Structure-Based Multi-Targeted Molecular Docking and Dynamic Simulation of Soybean-Derived Isoflavone Genistin as a Potential Breast Cancer Signaling Proteins Inhibitor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Ligands and Proteins
2.2. Molecular Docking
2.3. Selection of Positive and Negative Controls
2.4. Pharmacokinetics, ADMET, Drug-Likeness, and Radar Graph of Ligand Genistin
2.5. Molecular Dynamics (MD) Simulations Study
3. Results and Discussion
3.1. Molecular Docking
3.1.1. Ionization and Tautomerization of Genistin
3.1.2. Genistin, a Potential Inhibitor of ER Beta, Collapsin Response Mediator Protein 2 (CRMP2)
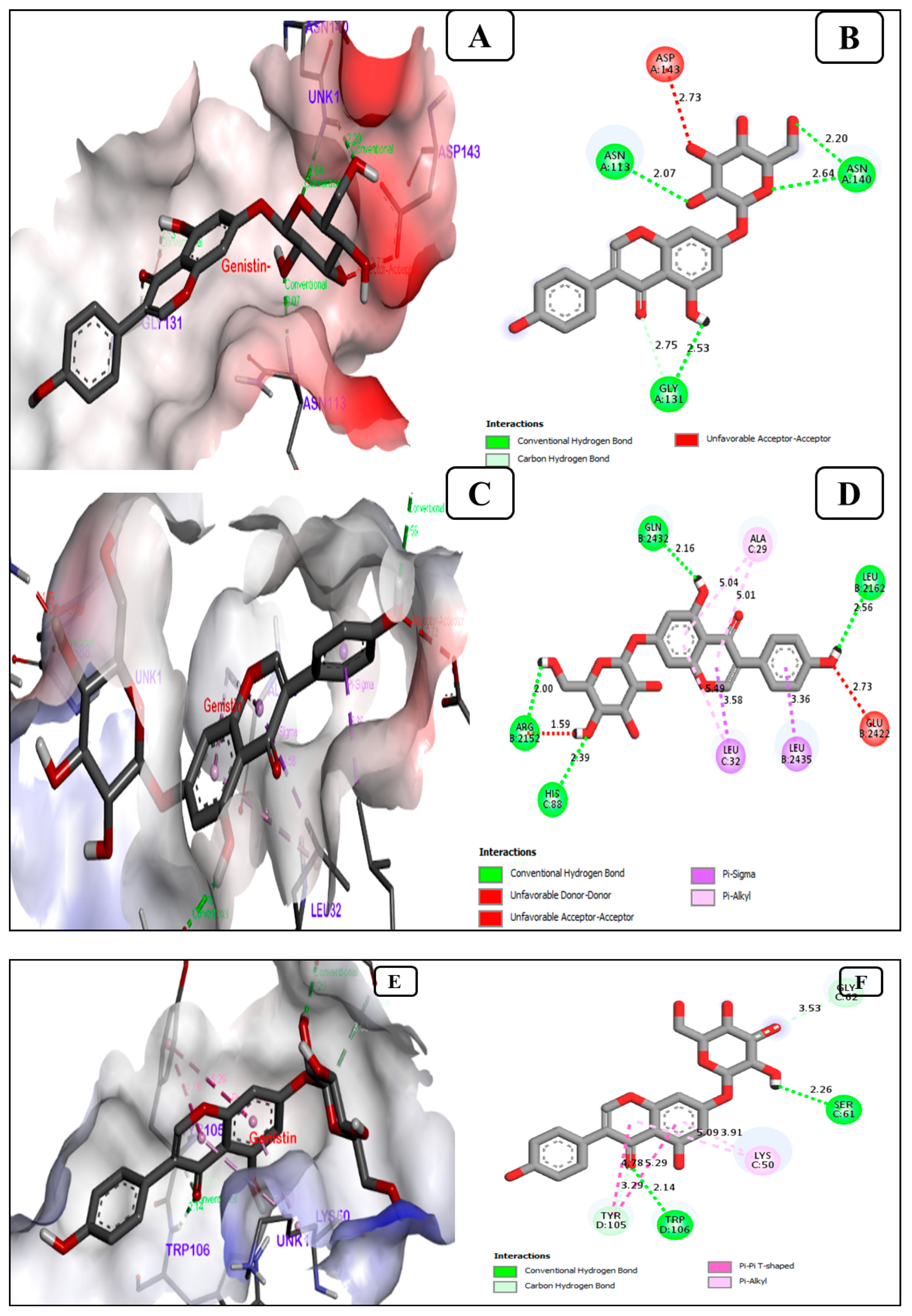
3.1.3. Genistin, a Potent Inhibitor of the Breast Cancer Antigen 15.3
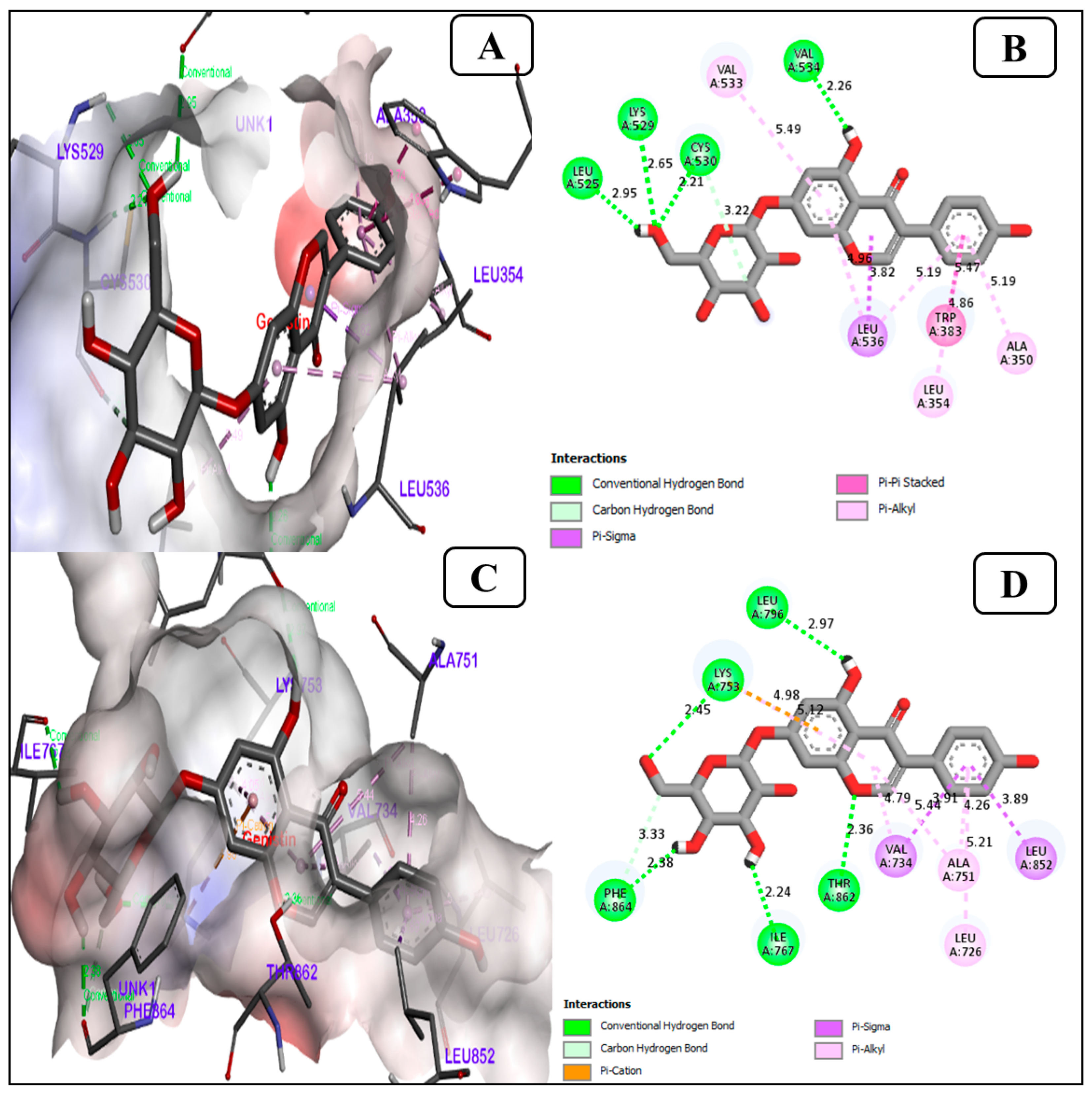
3.1.4. Genistin Is a Potent Inhibitor of the ER Alpha (ERα), Human Epidermal Growth Factor Receptor 2 (HER2)
3.2. In Silico Pharmacokinetics and ADMET Evaluation of Genistin
3.3. MD Simulation Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jha, V.; Devkar, S.; Gharat, K.; Kasbe, S.; Matharoo, D.K.; Pendse, S.; Bhosale, A.; Bhargava, A. Screening of Phytochemicals as Potential Inhibitors of Breast Cancer using Structure Based Multitargeted Molecular Docking Analysis. Phytomed. Plus 2022, 2, 100227. [Google Scholar] [CrossRef]
- Abd Elmoneim, O.; Al-Shammari, E.; Alam, M.J.; Alcantara, J.C.; Khan, M.A.; Eltoum, N.E.; Ashraf, S.A. Okra-Derived Dietary Carotenoid Lutein Against Breast Cancer, with an Approach towards Developing a Nutraceutical Product: A Meta-Analysis Study. J. Pharm. Res. Int. 2021, 33, 135–142. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar]
- Yuan, M.; Zhang, G.; Bai, W.; Han, X.; Li, C.; Bian, S. The Role of Bioactive Compounds in Natural Products Extracted from Plants in Cancer Treatment and Their Mechanisms Related to Anticancer Effects. Oxidative Med. Cell. Longev. 2022, 2022, 1429869. [Google Scholar] [CrossRef]
- Asma, S.T.; Acaroz, U.; Imre, K.; Morar, A.; Shah, S.R.; Hussain, S.Z.; Arslan-Acaroz, D.; Demirbas, H.; Hajrulai-Musliu, Z.; Istanbullugil, F.R.; et al. Natural Products/Bioactive Compounds as a Source of Anticancer Drugs. Cancers 2022, 14, 6203. [Google Scholar] [CrossRef]
- Sohrab, S.S.; Kamal, M.A. Screening, Docking, and Molecular Dynamics Study of Natural Compounds as an Anti-HER2 for the Management of Breast Cancer. Life 2022, 12, 1729. [Google Scholar] [PubMed]
- Acharya, R.; Chacko, S.; Bose, P.; Lapenna, A.; Pattanayak, S.P. Structure Based Multitargeted Molecular Docking Analysis of Selected Furanocoumarins against Breast Cancer. Sci. Rep. 2019, 9, 15743. [Google Scholar]
- Ashraf, S.A.; Elkhalifa, A.E.O. Multi-Targeted Molecular Docking, Pharmacokinetics, and Drug-Likeness Evaluation of Okra-Derived Ligand Abscisic Acid Targeting Signaling Proteins Involved in the Development of Diabetes. Molecules 2021, 26, 5957. [Google Scholar] [CrossRef]
- Veronesi, U.; Orecchia, R.; Zurrida, S.; Galimberti, V.; Luini, A.; Veronesi, P.; Gatti, G.; D’Aiuto, G.; Cataliotti, L.; Paolucci, R.; et al. Avoiding axillary dissection in breast cancer surgery: A randomized trial to assess the role of axillary radiotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2005, 16, 383–388. [Google Scholar] [CrossRef]
- Makki, J. Diversity of Breast Carcinoma: Histological Subtypes and Clinical Relevance. Clin. Med. Insights. Pathol. 2015, 8, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [Green Version]
- Tischkowitz, M.; Xia, B. PALB2/FANCN: Recombining cancer and Fanconi anemia. Cancer Res. 2010, 70, 7353–7359. [Google Scholar] [CrossRef] [Green Version]
- Costa, R.; Gill, N.; Rademaker, A.W.; Carneiro, B.A.; Chae, Y.K.; Kumthekar, P.; Gradishar, W.J.; Kurzrock, R.; Giles, F.J. Systematic analysis of early phase clinical studies for patients with breast cancer: Inclusion of patients with brain metastasis. Cancer Treat. Rev. 2017, 55, 10–15. [Google Scholar] [CrossRef]
- Harris, L.; Fritsche, H.; Mennel, R.; Norton, L.; Ravdin, P.; Taube, S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 5287–5312. [Google Scholar]
- Chen, W.; Zhang, Z.; Zhang, S.; Zhu, P.; Ko, J.K.; Yung, K.K. MUC1: Structure, Function, and Clinic Application in Epithelial Cancers. Int. J. Mol. Sci. 2021, 22, 6567. [Google Scholar] [PubMed]
- Deng, X.; Deng, J.; Yi, X.; Zou, Y.; Liu, H.; Li, C.; Deng, B.; Fan, H.; Hao, L. Ubiquitin-like protein FAT10 promotes osteosarcoma glycolysis and growth by upregulating PFKFB3 via stabilization of EGFR. Am. J. Cancer Res. 2020, 10, 2066–2082. [Google Scholar]
- Shimada, H. Is “liquid biopsy” useful for assessing HER2 status in gastric cancer? J. Gastroenterol. 2015, 50, 119–120. [Google Scholar] [CrossRef] [Green Version]
- Garg, S.; Lule, V.K.; Malik, R.K.; Tomar, S.K. Soy Bioactive Components in Functional Perspective: A Review. Int. J. Food Prop. 2016, 19, 2550–2574. [Google Scholar] [CrossRef] [Green Version]
- Bragagnolo, F.S.; Funari, C.S.; Ibáñez, E.; Cifuentes, A. Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds. Foods 2021, 10, 1308. [Google Scholar] [CrossRef]
- Pejčić, T.; Zeković, M.; Bumbaširević, U.; Kalaba, M.; Vovk, I.; Bensa, M.; Popović, L.; Tešić, Ž. The Role of Isoflavones in the Prevention of Breast Cancer and Prostate Cancer. Antioxidants 2023, 12, 368. [Google Scholar] [CrossRef] [PubMed]
- RCSB PDB. Available online: https://www.rcsb.org/ (accessed on 10 January 2023).
- PubChecm. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 10 January 2023).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed] [Green Version]
- SwissADME. Available online: http://www.swissadme.ch (accessed on 15 January 2023).
- Pharmacokinetic Properties. Available online: http://biosig.unimelb.edu.au/pkcsm/prediction (accessed on 15 January 2023).
- ADMET Evaluation. Available online: https://admetmesh.scbdd.com/service/evaluation/cal (accessed on 15 January 2023).
- Zoete, V.; Daina, A.; Bovigny, C.; Michielin, O. SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening. J. Chem. Inf. Model. 2016, 56, 1399–1404. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar]
- Awadelkareem, A.M.; Al-Shammari, E.; Elkhalifa, A.E.O.; Adnan, M.; Siddiqui, A.J.; Snoussi, M.; Khan, M.I.; Azad, Z.R.A.A.; Patel, M.; Ashraf, S.A. Phytochemical and In Silico ADME/Tox Analysis of Eruca sativa Extract with Antioxidant, Antibacterial and Anticancer Potential against Caco-2 and HCT-116 Colorectal Carcinoma Cell Lines. Molecules 2022, 27, 1409. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Sinha, M.; Ahmad, K. Study of Caspase 8 Inhibition for the Management of Alzheimer’s Disease: A Molecular Docking and Dynamics Simulation. Molecules 2020, 25, 2071. [Google Scholar] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar]
- Huang, S.Y.; Zou, X. Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins 2007, 66, 399–421. [Google Scholar] [PubMed]
- Jiménez, J.; Škalič, M.; Martínez-Rosell, G.; De Fabritiis, G. KDEEP: Protein–Ligand Absolute Binding Affinity Prediction via 3D-Convolutional Neural Networks. J. Chem. Inf. Model. 2018, 58, 287–296. [Google Scholar]
- Zheng, L.; Fan, J.; Mu, Y. OnionNet: A Multiple-Layer Intermolecular-Contact-Based Convolutional Neural Network for Protein-Ligand Binding Affinity Prediction. ACS Omega 2019, 4, 15956–15965. [Google Scholar]
- Son, J.; Kim, D. Development of a graph convolutional neural network model for efficient prediction of protein-ligand binding affinities. PLoS ONE 2021, 16, e0249404. [Google Scholar]
- Kyro, G.W.; Brent, R.I. HAC-Net: A Hybrid Attention-Based Convolutional Neural Network for Highly Accurate Protein-Ligand Binding Affinity Prediction. J. Chem. Inf. Model. 2023, 63, 1947–1960. [Google Scholar] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [PubMed] [Green Version]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar]
- Brozell, S.R.; Mukherjee, S.; Balius, T.E.; Roe, D.R.; Case, D.A.; Rizzo, R.C. Evaluation of DOCK 6 as a pose generation and database enrichment tool. J. Comput.-Aided Mol. Des. 2012, 26, 749–773. [Google Scholar] [PubMed] [Green Version]
- Gentile, F.; Yaacoub, J.C.; Gleave, J.; Fernandez, M.; Ton, A.-T.; Ban, F.; Stern, A.; Cherkasov, A. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat. Protoc. 2022, 17, 672–697. [Google Scholar]
- ten Brink, T.; Exner, T.E. Influence of protonation, tautomeric, and stereoisomeric states on protein-ligand docking results. J. Chem. Inf. Model. 2009, 49, 1535–1546. [Google Scholar] [PubMed]
- Bietz, S.; Urbaczek, S.; Schulz, B.; Rarey, M. Protoss: A holistic approach to predict tautomers and protonation states in protein-ligand complexes. J. Cheminformat. 2014, 6, 12. [Google Scholar]
- Zhou, Y.; Liu, X. The role of estrogen receptor beta in breast cancer. Biomark. Res. 2020, 8, 39. [Google Scholar] [PubMed]
- Mal, R.; Magner, A.; David, J.; Datta, J.; Vallabhaneni, M.; Kassem, M.; Manouchehri, J.; Willingham, N.; Stover, D.; Vandeusen, J.; et al. Estrogen Receptor Beta (ERβ): A Ligand Activated Tumor Suppressor. Front. Oncol. 2020, 10, 587386. [Google Scholar] [CrossRef] [PubMed]
- Cancer.Net. Available online: https://www.cancer.net/ (accessed on 28 January 2023).
- Lin, B.; Li, Y.; Wang, T.; Qiu, Y. CRMP2 is a therapeutic target that suppresses the aggressiveness of breast cancer cells by stabilizing RECK. Oncogene 2020, 39, 6024–6040. [Google Scholar] [CrossRef]
- Tan, F.; Thiele, C.J.; Li, Z. Collapsin response mediator proteins: Potential diagnostic and prognostic biomarkers in cancers (Review). Oncol. Lett. 2014, 7, 1333–1340. [Google Scholar]
- Fakhari, A.; Gharepapagh, E.; Dabiri, S.; Gilani, N. Correlation of cancer antigen 15-3 (CA15-3) serum level and bony metastases in breast cancer patients. Med. J. Islam. Repub. Iran 2019, 33, 142. [Google Scholar]
- Fang, C.; Cao, Y.; Liu, X.; Zeng, X.T.; Li, Y. Serum CA125 is a predictive marker for breast cancer outcomes and correlates with molecular subtypes. Oncotarget 2017, 8, 63963–63970. [Google Scholar] [PubMed] [Green Version]
- Gloucestershire Hospitals. Available online: https://www.gloshospitals.nhs.uk/our-services/services-we-offer/pathology/tests-and-investigations/ca-15-3-tumour-marker/ (accessed on 21 February 2023).
- Liu, Y.; Ma, H.; Yao, J. ERα, A Key Target for Cancer Therapy: A Review. OncoTargets Ther. 2020, 13, 2183–2191. [Google Scholar]
- Zhou, Z.; Qiao, J.X.; Shetty, A.; Wu, G.; Huang, Y.; Davidson, N.E.; Wan, Y. Regulation of estrogen receptor signaling in breast carcinogenesis and breast cancer therapy. Cell. Mol. Life Sci. CMLS 2014, 71, 1549. [Google Scholar]
- Xue, M.; Zhang, K.; Mu, K.; Xu, J.; Yang, H.; Liu, Y.; Wang, B.; Wang, Z.; Li, Z.; Kong, Q.; et al. Regulation of estrogen signaling and breast cancer proliferation by an ubiquitin ligase TRIM56. Oncogenesis 2019, 8, 30. [Google Scholar]
- Iqbal, N.; Iqbal, N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol. Biol. Int. 2014, 2014, 852748. [Google Scholar]
- Budi, H.S.; Ahmad, F.N.; Achmad, H.; Ansari, M.J.; Mikhailova, M.V.; Suksatan, W.; Chupradit, S.; Shomali, N.; Marofi, F. Human epidermal growth factor receptor 2 (HER2)-specific chimeric antigen receptor (CAR) for tumor immunotherapy; recent progress. Stem Cell Res. Ther. 2022, 13, 40. [Google Scholar] [CrossRef] [PubMed]




| Proteins and Their PDB IDs | GridPoint Dimensions (X × Y × Z) | Centre Grid Box (X × Y × Z Center) | Grid Spacing (Angstrom) |
|---|---|---|---|
| ER-Beta (5TOA) | 100 × 110 × 126 | 17.721 × 30.024 × 30.83 | 0.547 |
| Collapsin response mediator protein 2 (5LXX) | 122 × 124 × 122 | −36.223 × 17.428 × 23.675 | 0.703 |
| Breast cancer antigen 15.3 (1Y8X) | 92 × 82 × 116 | −1.86 × −8.38 × 22.858 | 0.664 |
| Ubiquitin-like protein activation complex (2NVU) | 122 × 126 × 80 | 88.819 × −26.425 × −9.079 | 0.972 |
| Glycoprotein Mucin 1 (5T6P) | 126 × 120 × 116 | 75.432 × 94.025 × 31.304 | 0.719 |
| ER-ALPHA (6CHZ) | 116 × 126 × 122 | −24.217 × 4.05 × −20.978 | 0.469 |
| Human epidermal growth factor receptor 2 (7PCD) | 100 × 126 × 126 | 2.245 × −11.712 × −16. 917 | 0.453 |
| S. No | Protein Name (PDB ID) | Total Structure Weight (kDa) | Name of Chains | (ΔG) Binding Energy (kcal/mol) of Genistin | (ΔG) Binding Energy (kcal/mol) of Positive Control (Everolimus) | (ΔG) Binding Energy (kcal/mol) of Positive Control (Lapatinib) | (ΔG) Binding Energy (kcal/mol) of Negative Control (Glycerol | No. of H-Bonds | H-Bond Forming Residues |
|---|---|---|---|---|---|---|---|---|---|
| 1. | ER-Beta (PDB ID-5TOA) | 56.6 | A, B | −8.3 | −7.4 | −7.7 | −3.8 | 2 | ARG(A)346, LYS(A)401 |
| 2. | Collapsin response mediator protein 2 (PDB ID-5LXX) | 111.09 | A, B | −9.6 | −10.0 | −7.8 | −4.6 | 6 | ASN(A)244, LYS(A)270, ARG(A)485, SER(A)486, SER(B)319, LYS(B)374 |
| 3. | Breast cancer antigen 15.3 (Ca 15.3) (PDB ID-1Y8X) | 29.34 | A, B | −7.0 | −7.1 | −8.0 | −3.7 | 4 | ASN(A)113, ASN(A)140, ASN(A)140, GLY(A)131 |
| 4. | ubiquitin-like protein activation complex (PDB ID-2NVU) | 188.89 | A, B, C, D, E | −9.5 | −9.9 | −10.1 | −4.1 | 4 | GLN(B)2432, LEU(B)2162, ARG(B)2152, HIS(C)88 |
| 5. | glycoprotein Mucin 1 (MUC1) (PDB ID-5T6P), | 95.8 | A, B, C, D, E, F | −8.8 | −8.9 | −7.1 | −3.8 | 2 | SER(C)61, TRP(D)106 |
| 6. | ER-ALPHA (PDB ID-6CHZ) | 30.69 | A | −8.8 | −7.7 | −7.2 | −3.8 | 4 | LEU(A)525, LYS(A) 529, CYS(A) 530, VAL(A) 534 |
| 7. | human epidermal growth factor receptor 2 (PDB ID-7PCD) | 37.62 | A | −9.7 | −6.8 | −7.4 | −3.4 | 5 | LEU(A)796, LYS(A)753, PHE(A)864, ILE(A)767, THR(A)862 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elkhalifa, A.E.O.; Al-Shammari, E.; Kuddus, M.; Adnan, M.; Sachidanandan, M.; Awadelkareem, A.M.; Qattan, M.Y.; Khan, M.I.; Abduljabbar, S.I.; Sarwar Baig, M.; et al. Structure-Based Multi-Targeted Molecular Docking and Dynamic Simulation of Soybean-Derived Isoflavone Genistin as a Potential Breast Cancer Signaling Proteins Inhibitor. Life 2023, 13, 1739. https://doi.org/10.3390/life13081739
Elkhalifa AEO, Al-Shammari E, Kuddus M, Adnan M, Sachidanandan M, Awadelkareem AM, Qattan MY, Khan MI, Abduljabbar SI, Sarwar Baig M, et al. Structure-Based Multi-Targeted Molecular Docking and Dynamic Simulation of Soybean-Derived Isoflavone Genistin as a Potential Breast Cancer Signaling Proteins Inhibitor. Life. 2023; 13(8):1739. https://doi.org/10.3390/life13081739
Chicago/Turabian StyleElkhalifa, Abd Elmoneim O., Eyad Al-Shammari, Mohammed Kuddus, Mohd Adnan, Manojkumar Sachidanandan, Amir Mahgoub Awadelkareem, Malak Yahia Qattan, Mohammad Idreesh Khan, Sanaa Ismael Abduljabbar, Mirza Sarwar Baig, and et al. 2023. "Structure-Based Multi-Targeted Molecular Docking and Dynamic Simulation of Soybean-Derived Isoflavone Genistin as a Potential Breast Cancer Signaling Proteins Inhibitor" Life 13, no. 8: 1739. https://doi.org/10.3390/life13081739
APA StyleElkhalifa, A. E. O., Al-Shammari, E., Kuddus, M., Adnan, M., Sachidanandan, M., Awadelkareem, A. M., Qattan, M. Y., Khan, M. I., Abduljabbar, S. I., Sarwar Baig, M., & Ashraf, S. A. (2023). Structure-Based Multi-Targeted Molecular Docking and Dynamic Simulation of Soybean-Derived Isoflavone Genistin as a Potential Breast Cancer Signaling Proteins Inhibitor. Life, 13(8), 1739. https://doi.org/10.3390/life13081739










