D.; Dworkin, J.P.; Sandford, S.A.; Bernstein, M.P.; Allamandola, L.J. The First Cell Membranes. Astrobiology 2002, 2, 371–382. [Google Scholar] [CrossRef] [PubMed]
- McCollom, T.M.; Ritter, G.; Simoneit, B.R.T. Lipid Synthesis Under Hydrothermal Conditions by Fischer- Tropsch-Type Reactions. Orig. Life Evol. Biosph. 1999, 29, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Rushdi, A.I.; Simoneit, B.R.T. Lipid Formation by Aqueous Fischer-Tropsch-Type Synthesis over a Temperature Range of 100 to 400 °C. Orig. Life Evol. Biosph. 2001, 31, 103–118. [Google Scholar] [CrossRef]
- Chen, I.A.; Walde, P. From Self-Assembled Vesicles to Protocells. Cold Spring Harb. Perspect. Biol. 2010, 2, a002170. [Google Scholar] [CrossRef] [PubMed]
- Mansy, S.S. Membrane Transport in Primitive Cells. Cold Spring Harb. Perspect. Biol. 2010, 2, a002188. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.A.; Szostak, J.W. A Kinetic Study of the Growth of Fatty Acid Vesicles. Biophys. J. 2004, 87, 988–998. [Google Scholar] [CrossRef] [Green Version]
- Hargreaves, W.R.; Deamer, D.W. Liposomes from ionic, single-chain amphiphiles. Biochemistry 1978, 17, 3759–3768. [Google Scholar] [CrossRef]
- Apel, C.L.; Deamer, D.W.; Mautner, M.N. Self-assembled vesicles of monocarboxylic acids and alcohols: Conditions for stability and for the encapsulation of biopolymers. Biochim. Biophys. Acta (BBA) Biomembr. 2002, 1559, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Maurer, S.E.; Deamer, D.W.; Boncella, J.M.; Monnard, P.-A. Chemical Evolution of Amphiphiles: Glycerol Monoacyl Derivatives Stabilize Plausible Prebiotic Membranes. Astrobiology 2009, 9, 979–987. [Google Scholar] [CrossRef]
- Groen, J.; Deamer, D.W.; Kros, A.; Ehrenfreund, P. Polycyclic Aromatic Hydrocarbons as Plausible Prebiotic Membrane Components. Orig. Life Evol. Biosph. 2012, 42, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansy, S.S.; Szostak, J.W. Thermostability of model protocell membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 13351–13355. [Google Scholar] [CrossRef]
- Mansy, S.S.; Schrum, J.P.; Krishnamurthy, M.; Tobé, S.; Treco, D.A.; Szostak, J.W. Template-directed synthesis of a genetic polymer in a model protocell. Nature 2008, 454, 122–125. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Dagar, S.; Verma, A.; Rajamani, S. Compositional heterogeneity confers selective advantage to model protocellular membranes during the origins of cellular life. Sci. Rep. 2020, 10, 4483. [Google Scholar] [CrossRef] [Green Version]
- Napier, J.; Yin, J. Formation of peptides in the dry state. Peptides 2006, 27, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Eisenreich, W.; Hecht, S.; Wächtershäuser, G. A Possible Primordial Peptide Cycle. Science 2003, 301, 938–940. [Google Scholar] [CrossRef] [PubMed]
- Zaia, D.A.M.; Zaia, C.T.B.V.; De Santana, H. Which Amino Acids Should Be Used in Prebiotic Chemistry Studies? Orig. Life Evol. Biosph. 2008, 38, 469–488. [Google Scholar] [CrossRef]
- Flegmann, A.W.; Tattersall, R. Energetics of peptide bond formation at elevated temperatures. J. Mol. Evol. 1979, 12, 349–355. [Google Scholar] [CrossRef]
- Rabinowitz, J.; Hampaï, A. Quantitative polyphosphate-induced “prebiotic” peptide formation in H2O by addition of certain azoles and ions. J. Mol. Evol. 1984, 21, 199–201. [Google Scholar] [CrossRef]
- Leman, L.; Orgel, L.; Ghadiri, M.R. Carbonyl Sulfide-Mediated Prebiotic Formation of Peptides. Science 2004, 306, 283–286. [Google Scholar] [CrossRef] [Green Version]
- Bao, P.; He, Y.-Q.; Li, G.-X.; Zhang, H.-E.; Xiao, K.-Q. A thermodynamic chemical reaction network drove autocatalytic prebiotic peptides formation. Geochim. Cosmochim. Acta 2022, 324, 102–116. [Google Scholar] [CrossRef]
- Mamajanov, I.; MacDonald, P.J.; Ying, J.; Duncanson, D.M.; Dowdy, G.R.; Walker, C.A.; Engelhart, A.E.; Fernández, F.M.; Grover, M.A.; Hud, N.V.; et al. Ester Formation and Hydrolysis during Wet–Dry Cycles: Generation of Far-from-Equilibrium Polymers in a Model Prebiotic Reaction. Macromolecules 2014, 47, 1334–1343. [Google Scholar] [CrossRef]
- Yu, S.-S.; Krishnamurthy, R.; Fernández, F.M.; Hud, N.V.; Schork, F.J.; Grover, M.A. Kinetics of prebiotic depsipeptide formation from the ester–amide exchange reaction. Phys. Chem. Chem. Phys. 2016, 18, 28441–28450. [Google Scholar] [CrossRef]
- Mayer, C.; Schreiber, U.; Dávila, M.J.; Schmitz, O.J.; Bronja, A.; Meyer, M.; Klein, J.; Meckelmann, S.W. Molecular Evolution in a Peptide-Vesicle System. Life 2018, 8, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohorille, A.; Schweighofer, K.; Wilson, M.A.; Lazzerini, P.E.; Capecchi, P.L.; Laghi-Pasini, F.; Thiel, G.; Greiner, T.; Dunigan, D.D.; Moroni, A.; et al. The Origin and Early Evolution of Membrane Channels. Astrobiology 2005, 5, 1–17. [Google Scholar] [CrossRef]
- Booth, I.R.; Miller, S.; Müller, A.; Lehtovirta-Morley, L. The evolution of bacterial mechanosensitive channels. Cell Calcium 2015, 57, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Westall, F.; Hickman-Lewis, K.; Hinman, N.; Gautret, P.; Campbell, K.; Bréhéret, J.; Foucher, F.; Hubert, A.; Sorieul, S.; Dass, A.; et al. A Hydrothermal-Sedimentary Context for the Origin of Life. Astrobiology 2018, 18, 259–293. [Google Scholar] [CrossRef]
- Camprubí, E.; de Leeuw, J.W.; House, C.H.; Raulin, F.; Russell, M.J.; Spang, A.; Tirumalai, M.R.; Westall, F. The Emergence of Life. Space Sci. Rev. 2019, 215, 56. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, U.; Locker-Grütjen, O.; Mayer, C. Hypothesis: Origin of Life in Deep-Reaching Tectonic Faults. Orig. Life Evol. Biosph. 2012, 42, 47–54. [Google Scholar] [CrossRef]
- Namani, T.; Deamer, D.W. Stability of Model Membranes in Extreme Environments. Orig. Life Evol. Biosph. 2008, 38, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible and Free. J. Comp. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water molecules. J. Comp. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Mackerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Thøgersen, L.; Schiøtt, B.; Vosegaard, T.; Nielsen, N.C.; Tajkhorshid, E. Peptide Aggregation and Pore Formation in a Lipid Bilayer: A Combined Coarse-Grained and All Atom Molecular Dynamics Study. Biophys. J. 2008, 95, 4337–4347. [Google Scholar] [CrossRef] [Green Version]
- Dávila, M.J.; Mayer, C. Membrane Structure Obtained in an Experimental Evolution Process. Life 2022, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Guixà-González, R.; Rodriguez-Espigares, I.; Ramírez-Anguita, J.M.; Carrió-Gaspar, P.; Martinez-Seara, H.; Giorgino, T.; Selent, J. MEMBPLUGIN: Studying membrane complexity in VMD. Bioinformatics 2014, 30, 1478–1480. [Google Scholar] [CrossRef] [PubMed]
- Gapsys, V.; De Groot, B.L.; Briones, R. Computational analysis of local membrane properties. J. Comput. Aided Mol. Des. 2013, 27, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Lindblom, G.; Orädd, G. Lipid lateral diffusion and membrane heterogeneity. Biochim. Biophys. Acta (BBA) Biomembr. 2009, 1788, 234–244. [Google Scholar] [CrossRef] [Green Version]
- Misuraca, L.; Matsuo, T.; Cisse, A.; LoRicco, J.; Caliò, A.; Zanotti, J.-M.; Demé, B.; Oger, P.; Peters, J. High temperature molecular motions within a model protomembrane architecture. Phys. Chem. Chem. Phys. 2022, 24, 15083–15090. [Google Scholar] [CrossRef]
- Vermeer, L.S.; de Groot, B.L.; Réat, V.; Milon, A.; Czaplicki, J. Acyl chain order parameter profiles in phospholipid bilayers: Computation from molecular dynamics simulations and comparison with 2H NMR experiments. Eur. Biophys. J. 2007, 36, 919–931. [Google Scholar] [CrossRef] [Green Version]
- Heller, H.; Schaefer, M.; Schulten, K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phase. J. Phys. Chem. 1993, 97, 8343–8360. [Google Scholar] [CrossRef]
- Tieleman, D.P.; Forrest, L.R.; Sansom, M.S.P.; Berendsen, H.J.C. Lipid Properties and the Orientation of Aromatic Residues in OmpF, Influenza M2, and Alamethicin Systems: Molecular Dynamics Simulations. Biochemistry 1998, 37, 17554–17561. [Google Scholar] [CrossRef] [PubMed]
- Bachar, M.; Becker, O.M. Protein-Induced Membrane Disorder: A Molecular Dynamics Study of Melittin in a Dipalmitoylphosphatidylcholine Bilayer. Biophys. J. 2000, 78, 1359–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doktorova, M.N. Biophysics of Asymmetric Membranes: Protocols and Revelations. Ph.D. Dissertation, Cornell University, Ithaca, NY, YSA, 2018. [Google Scholar]
- Allen, W.J.; Lemkul, J.A.; Bevan, D.R. GridMAT-MD: A grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 2009, 30, 1749–1958. [Google Scholar] [CrossRef]
- Tian, C.-A.; Chiu, C.-C. Importance of Hydrophilic Groups on Modulating the Structural, Mechanical, and Interfacial Properties of Bilayers: A Comparative Molecular Dynamics Study of Phosphatidylcholine and Ion Pair Amphiphile Membranes. Int. J. Mol. Sci. 2018, 19, 1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, O.S.; Koeppe, R.E. Bilayer Thickness and Membrane Protein Function: An Energetic Perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef] [Green Version]

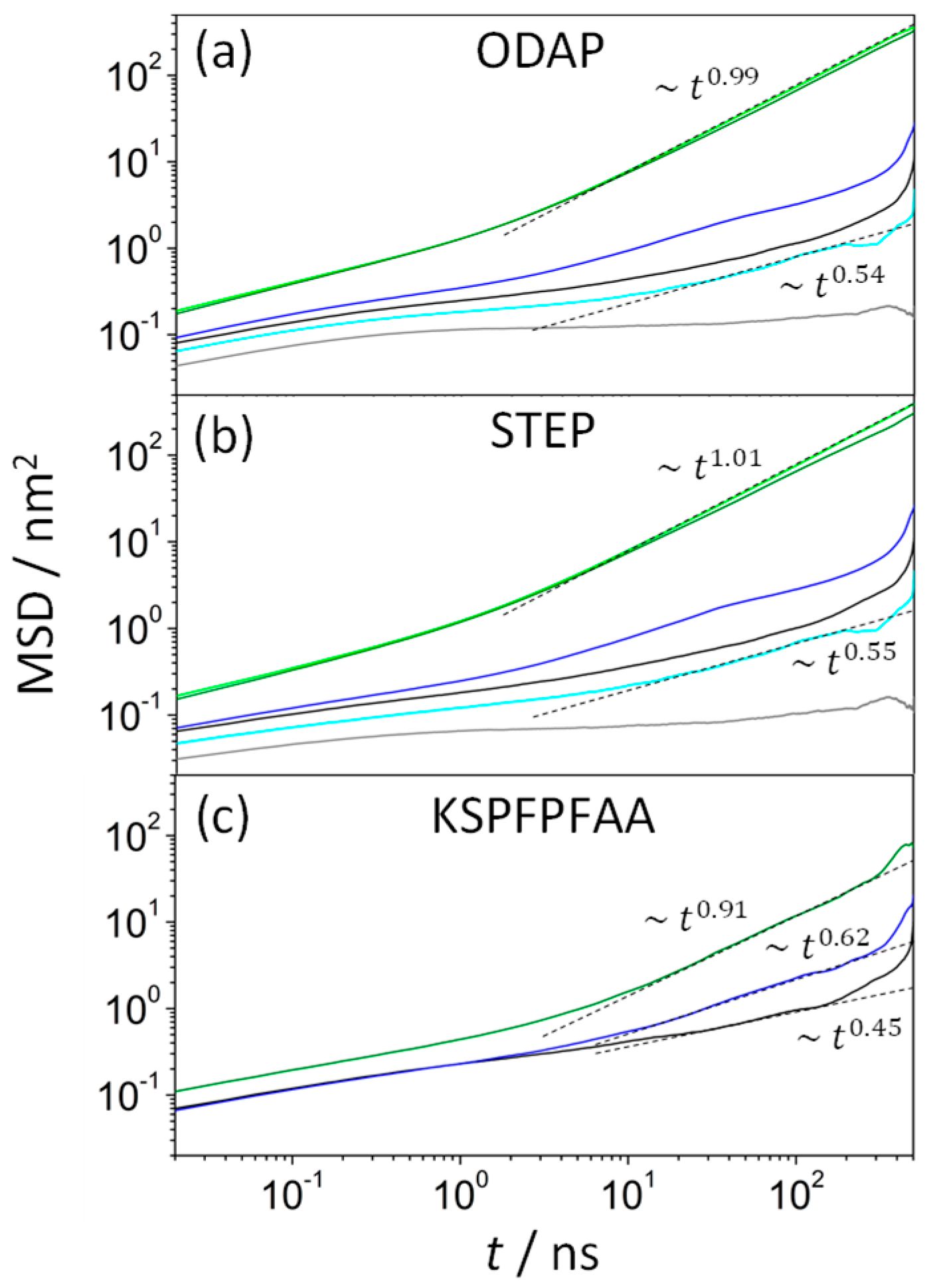

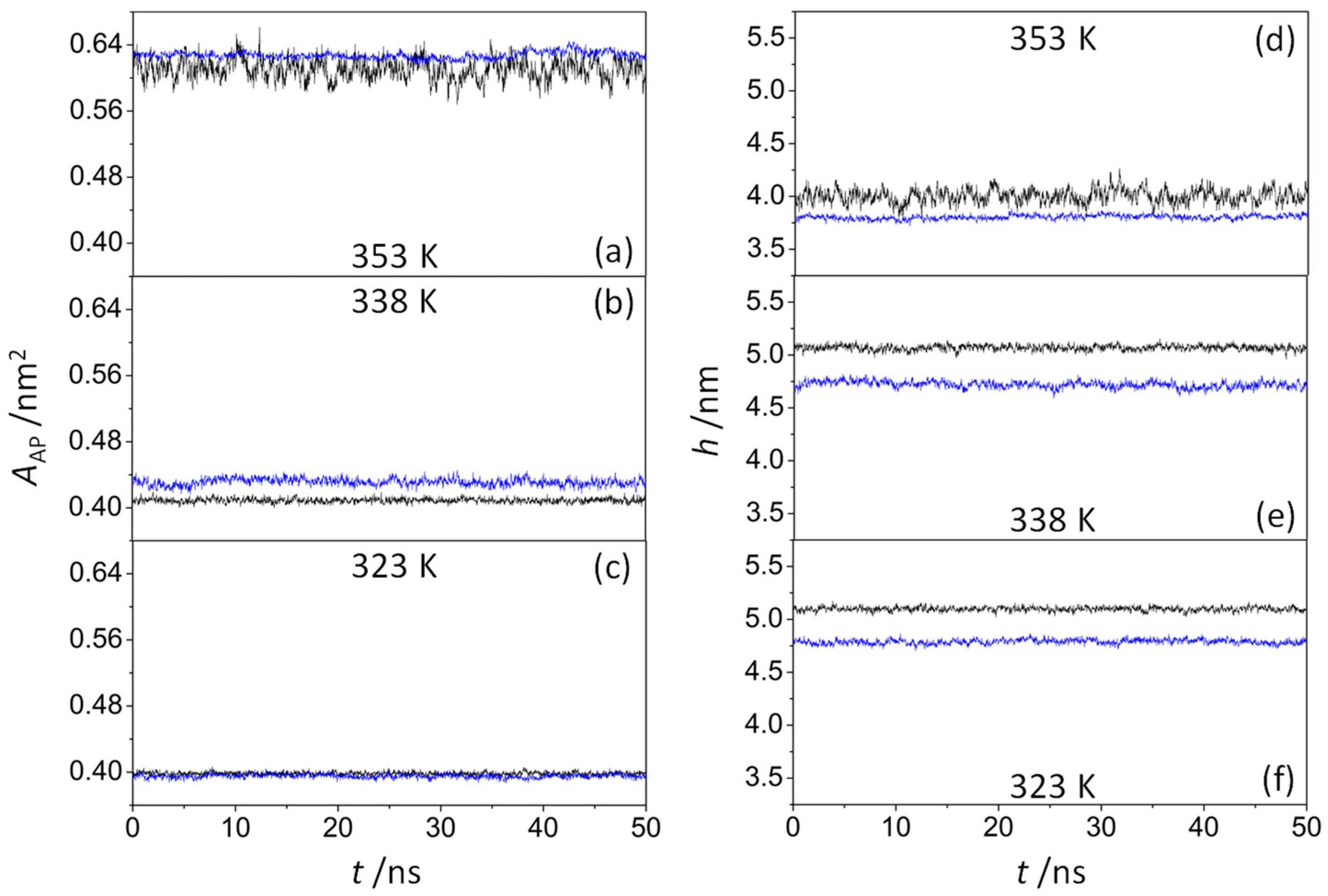

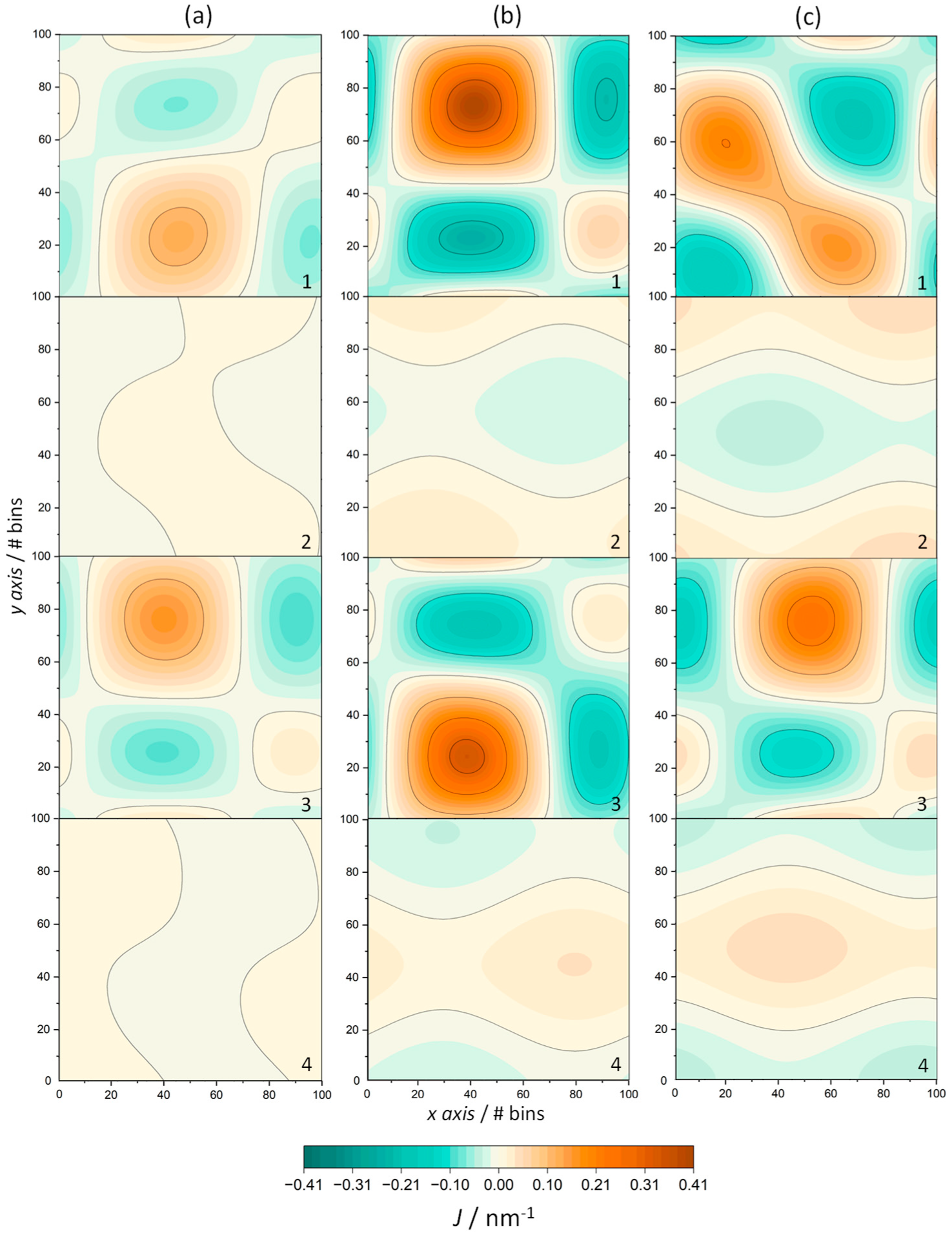

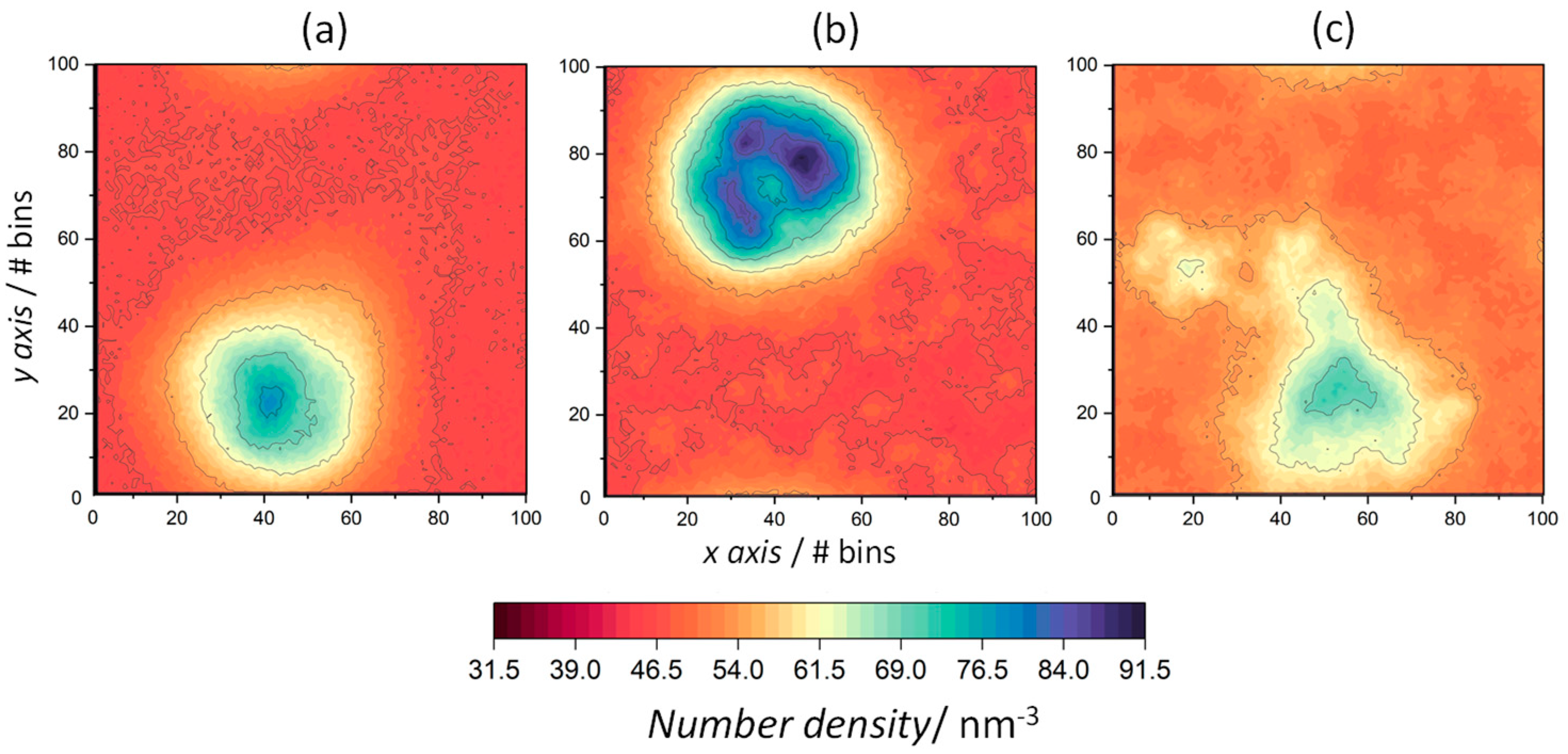
| System | T/K | Iρ | AAP/nm2 | h/nm |
|---|---|---|---|---|
| ODAP—STEP | 353 | 0.56 (0.06) | 0.610 (0.013) | 4.00 (0.07) |
| 338 | 0.20 (0.02) | 0.409 (0.003) | 5.07 (0.03) | |
| 323 | 0.19 (0.02) | 0.398 (0.002) | 5.10 (0.02) | |
| ODAP—STEP—KSPFPFAA | 353 | 0.60 (0.07) | 0.628 (0.004) | 3.80 (0.02) |
| 338 | 0.31 (0.02) | 0.431 (0.004) | 4.72 (0.03) | |
| 323 | 0.30 (0.02) | 0.395 (0.003) | 4.79 (0.02) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dávila, M.J.; Mayer, C. Structural Phenomena in a Vesicle Membrane Obtained through an Evolution Experiment: A Study Based on MD Simulations. Life 2023, 13, 1735. https://doi.org/10.3390/life13081735
Dávila MJ, Mayer C. Structural Phenomena in a Vesicle Membrane Obtained through an Evolution Experiment: A Study Based on MD Simulations. Life. 2023; 13(8):1735. https://doi.org/10.3390/life13081735
Chicago/Turabian StyleDávila, María J., and Christian Mayer. 2023. "Structural Phenomena in a Vesicle Membrane Obtained through an Evolution Experiment: A Study Based on MD Simulations" Life 13, no. 8: 1735. https://doi.org/10.3390/life13081735
APA StyleDávila, M. J., & Mayer, C. (2023). Structural Phenomena in a Vesicle Membrane Obtained through an Evolution Experiment: A Study Based on MD Simulations. Life, 13(8), 1735. https://doi.org/10.3390/life13081735








