Evaluation of the Precision of Kinetic Stem Cell (KSC) Counting for Specific Quantification of Human Mesenchymal Stem Cells in Heterogeneous Tissue Cell Preparations
Abstract
:1. Introduction
2. Materials and Methods
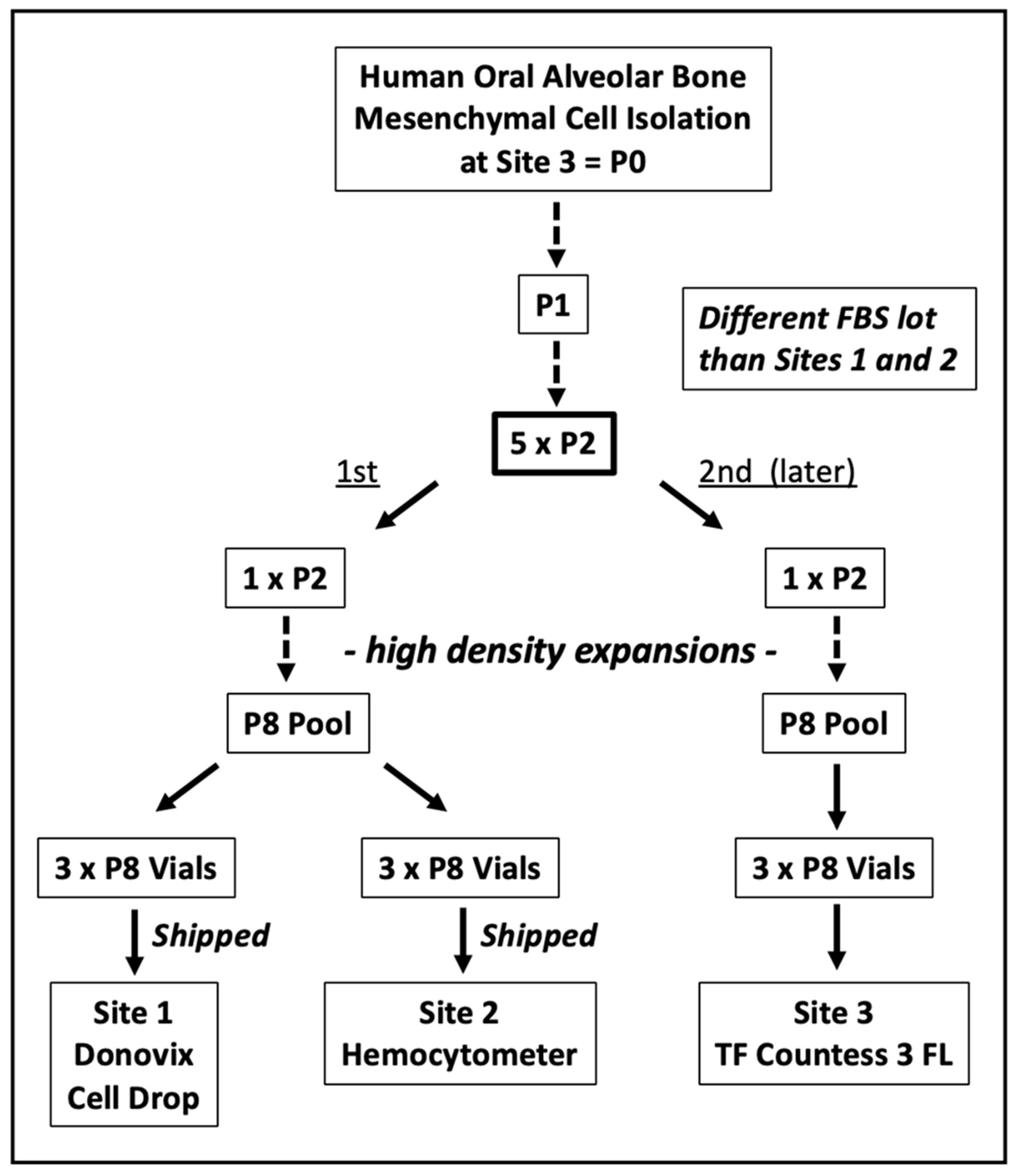
2.1. Oral Alveolar Bone Tissue Cell Derivation
2.2. Serial Cell Culture
2.3. Kinetic Stem Cell Counting Analyses
2.4. Statistical Methods
3. Results
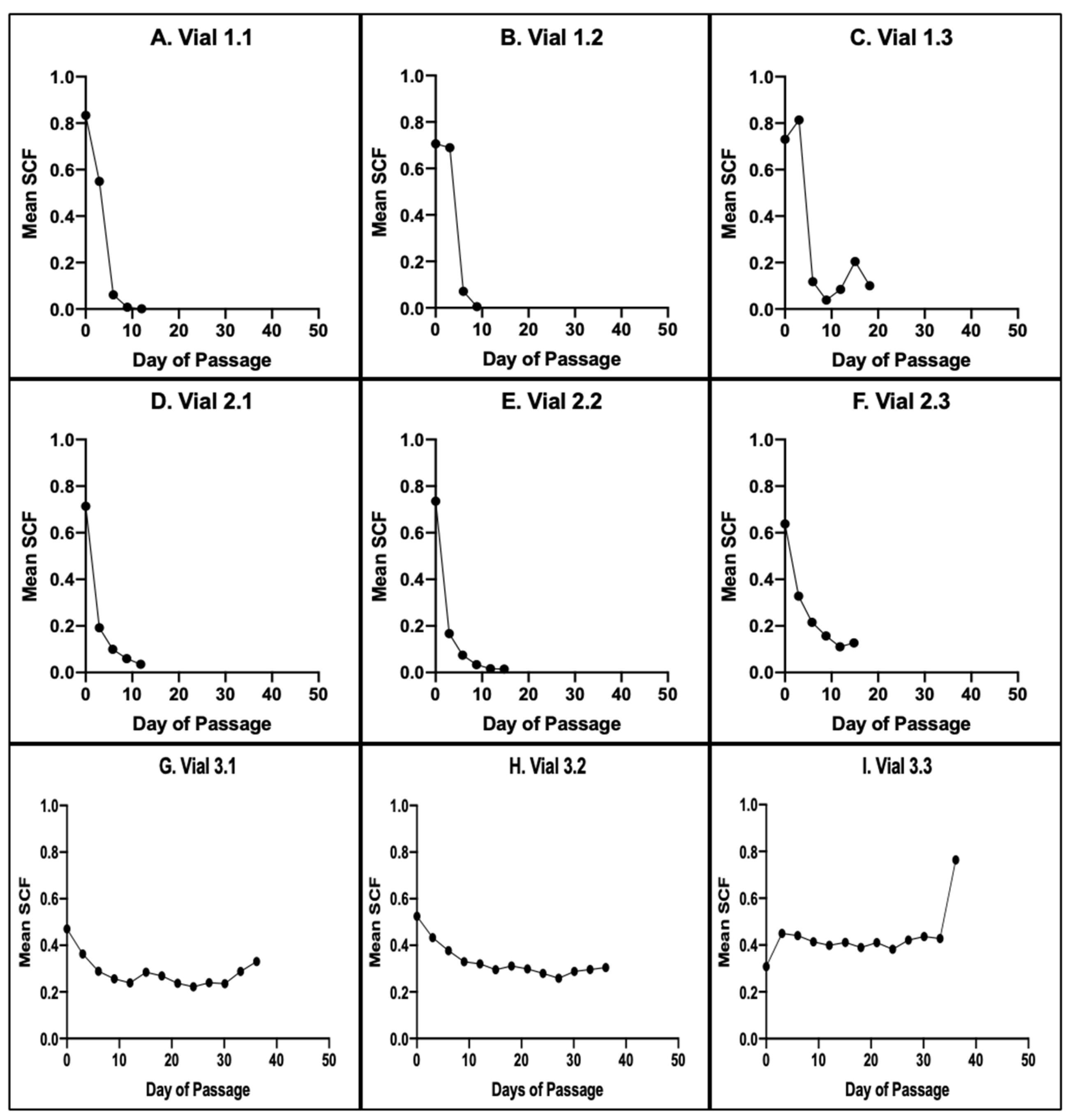
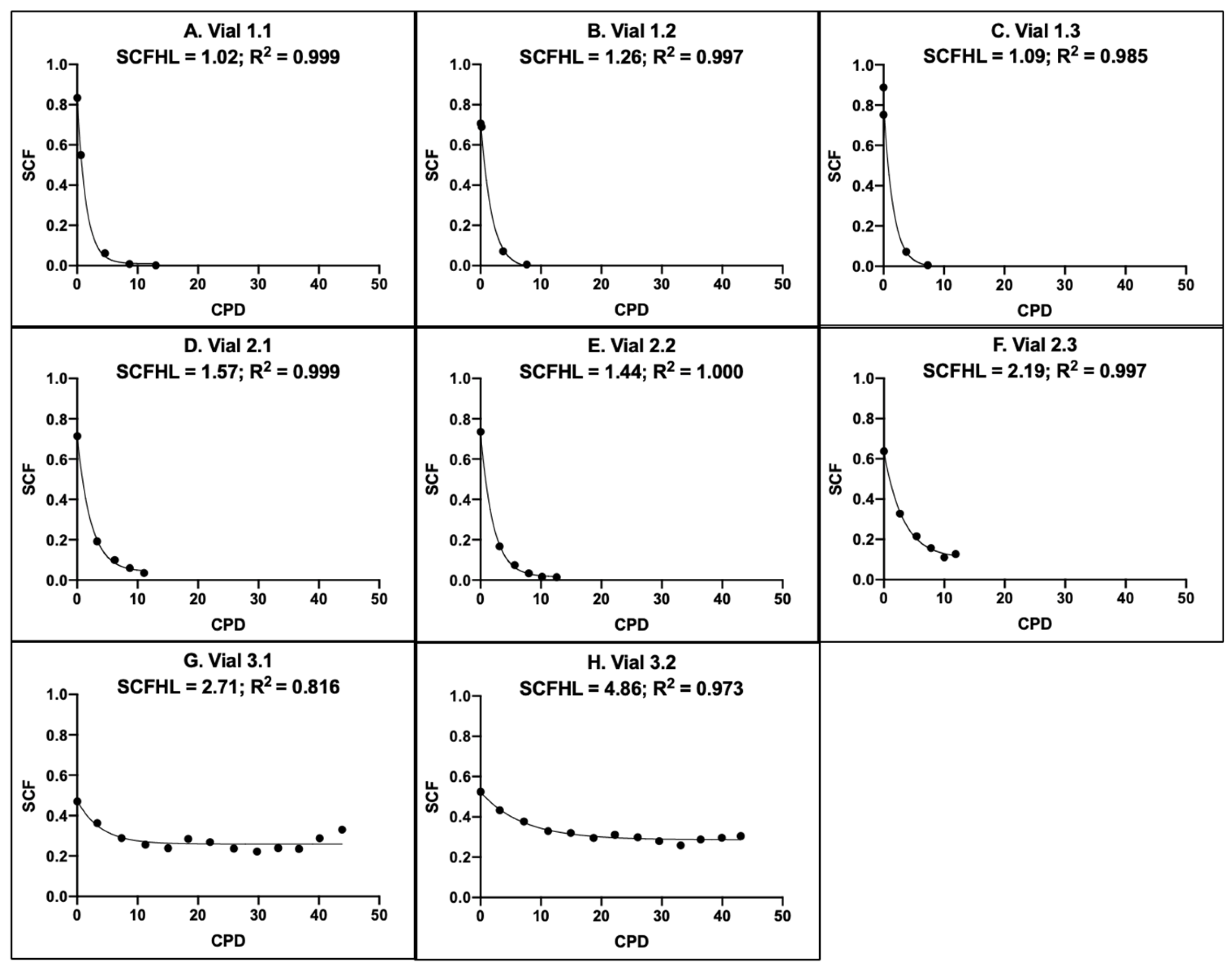
3.1. Intra-Site and Inter-Site Precision of MSC Fraction Determination
3.2. Evaluation of Other Initial Mean Cell Kinetics Factors
3.3. Evaluation of Changes in Cell Subtype-Specific Fractions during Serial Culture
3.4. Evaluation of the Precision of CPD Algorithms for Rapidly Counting Mesenchymal Stem Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dutton, R.; Abdi, F.; Minnetyan, L.; Sherley, J.L. A computational simulation technology for specific counting of perinatal and postnatal human tissue stem cells for transplantation medicine. OBM Transplant. 2020, 4, 24. [Google Scholar] [CrossRef]
- Sherley, J.L. A kinetic stem cell counting analysis of the specific of cell culture medium growth factors on adipose-derived mesenchymal stem cells. Life 2023, 13, 614. [Google Scholar] [CrossRef] [PubMed]
- Purton, L.E.; Scadden, D.T. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell 2007, 1, 263–270. [Google Scholar] [CrossRef] [PubMed]
- McDermott, S.P.; Eppert, K.; Lechman, E.R.; Doedens, M.; Dick, J.E. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood 2010, 116, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Ivanovic, Z. Hematopoietic stem cells in research and clinical applications: The “CD34 issue”. World J. Stem Cells 2010, 2, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Mantri, S.; Reinisch, A.; Dejene, B.T.; Lyell, D.J.; DiGiusto, D.L.; Agarwal-Hashmi, R.; Majeti, R.; Weinberg, K.I.; Porteus, M.H. CD34 expression does not correlate with immunophenotypic stem cell or progenitor content in human cord blood products. Blood Adv. 2020, 4, 5357–5361. [Google Scholar] [CrossRef] [PubMed]
- Olsson, R.; Remberger, M.; Schaffer, M.; Berggren, D.M.; Svahn, B.-M.; Mattsson, J.; Ringden, O. Graft failure in the modern era of allogeneic hematopoietic SCT. Bone Marrow Transp. 2013, 48, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Han, T.; Zhang, Y.; Cheng, Y.; Xu, Z.; Mo, X.; Wang, F.; Yan, C.; Sun, Y.; Wang, J.; et al. The incidence, outcomes, and risk factors of secondary poor graft function in haploidentical hematopoietic stem cell transplantation for acquired aplastic anemia. Front. Immunol. 2022, 9, 896034. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.; Lu, Z.; Yao, X.; Gong, Y.; Yang, T.; Wang, Y. Recent advancements in poor graft function following hematopoietic stem cell transplantation. Front. Immunol. 2022, 2, 911174. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-Q.; Wang, Y.; Zhang, X.-H.; Xu, L.-P.; Liu, K.-L.; Yan, C.-H.; Liu, Z.-Y.; Huang, X.-J. Virus reactivation and low dose of CD34+ cell, rather than haploidentical transplantation, were associated with secondary poor graft function within the first 100 days after allogeneic stem cell transplantation. Ann. Hematol. 2019, 98, 1877–1883. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, J.; Krampera, M. The challenge of defining mesenchymal stromal cell potency assays and their potential use as release criteria. Cytotherapy 2015, 17, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Ketterl, N.; Brachtl, G.; Schuh, C.; Bieback, K.; Schallmoser, K.; Reinisch, A.; Strunk, D. A robust potency assay highlights significant donor variation of human mesenchymal stem/progenitor cell immune modulatory capacity and extended radio-resistance. Stem Cell Res. Ther. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Webster, A.; Genever, P. Nomenclature and heterogeneity: Consequences for the use of mesenchymal stem cells in regenerative medicine. Regen. Med. 2019, 14, 595–611. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.; Tarle, S.; Osibin, W.; Kinfu, Y.; Kaigler, D. Standardization and safety of alveolar bone–derived stem cell isolation. J. Dental Res. 2014, 93, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Hoch, A.I.; Leach, J.K. Concise review: Optimizing expansion of bone marrow mesenchymal stem/stromal cells for clinical applications. Stem Cells Transl. Med. 2014, 3, 643–652, Erratum in: Stem Cells Transl. Med. 2015, 4, 412. [Google Scholar] [CrossRef] [PubMed]
- Moll, G.; Ankrum, J.A.; Olson, S.D.; Nolta, J.A. Improved MSC minimal criteria to maximize patient safety: A call to embrace tissue factor and hemocompatibility assessment of MSC products. Stem Cells Transl. Med. 2022, 11, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Lappin, T.; Cheng, T. An urgent need for standardization of stem cells and stem cell-derived products toward clinical applications. Stem Cells Transl. Med. 2021, 10, S1–S3. [Google Scholar] [CrossRef] [PubMed]
- Bieback, K.; Hecker, A.; Schlechter, T.; Hofmann, I.; Brousos, N.; Redmer, T.; Besser, D.; Klüter, H.; Müller, A.M.; Becker, M. Replicative aging and differentiation potential of human adipose tissue-derived mesenchymal stromal cells expanded in pooled human or fetal bovine serum. Cytotherapy 2012, 14, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Astori, G.; Amati, E.; Bambi, F.; Bernardi, M.; Chieregato, K.; Schäfer, R.; Sella, S.; Rodeghiero, F. Platelet lysate as a substitute for animal serum for the ex-vivo expansion of mesenchymal stem/stromal cells: Present and future. Stem Cell Res. Ther. 2016, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Guiotto, M.; Raffoul, W.; Hart, A.M.; Riehle, M.O.; di Summa, P.G. Human platelet lysate to substitute fetal bovine serum in hMSC expansion for translational applications: A systematic review. J. Transl. Med. 2020, 18, 351. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chopra, H.; Daley, M.P.; Kumar, A.; Sugai, J.; Dahlkemper, A.; Kaigler, D.; Sherley, J.L. Evaluation of the Precision of Kinetic Stem Cell (KSC) Counting for Specific Quantification of Human Mesenchymal Stem Cells in Heterogeneous Tissue Cell Preparations. Life 2024, 14, 51. https://doi.org/10.3390/life14010051
Chopra H, Daley MP, Kumar A, Sugai J, Dahlkemper A, Kaigler D, Sherley JL. Evaluation of the Precision of Kinetic Stem Cell (KSC) Counting for Specific Quantification of Human Mesenchymal Stem Cells in Heterogeneous Tissue Cell Preparations. Life. 2024; 14(1):51. https://doi.org/10.3390/life14010051
Chicago/Turabian StyleChopra, Hitesh, Michael P. Daley, Adhya Kumar, James Sugai, Alex Dahlkemper, Darnell Kaigler, and James L. Sherley. 2024. "Evaluation of the Precision of Kinetic Stem Cell (KSC) Counting for Specific Quantification of Human Mesenchymal Stem Cells in Heterogeneous Tissue Cell Preparations" Life 14, no. 1: 51. https://doi.org/10.3390/life14010051
APA StyleChopra, H., Daley, M. P., Kumar, A., Sugai, J., Dahlkemper, A., Kaigler, D., & Sherley, J. L. (2024). Evaluation of the Precision of Kinetic Stem Cell (KSC) Counting for Specific Quantification of Human Mesenchymal Stem Cells in Heterogeneous Tissue Cell Preparations. Life, 14(1), 51. https://doi.org/10.3390/life14010051





