Applications of Deep Learning Algorithms to Ultrasound Imaging Analysis in Preclinical Studies on In Vivo Animals
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Searches
2.2. Eligibility Criteria
- Studies on preclinical/animal models with in vivo US acquisitions and developed or tested DL-based algorithms on US images or features extracted from the images;
- No restriction on the animal species used;
- No restriction on the DL architecture adopted in the studies and/or on their tasks;
- Studies using in vivo preclinical US images only for testing DL model performance.
- Studies performing US acquisitions on phantoms/ex vivo models/humans only;
- Studies proposing AI-based methods but not properly deep architectures;
- Publications not in the English language;
- Non-peer-reviewed original articles or conference proceedings.
2.3. Data Extraction and Analysis
3. Results
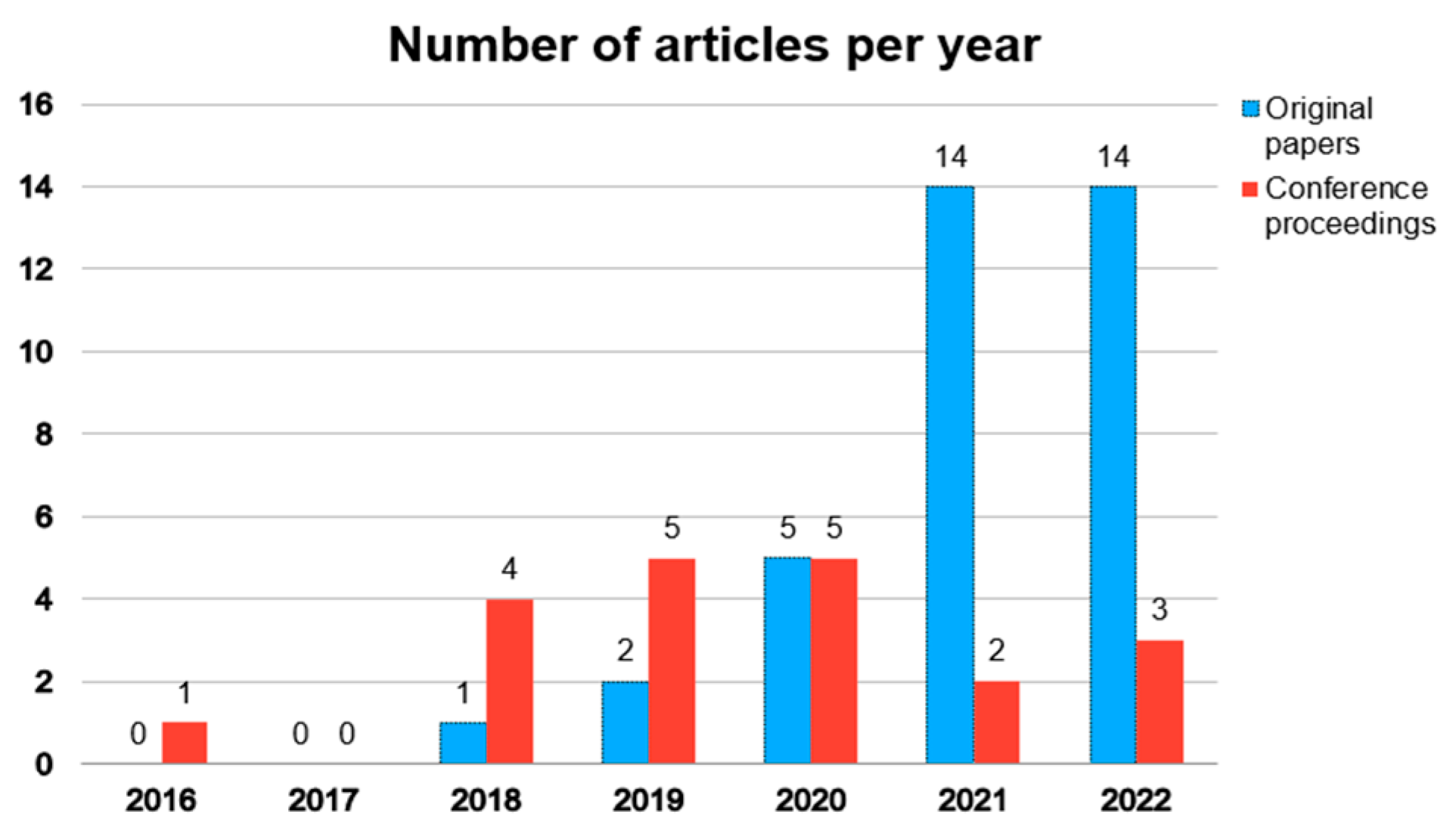
3.1. Search Results
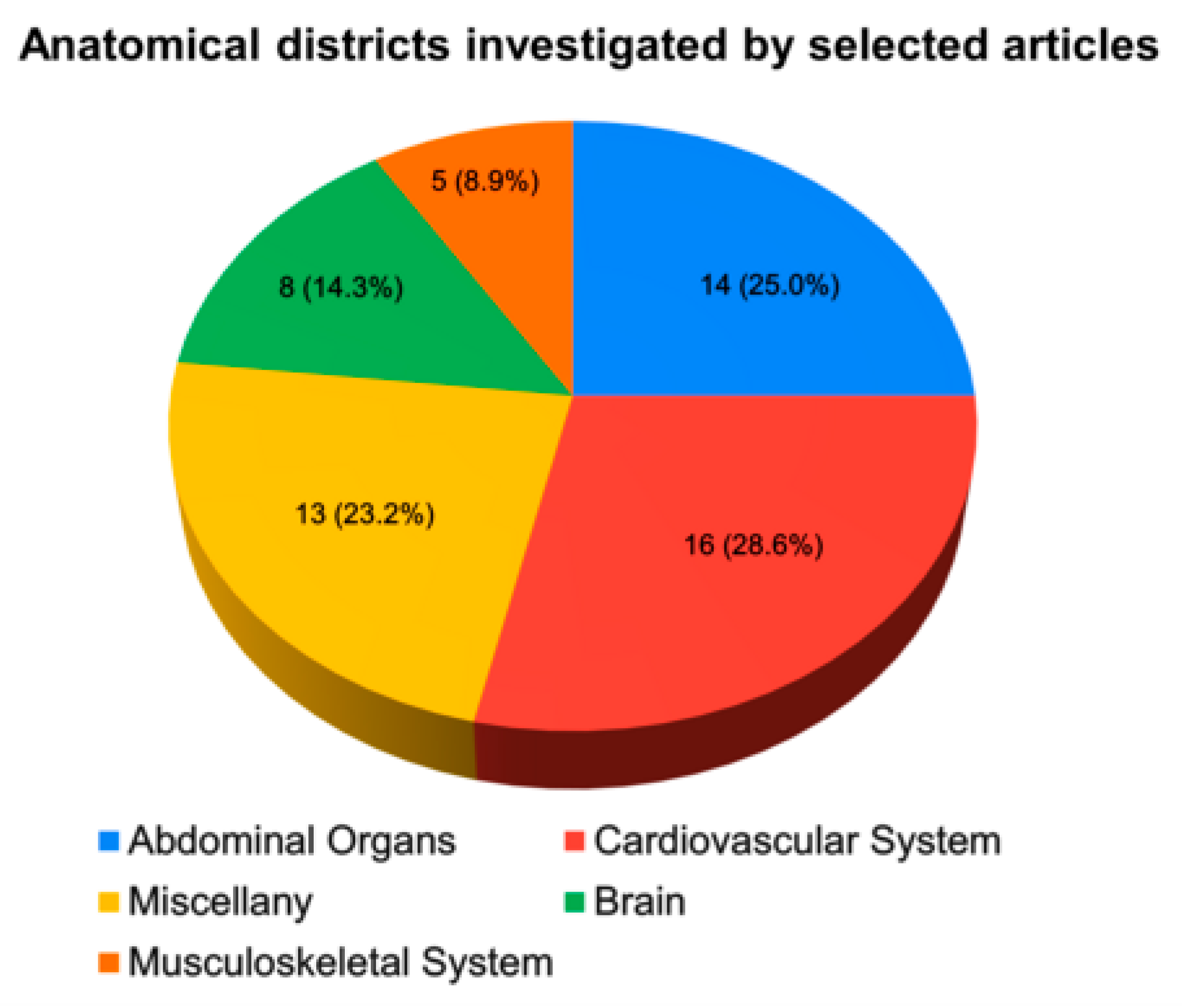
3.2. Cardiovascular System
3.3. Abdominal Organs
3.4. Musculoskeletal System
3.5. Brain
3.6. Miscellany
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moran, C.M.; Thomson, A.J.W. Preclinical Ultrasound Imaging—A Review of Techniques and Imaging Applications. Front. Phys. 2020, 8, 124. [Google Scholar] [CrossRef]
- Klibanov, A.L.; Hossack, J.A. Ultrasound in Radiology. Investig. Radiol. 2015, 50, 657–670. [Google Scholar] [CrossRef]
- Singh, R.; Culjat, M. Medical ultrasound devices. In Medical Devices: Surgical and Image-Guided Technologies; Culjat, M., Singh, R., Lee, H., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; Chapter 14; pp. 303–339. [Google Scholar]
- Powles, A.E.; Martin, D.J.; Wells, I.T.; Goodwin, C.R. Physics of ultrasound. Anaesth. Intensive Care Med. 2018, 19, 202–205. [Google Scholar] [CrossRef]
- Shriki, J. Ultrasound physics. Crit. Care Clin. 2014, 30, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.J.; Pilcher, J.; Richenberg, J.; Patel, U.; Frauscher, F. Applications of transrectal ultrasound in prostate cancer. Br. J. Radiol. 2012, 85, S3–S17. [Google Scholar] [CrossRef]
- Kumar, A.; Chuan, A. Ultrasound guided vascular access: Efficacy and safety. Best Pract. Res. Clin. Anaesthesiol. 2009, 23, 299–311. [Google Scholar] [CrossRef]
- Lindsey, M.L.; Kassiri, Z.; Virag, J.A.I.; Brás, L.E.d.C.; Scherrer-Crosbie, M.; Zhabyeyev, P.; Gheblawi, M.; Oudit, G.Y.; Onoue, T.; Iwataki, M.; et al. Guidelines for measuring cardiac physiology in mice. Am. J. Physiol. Circ. Physiol. 2018, 314, H733–H752. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, M.L.; Brunt, K.R.; Kirk, J.A.; Kleinbongard, P.; Calvert, J.W.; Brás, L.E.d.C.; DeLeon-Pennell, K.Y.; Del Re, D.P.; Frangogiannis, N.G.; Frantz, S.; et al. Guidelines for in vivo mouse models of myocardial infarction. Am. J. Physiol. Circ. Physiol. 2021, 321, H1056–H1073. [Google Scholar] [CrossRef]
- Tanter, M.; Fink, M. Ultrafast imaging in biomedical ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2014, 61, 102–119. [Google Scholar] [CrossRef]
- Macé, E.; Montaldo, G.; Cohen, I.; Baulac, M.; Fink, M.; Tanter, M. Functional ultrasound imaging of the brain. Nat. Methods 2011, 8, 662–664. [Google Scholar] [CrossRef]
- Errico, C.; Pierre, J.; Pezet, S.; Desailly, Y.; Lenkei, Z.; Couture, O.; Tanter, M. Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature 2015, 527, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Jeffries, K.; Couture, O.; Dayton, P.A.; Eldar, Y.C.; Hynynen, K.; Kiessling, F.; O’Reilly, M.; Pinton, G.F.; Schmitz, G.; Tang, M.-X.; et al. Super-resolution Ultrasound Imaging. Ultrasound Med. Biol. 2020, 46, 865–891. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, B.B.; Liu, J.-B.; Forsberg, F. Ultrasound contrast agents: A review. Ultrasound Med. Biol. 1994, 20, 319–333. [Google Scholar] [CrossRef] [PubMed]
- van Sloun, R.J.; Demi, L.; Postema, A.W.; de la Rosette, J.J.; Wijkstra, H.; Mischi, M. Ultrasound-contrast-agent dispersion and velocity imaging for prostate cancer localization. Med. Image Anal. 2017, 35, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Qiao, H.; Dai, Q.; Ma, C. Deep learning in photoacoustic imaging: A review. J. Biomed. Opt. 2021, 26, 040901. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, I.; Huland, D.M.; Vermesh, O.; Frostig, H.E.; Tummers, W.S.; Gambhir, S.S. Photoacoustic clinical imaging. Photoacoustics 2019, 14, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Subochev, P.; Smolina, E.; Sergeeva, E.; Kirillin, M.; Orlova, A.; Kurakina, D.; Emyanov, D.; Razansky, D. Toward whole-brain in vivo optoacoustic angiography of rodents: Modeling and experimental observations. Biomed. Opt. Express 2020, 11, 1477–1488. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, Q.; Rebling, J.; Razansky, D. Cortex-wide microcirculation mapping with ultrafast large-field multifocal illumination microscopy. J. Biophotonics 2020, 13, e202000198. [Google Scholar] [CrossRef]
- Rebling, J.; Estrada, H.; Gottschalk, S.; Sela, G.; Zwack, M.; Wissmeyer, G.; Ntziachristos, V.; Razansky, D. Dual-wavelength hybrid optoacoustic-ultrasound biomicroscopy for functional imaging of large-scale cerebral vascular networks. J. Biophotonics 2018, 11, e201800057. [Google Scholar] [CrossRef]
- Yao, J.; Wang, L.; Yang, J.-M.; Maslov, K.I.; Wong, T.T.W.; Li, L.; Huang, C.-H.; Zou, J.; Wang, L.V. High-speed label-free functional photoacoustic microscopy of mouse brain in action. Nat. Methods 2015, 12, 407–410. [Google Scholar] [CrossRef]
- Aboofazeli, M.; Abolmaesumi, P.; Fichtinger, G.; Mousavi, P. Tissue characterization using multiscale products of wavelet transform of ultrasound radio frequency echoes. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2009, 2009, 479–482. [Google Scholar] [CrossRef]
- Bercoff, J.; Tanter, M.; Fink, M. Supersonic shear imaging: A new technique for soft tissue elasticity mapping. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2004, 51, 396–409. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Cho, K.; van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1724–1734. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Aristizábal, O.; Qiu, Z.; Gallego, E.; Aristizábal, M.; Mamou, J.; Wang, Y.; Ketterling, J.A.; Turnbull, D.H. Longitudinal in Utero Analysis of Engrailed-1 Knockout Mouse Embryonic Phenotypes Using High-Frequency Ultrasound. Ultrasound Med. Biol. 2023, 49, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Banzato, T.; Bonsembiante, F.; Aresu, L.; Gelain, M.; Burti, S.; Zotti, A. Use of transfer learning to detect diffuse degenerative hepatic diseases from ultrasound images in dogs: A methodological study. Veter. J. 2018, 233, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Blons, M.; Deffieux, T.; Osmanski, B.-F.; Tanter, M.; Berthon, B. PerceptFlow: Real-Time Ultrafast Doppler Image Enhancement Using Deep Convolutional Neural Network and Perceptual Loss. Ultrasound Med. Biol. 2023, 49, 225–236. [Google Scholar] [CrossRef]
- Brattain, L.J.; Pierce, T.T.; Gjesteby, L.A.; Johnson, M.R.; DeLosa, N.D.; Werblin, J.S.; Gupta, J.F.; Ozturk, A.; Wang, X.; Li, Q.; et al. AI-Enabled, Ultrasound-Guided Handheld Robotic Device for Femoral Vascular Access. Biosensors 2021, 11, 522. [Google Scholar] [CrossRef]
- Brown, K.G.; Ghosh, D.; Hoyt, K. Deep Learning of Spatiotemporal Filtering for Fast Super-Resolution Ultrasound Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 67, 1820–1829. [Google Scholar] [CrossRef]
- Brown, K.G.; Waggener, S.C.; Redfern, A.D.; Hoyt, K. Faster super-resolution ultrasound imaging with a deep learning model for tissue decluttering and contrast agent localization. Biomed. Phys. Eng. Express 2021, 7, 065035. [Google Scholar] [CrossRef]
- Cao, Y.; Xiao, X.; Liu, Z.; Yang, M.; Sun, D.; Guo, W.; Cui, L.; Zhang, P. Detecting vulnerable plaque with vulnerability index based on convolutional neural networks. Comput. Med. Imaging Graph. 2020, 81, 101711. [Google Scholar] [CrossRef]
- Carson, T.D.; Ghoshal, G.; Cornwall, G.B.P.; Tobias, R.; Schwartz, D.G.M.; Foley, K.T. Artificial Intelligence-enabled, Real-time Intraoperative Ultrasound Imaging of Neural Structures Within the Psoas: Validation in a Porcine Spine Model. Spine 2021, 46, E146–E152. [Google Scholar] [CrossRef]
- Chen, X.; Lowerison, M.R.; Dong, Z.; Han, A.; Song, P. Deep Learning-Based Microbubble Localization for Ultrasound Localization Microscopy. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2022, 69, 1312–1325. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Dai, M.; Xiao, T.; Fu, T.; Han, H.; Wang, Y.; Wang, W.; Ding, H.; Yu, J. Quantitative evaluation of liver fibrosis based on ultrasound radio frequency signals: An animal experimental study. Comput. Methods Programs Biomed. 2021, 199, 105875. [Google Scholar] [CrossRef] [PubMed]
- Chifor, R.; Li, M.; Nguyen, K.-C.T.; Arsenescu, T.; Chifor, I.; Badea, A.F.; Badea, M.E.; Hotoleanu, M.; Major, P.W.; Le, L.H. Three-dimensional periodontal investigations using a prototype handheld ultrasound scanner with spatial positioning reading sensor. Med. Ultrason. 2021, 23, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yang, J.; Lee, S.Y.; Kim, J.; Lee, J.; Kim, W.J.; Lee, S.; Kim, C. Deep Learning Enhances Multiparametric Dynamic Volumetric Photoacoustic Computed Tomography In Vivo (DL-PACT). Adv. Sci. 2022, 10, e2202089. [Google Scholar] [CrossRef]
- Dai, M.; Li, S.; Wang, Y.; Zhang, Q.; Yu, J. Post-processing radio-frequency signal based on deep learning method for ultrasonic microbubble imaging. Biomed. Eng. Online 2019, 18, 95. [Google Scholar] [CrossRef]
- Di Ianni, T.; Airan, R.D. Deep-fUS: A Deep Learning Platform for Functional Ultrasound Imaging of the Brain Using Sparse Data. IEEE Trans. Med. Imaging 2022, 41, 1813–1825. [Google Scholar] [CrossRef]
- Du, G.; Zhan, Y.; Zhang, Y.; Guo, J.; Chen, X.; Liang, J.; Zhao, H. Automated segmentation of the gastrocnemius and soleus in shank ultrasound images through deep residual neural network. Biomed. Signal Process. Control 2022, 73, 103447. [Google Scholar] [CrossRef]
- Duan, C.; Montgomery, M.K.; Chen, X.; Ullas, S.; Stansfield, J.; McElhanon, K.; Hirenallur-Shanthappa, D. Fully automated mouse echocardiography analysis using deep convolutional neural networks. Am. J. Physiol. Circ. Physiol. 2022, 323, H628–H639. [Google Scholar] [CrossRef]
- Gulenko, O.; Yang, H.; Kim, K.; Youm, J.Y.; Kim, M.; Kim, Y.; Jung, W.; Yang, J.-M. Deep-Learning-Based Algorithm for the Removal of Electromagnetic Interference Noise in Photoacoustic Endoscopic Image Processing. Sensors 2022, 22, 3961. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Du, G.-Q.; Shen, W.-Q.; Du, C.; He, P.-N.; Siuly, S. Automatic myocardial infarction detection in contrast echocardiography based on polar residual network. Comput. Methods Programs Biomed. 2021, 198, 105791. [Google Scholar] [CrossRef]
- Hyun, D.; Abou-Elkacem, L.; Bam, R.; Brickson, L.L.; Herickhoff, C.D.; Dahl, J.J. Nondestructive Detection of Targeted Microbubbles Using Dual-Mode Data and Deep Learning for Real-Time Ultrasound Molecular Imaging. IEEE Trans. Med. Imaging 2020, 39, 3079–3088. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Luo, Y.; He, X.; Wang, K.; Song, W.; Ye, Q.; Feng, L.; Wang, W.; Hu, X.; Li, H. Development and validation of the diagnostic accuracy of artificial intelligence-assisted ultrasound in the classification of splenic trauma. Ann. Transl. Med. 2022, 10, 1060. [Google Scholar] [CrossRef]
- Kim, T.; Hedayat, M.; Vaitkus, V.V.; Belohlavek, M.; Krishnamurthy, V.; Borazjani, I. Automatic segmentation of the left ventricle in echocardiographic images using convolutional neural networks. Quant. Imaging Med. Surg. 2021, 11, 1763–1781. [Google Scholar] [CrossRef]
- Lafci, B.; Mercep, E.; Morscher, S.; Dean-Ben, X.L.; Razansky, D. Deep Learning for Automatic Segmentation of Hybrid Optoacoustic Ultrasound (OPUS) Images. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2021, 68, 688–696. [Google Scholar] [CrossRef]
- Liu, Q.; Liua, Z.; Xu, W.; Wen, H.; Dai, M.; Chen, X. Diagnosis of Significant Liver Fibrosis by Using a DCNN Model with Fusion of Features from US B-Mode Image and Nakagami Parametric Map: An Animal Study. IEEE Access 2021, 9, 89300–89310. [Google Scholar] [CrossRef]
- Milecki, L.; Poree, J.; Belgharbi, H.; Bourquin, C.; Damseh, R.; Delafontaine-Martel, P.; Lesage, F.; Gasse, M.; Provost, J. A Deep Learning Framework for Spatiotemporal Ultrasound Localization Microscopy. IEEE Trans. Med. Imaging 2021, 40, 1428–1437. [Google Scholar] [CrossRef]
- Mitra, J.; Qiu, J.; MacDonald, M.; Venugopal, P.; Wallace, K.; Abdou, H.; Richmond, M.; Elansary, N.; Edwards, J.; Patel, N.; et al. Automatic hemorrhage detection from color Doppler ultrasound using a Generative Adversarial Network (GAN)-based anomaly detection method. IEEE J. Transl. Eng. Health Med. 2022, 10, 1800609. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Podkowa, A.S.; Park, T.H.; Miller, R.J.; Do, M.N.; Oelze, M.L. Use of a convolutional neural network and quantitative ultrasound for diagnosis of fatty liver. Ultrasound Med. Biol. 2021, 47, 556–568. [Google Scholar] [CrossRef]
- Olszynski, P.; Marshall, R.A.; Olver, T.D.; Oleniuk, T.; Auser, C.; Wilson, T.; Atkinson, P.; Woods, R. Performance of an automated ultrasound device in identifying and tracing the heart in porcine cardiac arrest. Ultrasound J. 2022, 14, 1. [Google Scholar] [CrossRef]
- Pan, Y.-C.; Chan, H.-L.; Kong, X.; Hadjiiski, L.M.; Kripfgans, O.D. Multi-class deep learning segmentation and automated measurements in periodontal sonograms of a porcine model. Dentomaxillofacial Radiol. 2022, 51, 20210363. [Google Scholar] [CrossRef]
- Park, J.H.; Seo, E.; Choi, W.; Lee, S.J. Ultrasound deep learning for monitoring of flow–vessel dynamics in murine carotid artery. Ultrasonics 2022, 120, 106636. [Google Scholar] [CrossRef]
- Qiu, Z.; Xu, T.; Langerman, J.; Das, W.; Wang, C.; Nair, N.; Aristizabal, O.; Mamou, J.; Turnbull, D.H.; Ketterling, J.A.; et al. A Deep Learning Approach for Segmentation, Classification, and Visualization of 3-D High-Frequency Ultrasound Images of Mouse Embryos. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2021, 68, 2460–2471. [Google Scholar] [CrossRef]
- Sharma, A.; Pramanik, M. Convolutional neural network for resolution enhancement and noise reduction in acoustic resolution photoacoustic microscopy. Biomed. Opt. Express 2020, 11, 6826–6839. [Google Scholar] [CrossRef] [PubMed]
- Solomon, O.; Cohen, R.; Zhang, Y.; Yang, Y.; He, Q.; Luo, J.; van Sloun, R.J.G.; Eldar, Y.C. Deep Unfolded Robust PCA With Application to Clutter Suppression in Ultrasound. IEEE Trans. Med. Imaging 2020, 39, 1051–1063. [Google Scholar] [CrossRef]
- Song, J.; Yin, H.; Huang, J.; Wu, Z.; Wei, C.; Qiu, T.; Luo, Y. Deep learning for assessing liver fibrosis based on acoustic nonlinearity maps: An in vivo study of rabbits. Comput. Assist. Surg. 2022, 27, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yang, X.; Shajudeen, P.; Sears, C.; Taraballi, F.; Weiner, B.; Tasciotti, E.; Dollahon, D.; Park, H.; Righetti, R. A CNN-based method to reconstruct 3-D spine surfaces from US images in vivo. Med. Image Anal. 2021, 74, 102221. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jiang, J.; Zhang, J.; Wang, Y.; Wang, B. Acoustic-resolution-based spectroscopic photoacoustic endoscopy towards molecular imaging in deep tissues. Opt. Express 2022, 30, 35014–35028. [Google Scholar] [CrossRef]
- Zhu, X.; Huang, Q.; DiSpirito, A.; Vu, T.; Rong, Q.; Peng, X.; Sheng, H.; Shen, X.; Zhou, Q.; Jiang, L.; et al. Real-time whole-brain imaging of hemodynamics and oxygenation at micro-vessel resolution with ultrafast wide-field photoacoustic microscopy. Light Sci. Appl. 2022, 11, 138. [Google Scholar] [CrossRef]
- Ahn, S.S.; Ta, K.; Lu, A.; Stendahl, J.C.; Sinusas, A.J.; Duncan, J.S. Unsupervised motion tracking of left ventricle in echocardiography. Proc. SPIE Int. Soc. Opt. Eng. 2020, 11319, 113190Z. [Google Scholar] [CrossRef]
- Allman, D.; Assis, F.; Chrispin, J.; Bell, M.A.L. Deep learning to detect catheter tips in vivo during photoacoustic-guided catheter interventions: Invited Presentation. In Proceedings of the 2019 53rd Annual Conference on Information Sciences and Systems, CISS 2019, Baltimore, MD, USA, 20–22 March 2019. [Google Scholar] [CrossRef]
- Allman, D.; Bell, M.A.L.; Chrispin, J.; Assis, F. A deep learning-based approach to identify in vivo catheter tips during photoacoustic-guided cardiac interventions. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2019, San Francisco, CA, USA, 3–6 February 2019; Volume 10878. [Google Scholar] [CrossRef]
- Cai, Q.; Yin, H.; Liu, D.C.; Liu, P. Using Learnt Nakagami parametric mapping to classify fatty liver in rabbits. In The Fourth International Symposium on Image Computing and Digital Medicine; Association for Computing Machinery: New York, NY, USA, 2020; pp. 182–186. [Google Scholar] [CrossRef]
- Cohen, R.; Zhang, Y.; Solomon, O.; Toberman, D.; Taieb, L.; Sloun, R.V.; Eldar, Y.C. Deep Convolutional Robust PCA with Application to Ultrasound Imaging. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 3212–3216. [Google Scholar] [CrossRef]
- Hyun, D.; Brickson, L.L.; Abou-Elkacem, L.; Bam, R.; Dahl, J.J. Nondestructive Targeted Microbubble Detection Using a Dual-Frequency Beamforming Deep Neural Network. In Proceedings of the 2018 IEEE International Ultrasonics Symposium (IUS), Kobe, Japan, 22–25 October 2018; pp. 1–9. [Google Scholar] [CrossRef]
- Kim, J.; Dong, Z.; Lowerison, M.R.; Sekaran, N.V.C.; You, Q.; Llano, D.A.; Song, P. Deep Learning-based 3D Beamforming on a 2D Row Column Addressing (RCA) Array for 3D Super-resolution Ultrasound Localization Microscopy. In Proceedings of the IUS 2022—IEEE International Ultrasonics Symposium, Venice, Italy, 10–13 October 2022. [Google Scholar] [CrossRef]
- Kulhare, S.; Zheng, X.; Mehanian, C.; Gregory, C.; Zhu, M.; Gregory, K.; Xie, H.; Jones, J.M.; Wilson, B. Ultrasound-Based Detection of Lung Abnormalities Using Single Shot Detection Convolutional Neural Networks. In Proceedings of the Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation, Granada, Spain, 16–20 September 2018; pp. 65–73. [Google Scholar] [CrossRef]
- Lafci, B.; Mercep, E.; Morscher, S.; Deán-Ben, X.L.; Razansky, D. Efficient segmentation of multi-modal optoacoustic and ultrasound images using convolutional neural networks. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2020, San Francisco, CA, USA, 2–5 February 2020; Volume 11240. [Google Scholar] [CrossRef]
- Lee, B.C.; Vaidya, K.; Jain, A.K.; Chen, A. Guidewire Segmentation in 4D Ultrasound Sequences Using Recurrent Fully Convolutional Networks. In Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis; ASMUS 2020 PIPPI 2020, Lecture Notes in Computer Science; Hu, Y., Ed.; Springer: Cham, Switzerland, 2020; Volume 12437. [Google Scholar] [CrossRef]
- Mehanian, C.; Kulhare, S.; Millin, R.; Zheng, X.; Gregory, C.; Zhu, M.; Xie, H.; Jones, J.; Lazar, J.; Halse, A.; et al. Deep Learning-Based Pneumothorax Detection in Ultrasound Videos. In Smart Ultrasound Imaging and Perinatal, Preterm and Paediatric Image Analysis; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11798. [Google Scholar] [CrossRef]
- Mitra, J.; MacDonald, M.; Venugopal, P.; Wallace, K.; Abdou, H.; Richmond, M.; Elansary, N.; Edwards, J.; Patel, N.; Morrison, J.; et al. Integrating artificial intelligence and color Doppler US for automatic hemorrhage detection. In Proceedings of the 2021 IEEE International Ultrasonics Symposium (IUS), Virtual Symposium, 11–16 September 2021. [Google Scholar] [CrossRef]
- Nguyen, T.; Do, M.; Oelze, M.L. Sensitivity Analysis of Reference-Free Quantitative Ultrasound Tissue Classification. In Proceedings of the 2018 IEEE International Ultrasonics Symposium, IUS 2018, Kobe, Japan, 22–25 October 2018. [Google Scholar] [CrossRef]
- Nisar, H.; Carnahan, P.K.; Fakim, D.; Akhuanzada, H.; Hocking, D.; Peters, T.M.; Chen, E.C.S. Towards ultrasound-based navigation: Deep learning based IVC lumen segmentation from intracardiac echocardiography. Med. Imaging 2022, 12034, 467–476. [Google Scholar] [CrossRef]
- Ossenkoppele, B.W.; Wei, L.; Luijten, B.; Vos, H.J.; De Jong, N.; Van Sloun, R.J.; Verweij, M.D. 3-D contrast enhanced ultrasound imaging of an in vivo chicken embryo with a sparse array and deep learning based adaptive beamforming. In Proceedings of the 2022 IEEE International Ultrasonics Symposium (IUS), Institute of Electrical and Electronics Engineers, Venice, Italy, 10–13 October 2022. [Google Scholar] [CrossRef]
- Qiu, Z.; Nair, N.; Langerman, J.; Aristizabal, O.; Mamou, J.; Turnbull, D.H.; Ketterling, J.A.; Wang, Y. Automatic Mouse Embryo Brain Ventricle & Body Segmentation and Mutant Classification from Ultrasound Data Using Deep Learning. In Proceedings of the 2019 IEEE International Ultrasonics Symposium (IUS), Glasgow, UK, 6–9 October 2019; pp. 12–15. [Google Scholar] [CrossRef][Green Version]
- Qiu, Z.; Langerman, J.; Nair, N.; Aristizabal, O.; Mamou, J.; Turnbull, D.H.; Ketterling, J.; Wang, Y. Deep Bv: A Fully Automated System for Brain Ventricle Localization and Segmentation In 3D Ultrasound Images of Embryonic Mice. IEEE Signal Process Med. Biol. Symp. 2018, 2018, 1–6. [Google Scholar] [CrossRef]
- Xu, J.; Xue, L.; Yu, J.; Ding, H. Evaluation of liver fibrosis based on ultrasound radio frequency signals. In Proceedings of the 2021 14th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 23–25 October 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Xu, T.; Qiu, Z.; Das, W.; Wang, C.; Langerman, J.; Nair, N.; Aristizabal, O.; Mamou, J.; Turnbull, D.H.; Ketterling, J.A.; et al. Deep Mouse: An End-to-End Auto-Context Refinement Framework for Brain Ventricle & Body Segmentation in Embryonic Mice Ultrasound Volumes. Proc. IEEE Int. Symp. Biomed. Imaging 2020, 2020, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Li, W.; Jing, J.; Yu, J.; Yi, S.; Yan, W. Automatic segmentation of the Epicardium and Endocardium using convolutional neural network. In Proceedings of the International Conference on Signal Processing Proceedings, ICSP, Chengdu, China, 6–10 November 2016. [Google Scholar] [CrossRef]
- Crick, S.J.; Sheppard, M.N.; Ho, S.Y.; Gebstein, L.; Anderson, R.H. Anatomy of the pig heart: Comparisons with normal human cardiac structure. J. Anat. 1998, 193, 105–119. [Google Scholar] [CrossRef]
- Xue, Y.; Xu, T.; Zhang, H.; Long, L.R.; Huang, X. SegAN: Adversarial Network with Multi-scale L1 Loss for Medical Image Segmentation. Neuroinformatics 2018, 16, 383–392. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 386–397. [Google Scholar] [CrossRef]
- Abdulla, W. Mask R-CNN for Object Detection and Instance Segmentation on Keras and TensorFlow. 2017. Available online: https://github.com/matterport/MaskRCNN (accessed on 21 July 2023).
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]

| Ref | Animal Model | Anatomical District | Aim of Study 1 | DL Network Task 1 | DL Architecture 1 | Main Result 1 |
|---|---|---|---|---|---|---|
| [28] | mouse | Embryo | Segmentation of Embryo body | Segmentation | FCN | no significant changes between control and mutant mice embryos |
| [29] | dog | Liver | Binary classification of degenerative hepatic disease | Classification | DNN | AUC = 0.91; Se = 100%; Sp = 82.8%; PLR = 5.25; NLR = 0.0 |
| [30] | mouse | Brain Vasculature | Vessel visualisation improvement | Image Quality Improvement | CNNs | CNR ↑ 56%; spatial resolution ≃ 100µm |
| [31] | pig | Femoral Artery | Needle detection to create femoral vascular access | Needle Detection | CNN | Precision = 0.97–0.94; Recall = 0.96–0.89 in artery and vein detection, respectively |
| [32] | rat | Breast Tumour Vasculature | MB segmentation and localisation through a spatiotemporal filter | MB Localisation | 3D-CNN | Acc = 88.0% Se = 82.9% Sp = 93.0% |
| [33] | rat | Hind Limb Vasculature | Tissue decluttering and contrast agent localisation | Contrast Agent Localisation | 3D-CNN | Qualitative results |
| [34] | rabbit | Plaque | Classification vulnerability of atherosclerosis plaques | Classification | CNN | AUC = 0.714; Acc = 73.5%; Se = 76.92% and Sp = 71.42% |
| [35] | pig | Psoas Muscle | Classification of bone and muscle regions | Segmentation + Classification | CNNs | DSC = 92%; Acc > 95% for nerve detection; DSC > 95% for bone and muscle |
| [36] | chicken | Embryo Chorioallantoic Membrane | MB localisation for real-time visualisation of the high-resolution microvasculature | MB Localisation | CNN | faster localisation than the conventional method to reach 90% vessel saturation; >20% faster than MB separation |
| [37] | rat | Liver | Classification of liver fibrosis severity (F0-F4) | Classification | RNN | Acc = 0.83–0.80; AUC = 0.95–0.93 in train and validation tests, respectively |
| [38] | pig | Tooth, Bone and Gingiva | Segmentation and 3D reconstruction | Segmentation | CNN | mean accuracy precision (mAP) > 90% |
| [39] | rat | Brain and Whole Body | Improvement of image quality using image fusion (PA + CT) | Image Quality Improvement | 3D-CNN | ↑ static structural quality/dynamic contrast-enhanced whole-body/dynamic functional brain acquisitions |
| [40] | rabbit | Abdominal Artery | Differentiation of MB from tissue on RF signals | MB Localisation | CNN/RNN | ↑CTR and CNR by 22.3 dB and 42.8 dB, respectively |
| [41] | rat | Brain Vasculature | Brain vasculature reconstruction | PD Reconstruction | 3D-CNN | PSNR = 28.8; NMSE = 0.05 and MAE = 0.1193, with an 85% compression factor |
| [42] | rat | Shank Muscle | Segmentation of the shank muscle | Segmentation | CNN | DSC = 94.82% and 90.72% for Gas and Sol muscles, respectively |
| [43] | mouse | Heart Left Ventricle | Segmentation of left ventricle | Segmentation | Deep CNN | time analysis reduction > 92%; Pearson’s r = 0.85–0.99 |
| [44] | rat and rabbit | Colorectum and Urethra | Removing EMI Noise | Image Quality Improvement | CNNs | U-Net modified outperforming in EMI noise removal vs. others |
| [45] | mouse | Heart | Identification and classification of myocardial regions (health/infarction) | Classification | RNN | Precision = 99.6% and 98.7%, AUC = 0.999 and 0.996 on two test sets, respectively |
| [46] | mouse | Breast Tumour Vasculature | Nondestructive detection of adherent MB signatures | MB Localisation | FCN | DSC = 0.45; AUC = 0.90 |
| [47] | pig | Spleen | Classification of splenic trauma | Classification | CNNs | Acc = 0.85; Se = 0.82; Sp = 0.88; PPV = 0.87; NPV = 0.83 |
| [48] | pig | Heart Left Ventricle | Segmentation of left ventricle | Segmentation | CNNs | DSC = 0.90 and 0.91 for U-Net and segAN, respectively |
| [49] | mouse | Brain, Liver and Kidney | Segmentation of whole-body, liver and kidney | Segmentation | CNN | DSC = 0.91/0.96/0.97 for brain/liver/kidney, respectively |
| [50] | rat | Liver | Liver fibrosis assessment by features extraction and integration | Features Extraction | DCNN | Acc = 0.83; Se = 0.82; Sp = 0.84; AUC = 0.87 for several livers fibrosis recognition |
| [51] | rat | Brain Vasculature | MB tracking for mouse brain perfusion | MB Localisation | 3D-CNN | ↑ in resolving 10 µm micro-vessels vs. conventional approach |
| [52] | pig | Femoral Artery | Haemorrhage identification by exploring blood flow anomalies | Anomaly Detection | DCGAN | AUC = 0.90/0.87/0.62 immediately/10 min/30 min post-injury, respectively |
| [53] | rabbit | Liver | Classification of fatty liver state | Classification | CNN | Acc = 74% and 81% in testing and training data, respectively |
| [54] | pig | Heart | Segmentation of the heart during a cardiac arrest | Segmentation | n.a. | Borders’ recognition and tracing in porcine hearts |
| [55] | pig | Tooth | Identification of periodontal structures and assessment of their diagnostic dimensions | Segmentation | CNN | DSC ≥ 90 ± 7.2%; ≥78.6 ± 13.2% and ≥62.6 ± 17.7% in two test sets, for soft tissue, bone, and crown segmentation, respectively |
| [56] | rat | Carotid Artery | Measuring blood flow vessels with high resolution | Blood Flow Measure | CNN | ↑ performance in measuring vascular stiffness and complicated flow–vessel dynamics vs. conventional techniques |
| [57] | mouse | Embryo | 3D Segmentation and classification of embryos in normal/mutant | Segmentation + Classification | 3D-CNN | DSC = 0.924/0.887 for body and BV, respectively |
| [58] | rat | Sentinel Lymph Node Vasculature | Improvement of lateral resolution of PA microscopy | Improvement Image Quality | CNN | ↑ in resolution and signal strength and ↓ in background signal |
| [59] | rat | Brain Vasculature | Improving convergence rate and image reconstruction quality | Pattern Recognition | CNN | ↑ performance of proposed method vs. ResNet |
| [60] | rabbit | Liver | Classification of liver fibrosis stages | Classification | CNN | AUC = 0.82/0.88/0.90; Se = 0.83/0.8/0.83; Sp = 0.66/0.86/0.92; Acc = 0.75/0.84/0.90 for significant fibrosis/advanced fibrosis/cirrhosis, respectively |
| [61] | rabbit | Spine Surface | Segmentation and 3D Reconstruction of spine surface | Segmentation | CNN | overall MAE = 0.24 ± 0.29 mm; MAE ↓ 26.28% and the number of US surface points across the lumbar region ↑ 21.61% |
| [62] | rabbit | Near Rectum | Removing electrical noise from the step motor to reduce scanning time | Improvement Image Quality | CNN | Good denoising |
| [63] | mouse | Brain Vasculature | Image Upsampling | Image Upsampling | FCN | smoother vessel boundaries, ↓ artefacts, more consistent vessel intensity and vessel profile vs. undersampled images |
| Ref | Animal Model | Anatomical District | Aim of Study 1 | DL Network Task 1 | DL Architecture 1 | Main Result 1 |
|---|---|---|---|---|---|---|
| [64] | dog | Left Ventricle | Tracking of left ventricle motion | Segmentation | CNN | good performance in tracking LV concerning conventional methods |
| [65] | pig | Femoral Vein | Detection of catheter tips | Object Detection | CNNs | classification rates of 88.8% and 91.4% and MAE = 0.279 mm and 0.478 mm for linear and phased arrays, respectively |
| [66] | pig | Femoral Vein | Detection of catheter tips | Object Detection | CNN | a classification rate of 91.4% and a misclassification rate of 7.86% |
| [67] | rabbit | Liver | Classification of fatty liver disease stages | Classification | CNN | Acc = 85.48%; Se = 91.52%; Sp = 76.67%; F1-Score = 0.89; Precision = 85.84% |
| [68] | rat | Brain | Visualisation of blood vessels | Improving image quality | DNN | better contrast in vascular visualisation than common methods |
| [69] | mouse | Liver (Hepatocellular Carcinoma) | Nondestructive detection of adherent MBs signatures | MBs detection | FCN | AUC = 0.91 and DSC = 0.56 |
| [70] | mouse | Brain | Detection of microvessel networks | MBs detection | FCN | significant improvement in image reconstruction concerning conventional beamforming methods |
| [71] | pig | Lung | Detection of five lung abnormalities | Classification | CNN | Se and SP > 85% for all features except for B-lines detection |
| [72] | mouse | Brain, Liver and Kidney | Segmentation of whole-body, liver and kidney | Segmentation | CNN | DSC = 0.98/0.96/0.97 for brain/liver/kidney, respectively |
| [73] | pig | Heart | Guidewire segmentation in cardiac intervention | Segmentation | 3D-CNN | MHD = 4.1; DSC = 0.56 |
| [74] | pig | Lung | Pneumothorax detection | Feature extraction | CNN + RNN | Se = 84%; Sp = 82%; AUC = 0.88 |
| [75] | pig | Femoral Artery | Haemorrhage identification by exploring blood flow anomalies | Anomaly Detection | GAN | Sp = 70% and Se = 81–64% immediately and 10 min post-injury, respectively |
| [76] | rabbit | Liver | Classification of fatty liver state | Classification | CNN | Acc = 73% on testing data compared to 60% with conventional QUS |
| [77] | pig | Inferior Vena Cava | Vessel Lumen Segmentation | Segmentation | CNN | DSC = 0.90; TP = 57.80; TN = 31.06; FP = 6.04; FN = 5.11 post-processing |
| [78] | chicken | Embryo | Improvement of image quality | Beamforming | CNN | qualitative improvements in image quality |
| [79] | mouse | Embryo | 3D Segmentation and classification of embryos in normal/mutant | Segmentation + Classification | 3D-CNN | DSC = 0.925/0.896 for body and BV, respectively |
| [80] | mouse | Embryo | 3D Segmentation of embryo brain ventricle | Segmentation | 3D-CNN | DSC = 0.896 in testing |
| [81] | rat | Liver | Classification of liver fibrosis severity (S0–S3) | Classification | RNN | Acc = 87.5/81.3/93.7/87.5%; AUC = 0.90/0.94/0.92/0.93, for S0/S1/S2/S3, respectively |
| [82] | mouse | Embryo | 3D Segmentation of embryos body and brain ventricle | Segmentation | 3D-CNN | DSC = 0.934/0.906 for body and BV, respectively |
| [83] | rat | Heart | Obtaining the position of the Epicardium and Endocardium | Segmentation | CNN | Accuracy from 82.26% to 85.03% by comparing semi-automatic with automatic segmentation method |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Rosa, L.; L’Abbate, S.; Kusmic, C.; Faita, F. Applications of Deep Learning Algorithms to Ultrasound Imaging Analysis in Preclinical Studies on In Vivo Animals. Life 2023, 13, 1759. https://doi.org/10.3390/life13081759
De Rosa L, L’Abbate S, Kusmic C, Faita F. Applications of Deep Learning Algorithms to Ultrasound Imaging Analysis in Preclinical Studies on In Vivo Animals. Life. 2023; 13(8):1759. https://doi.org/10.3390/life13081759
Chicago/Turabian StyleDe Rosa, Laura, Serena L’Abbate, Claudia Kusmic, and Francesco Faita. 2023. "Applications of Deep Learning Algorithms to Ultrasound Imaging Analysis in Preclinical Studies on In Vivo Animals" Life 13, no. 8: 1759. https://doi.org/10.3390/life13081759
APA StyleDe Rosa, L., L’Abbate, S., Kusmic, C., & Faita, F. (2023). Applications of Deep Learning Algorithms to Ultrasound Imaging Analysis in Preclinical Studies on In Vivo Animals. Life, 13(8), 1759. https://doi.org/10.3390/life13081759











