The Microbiota–Gut–Brain Axis and Neurological Disorders: A Comprehensive Review
Abstract
:1. Introduction
2. The Bidirectional Relationships between MGBA and Neurological Disorders
2.1. The MGBA and Developmental Disabilities and Metabolic Disorders
2.1.1. Rett Syndrome
2.1.2. ASD
2.1.3. ADHD
The MGBA and NDDs
2.1.4. NDD with Cognitive Syndrome
AD
- Gut–brain link to childhood dementia
- 2.
- The bidirectional communication between AD and gut microbiota
- 3.
- Apolipoprotein E (APOE) influence the composition of the gut microbiota
Non-Alzheimer’s Neurodegeneration and Dementias
FTLD
Prion Disease (Creutzfeldt–Jakob Disease)
2.1.5. NDD with Movement Disorders
WD
2.1.6. Neurodegeneration with Cognitive and Movement Syndromes
MSA
Huntington’s Chorea (HC)
PD
2.2. Immune-Mediated Nervous System Diseases
2.2.1. MS
The Function of Astrocytes in MS
Pathological Alterations of Synaptic Structure and Function in MS
MS and the Commensal Microbiota
2.2.2. ALS
2.3. Non-Communicable Neurological Disorders
TLE
2.4. Mental (Behavioural) Disorders
2.4.1. Depression
2.4.2. SCZ
3. Therapeutics and Their Bidirectional Correlation with the Gut–Brain Axis
3.1. Psychotropic Agents
3.1.1. Can the Gut Microbiome Be the New Marker for Safety and Efficacy of Neuro/Psychotropic Drugs?
Psychotropic Agents and Gut Microbiome
- 1.
- Antidepressants
- 2.
- Antipsychotics
Neurologic Drugs and Gut Microbiome
- 1.
- AD pharmacotherapies
- 2.
- Parkinson’s disease pharmacotherapy
Antiseizure Medications (ASMs)
3.1.2. Can Interventions Replenish Gut Microbiome Alter Response to CNS Drugs?
3.1.3. The Effect of Antibiotic-Induced Dysbiosis
4. Other Factors Influencing MGBA
4.1. Mode of Delivery
4.2. Exercise
4.3. Stress
4.4. Genetics and Epigenetics
Evidence on the Genetic Links between GI and Neuropsychiatric Conditions: Common Genetic Determinants to Common Therapies?
5. Conclusions
6. Future and Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ADHD | Attention-deficit/hyperactivity disorder |
| ALS | Amyotrophic lateral sclerosis |
| APOE | Apolipoprotein E |
| ASD | Autism spectrum disorder |
| ASMs | Antiseizure medications |
| BBB | Blood–brain barrier |
| BDNF | Brain-derived neurotrophic factor |
| CJD | Creutzfeldt–Jakob disease |
| CNS | Central nervous system |
| C-sections | Caesarean section |
| ENS | Enteric nervous system |
| FMT | Faecal microbiota transplantation |
| FTLD | Fronto-temporal lobe Dementia |
| GABA | Gamma-aminobutyric acid |
| GF | Germ-free |
| GI | Gastrointestinal |
| HPA-axis | Hypothalamic-pituitary-adrenal axis |
| IL | Interleukin |
| MDD | Major depressive disorder |
| MGBA | Microbiota–gut–brain axis |
| MHC | Major histocompatibility complex |
| MS | Multiple sclerosis |
| MSA | Multisystem atrophy |
| NDDs | Neurodegenerative diseases |
| PD | Parkinson’s disease |
| PFC | Prefrontal cortex |
| SCFAs | Short-chain fatty acids |
| SCZ | Schizophrenia |
| SGA | Second-generation antipsychotics |
| TLE | Temporal lobe epilepsy |
| WD | Wilson–Konovalov disease |
References
- Rhee, S.H.; Pothoulakis, C.; Mayer, E.A. Principles and Clinical Implications of the Brain-Gut-Enteric Microbiota Axis. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Mahony, S.M. The Microbiome-Gut-Brain Axis: From Bowel to Behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Collins, S.M.; Bercik, P. The Microbiota-Gut-Brain Axis in Functional Gastrointestinal Disorders. Gut Microbes 2014, 5, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.; Kubo, C.; Koga, Y. Postnatal Microbial Colonization Programs the Hypothalamic–Pituitary–Adrenal System for Stress Response in Mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Wine, E.; Rodrigues, D.M.; Cho, J.H.; Whary, M.T.; Philpott, D.J.; MacQueen, G.; Sherman, P.M. Bacterial Infection Causes Stress-Induced Memory Dysfunction in Mice. Gut 2011, 60, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, K.M.; Kang, N.; Bienenstock, J.; Foster, J.A. Reduced Anxiety-like Behavior and Central Neurochemical Change in Germ-Free Mice: Behavior in Germ-Free Mice. Neurogastroenterol. Motil. 2011, 23, 255-e119. [Google Scholar] [CrossRef]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The Microbiome-Gut-Brain Axis during Early Life Regulates the Hippocampal Serotonergic System in a Sex-Dependent Manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef]
- Bercik, P.; Denou, E.; Collins, J.; Jackson, W.; Lu, J.; Jury, J.; Deng, Y.; Blennerhassett, P.; Macri, J.; McCoy, K.D.; et al. The Intestinal Microbiota Affect Central Levels of Brain-Derived Neurotropic Factor and Behavior in Mice. Gastroenterology 2011, 141, 599–609.e3. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus Strain Regulates Emotional Behavior and Central GABA Receptor Expression in a Mouse via the Vagus Nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef]
- Savignac, H.M.; Kiely, B.; Dinan, T.G.; Cryan, J.F. B Ifidobacteria Exert Strain-specific Effects on Stress-related Behavior and Physiology in BALB/c Mice. Neurogastroenterol. Motil. 2014, 26, 1615–1627. [Google Scholar] [CrossRef]
- Desbonnet, L.; Clarke, G.; Traplin, A.; O’Sullivan, O.; Crispie, F.; Moloney, R.D.; Cotter, P.D.; Dinan, T.G.; Cryan, J.F. Gut Microbiota Depletion from Early Adolescence in Mice: Implications for Brain and Behaviour. Brain Behav. Immun. 2015, 48, 165–173. [Google Scholar] [CrossRef]
- Bharwani, A.; Mian, M.F.; Surette, M.G.; Bienenstock, J.; Forsythe, P. Oral Treatment with Lactobacillus Rhamnosus Attenuates Behavioural Deficits and Immune Changes in Chronic Social Stress. BMC Med. 2017, 15, 7. [Google Scholar] [CrossRef]
- Lyte, M.; Varcoe, J.J.; Bailey, M.T. Anxiogenic Effect of Subclinical Bacterial Infection in Mice in the Absence of Overt Immune Activation. Physiol. Behav. 1998, 65, 63–68. [Google Scholar] [CrossRef]
- O’Mahony, S.M.; Felice, V.D.; Nally, K.; Savignac, H.M.; Claesson, M.J.; Scully, P.; Woznicki, J.; Hyland, N.P.; Shanahan, F.; Quigley, E.M.; et al. Disturbance of the Gut Microbiota in Early-Life Selectively Affects Visceral Pain in Adulthood without Impacting Cognitive or Anxiety-Related Behaviors in Male Rats. Neuroscience 2014, 277, 885–901. [Google Scholar] [CrossRef]
- Guzzetta, K.E.; Cryan, J.F.; O’Leary, O.F. Microbiota-Gut-Brain Axis Regulation of Adult Hippocampal Neurogenesis. Brain Plast. 2022, 8, 97–119. [Google Scholar] [CrossRef]
- Kasarello, K.; Cudnoch-Jedrzejewska, A.; Czarzasta, K. Communication of Gut Microbiota and Brain via Immune and Neuroendocrine Signaling. Front. Microbiol. 2023, 14, 1118529. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Peng, J.; Gerner, S.T.; Yin, S.; Jiang, Y. Gut Microbiota-Brain Interaction: An Emerging Immunotherapy for Traumatic Brain Injury. Exp. Neurol. 2021, 337, 113585. [Google Scholar] [CrossRef]
- Loh, J.S.; Mak, W.Q.; Tan, L.K.S.; Ng, C.X.; Chan, H.H.; Yeow, S.H.; Foo, J.B.; Ong, Y.S.; How, C.W.; Khaw, K.Y. Microbiota-Gut-Brain Axis and Its Therapeutic Applications in Neurodegenerative Diseases. Signal Transduct. Target. Ther. 2024, 9, 37. [Google Scholar] [CrossRef]
- Sorboni, S.G.; Moghaddam, H.S.; Jafarzadeh-Esfehani, R.; Soleimanpour, S. A Comprehensive Review on the Role of the Gut Microbiome in Human Neurological Disorders. Clin. Microbiol. Rev. 2022, 35, e0033820. [Google Scholar] [CrossRef]
- Socała, K.; Doboszewska, U.; Szopa, A.; Serefko, A.; Włodarczyk, M.; Zielińska, A.; Poleszak, E.; Fichna, J.; Wlaź, P. The Role of Microbiota-Gut-Brain Axis in Neuropsychiatric and Neurological Disorders. Pharmacol. Res. 2021, 172, 105840. [Google Scholar] [CrossRef]
- Góralczyk-Bińkowska, A.; Szmajda-Krygier, D.; Kozłowska, E. The Microbiota-Gut-Brain Axis in Psychiatric Disorders. Int. J. Mol. Sci. 2022, 23, 11245. [Google Scholar] [CrossRef]
- Zigmond, M.J.; Wiley, C.A.; Chesselet, M.F. (Eds.) Neurobiology of Brain Disorders: Biological Basis of Neurological and Psychiatric Disorders, 2nd ed.; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Ip, J.P.K.; Mellios, N.; Sur, M. Rett Syndrome: Insights into Genetic, Molecular and Circuit Mechanisms. Nat. Rev. Neurosci. 2018, 19, 368–382. [Google Scholar] [CrossRef]
- Morello, N.; Schina, R.; Pilotto, F.; Phillips, M.; Melani, R.; Plicato, O.; Pizzorusso, T.; Pozzo-Miller, L.; Giustetto, M. Loss of Mecp2 Causes Atypical Synaptic and Molecular Plasticity of Parvalbumin-Expressing Interneurons Reflecting Rett Syndrome-Like Sensorimotor Defects. eNeuro 2018, 5, ENEURO.0086-18.2018. [Google Scholar] [CrossRef]
- Borghi, E.; Borgo, F.; Severgnini, M.; Savini, M.N.; Casiraghi, M.C.; Vignoli, A. Rett Syndrome: A Focus on Gut Microbiota. Int. J. Mol. Sci. 2017, 18, 344. [Google Scholar] [CrossRef]
- Shulyakova, N.; Andreazza, A.C.; Mills, L.R.; Eubanks, J.H. Mitochondrial Dysfunction in the Pathogenesis of Rett Syndrome: Implications for Mitochondria-Targeted Therapies. Front. Cell Neurosci. 2017, 11, 58. [Google Scholar] [CrossRef]
- Musokhranova, U.; Grau, C.; Vergara, C.; Rodríguez-Pascau, L.; Xiol, C.; Castells, A.A.; Alcántara, S.; Rodríguez-Pombo, P.; Pizcueta, P.; Martinell, M.; et al. Mitochondrial Modulation with Leriglitazone as a Potential Treatment for Rett Syndrome. J. Transl. Med. 2023, 21, 756. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Pindo, M.; Renzi, D.; et al. Altered Gut Microbiota in Rett Syndrome. Microbiome 2016, 4, 41. [Google Scholar] [CrossRef]
- Thapa, S.; Venkatachalam, A.; Khan, N.; Naqvi, M.; Balderas, M.; Runge, J.K.; Haag, A.; Hoch, K.M.; Glaze, D.G.; Luna, R.A.; et al. Assessment of the Gut Bacterial Microbiome and Metabolome of Girls and Women with Rett Syndrome. PLoS ONE 2021, 16, e0251231. [Google Scholar] [CrossRef]
- Caputi, V.; Hill, L.; Figueiredo, M.; Popov, J.; Hartung, E.; Margolis, K.G.; Baskaran, K.; Joharapurkar, P.; Moshkovich, M.; Pai, N. Functional Contribution of the Intestinal Microbiome in Autism Spectrum Disorder, Attention Deficit Hyperactivity Disorder, and Rett Syndrome: A Systematic Review of Pediatric and Adult Studies. Front. Neurosci. 2024, 18, 1341656. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/autism/about/?CDC_AAref_Val=https://www.cdc.gov/ncbddd/autism/facts.html (accessed on 19 June 2024).
- Hashem, S.; Nisar, S.; Bhat, A.A.; Yadav, S.K.; Azeem, M.W.; Bagga, P.; Fakhro, K.; Reddy, R.; Frenneaux, M.P.; Haris, M. Genetics of Structural and Functional Brain Changes in Autism Spectrum Disorder. Transl. Psychiatry 2020, 10, 229. [Google Scholar] [CrossRef]
- Taniya, M.A.; Chung, H.-J.; Al Mamun, A.; Alam, S.; Aziz, A.; Emon, N.U.; Islam, M.; Hong, S.-T.S.; Podder, B.R.; Mimi, A.A.; et al. Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front. Cell. Infect. Microbiol. 2022, 12, 915701. [Google Scholar] [CrossRef]
- Rubia, K. Neuro-Anatomic Evidence for the Maturational Delay Hypothesis of ADHD. Proc. Natl. Acad. Sci. USA 2007, 104, 19663–19664. [Google Scholar] [CrossRef]
- Shaw, P.; Eckstrand, K.; Sharp, W.; Blumenthal, J.; Lerch, J.P.; Greenstein, D.; Clasen, L.; Evans, A.; Giedd, J.; Rapoport, J.L. Attention-Deficit/Hyperactivity Disorder Is Characterized by a Delay in Cortical Maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 19649–19654. [Google Scholar] [CrossRef]
- Vaidya, C.J. Neurodevelopmental Abnormalities in ADHD. In Behavioral Neuroscience of Attention Deficit Hyperactivity Disorder and Its Treatment; Stanford, C., Tannock, R., Eds.; Current Topics in Behavioral Neurosciences; Springer: Berlin/Heidelberg, Germany, 2011; Volume 9, pp. 49–66. ISBN 978-3-642-24611-1. [Google Scholar]
- Dark, C.; Homman-Ludiye, J.; Bryson-Richardson, R.J. The Role of ADHD Associated Genes in Neurodevelopment. Dev. Biol. 2018, 438, 69–83. [Google Scholar] [CrossRef]
- Shaw, P.; Lerch, J.; Greenstein, D.; Sharp, W.; Clasen, L.; Evans, A.; Giedd, J.; Castellanos, F.X.; Rapoport, J. Longitudinal Mapping of Cortical Thickness and Clinical Outcome in Children and Adolescents with Attention-Deficit/Hyperactivity Disorder. Arch. Gen. Psychiatry 2006, 63, 540. [Google Scholar] [CrossRef]
- Ystrom, E.; Gustavson, K.; Brandlistuen, R.E.; Knudsen, G.P.; Magnus, P.; Susser, E.; Davey Smith, G.; Stoltenberg, C.; Surén, P.; Håberg, S.E.; et al. Prenatal Exposure to Acetaminophen and Risk of ADHD. Pediatrics 2017, 140, e20163840. [Google Scholar] [CrossRef]
- Pärtty, A.; Kalliomäki, M.; Wacklin, P.; Salminen, S.; Isolauri, E. A Possible Link between Early Probiotic Intervention and the Risk of Neuropsychiatric Disorders Later in Childhood: A Randomized Trial. Pediatr. Res. 2015, 77, 823–828. [Google Scholar] [CrossRef]
- Checa-Ros, A.; Jeréz-Calero, A.; Molina-Carballo, A.; Campoy, C.; Muñoz-Hoyos, A. Current Evidence on the Role of the Gut Microbiome in ADHD Pathophysiology and Therapeutic Implications. Nutrients 2021, 13, 249. [Google Scholar] [CrossRef]
- Dolina, S.; Margalit, D.; Malitsky, S.; Rabinkov, A. Attention-Deficit Hyperactivity Disorder (ADHD) as a Pyridoxine-Dependent Condition: Urinary Diagnostic Biomarkers. Med. Hypotheses 2014, 82, 111–116. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the Human Large Intestine: Its Physiologic Consequences and the Potential Contribution of Prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [PubMed]
- Prehn-Kristensen, A.; Zimmermann, A.; Tittmann, L.; Lieb, W.; Schreiber, S.; Baving, L.; Fischer, A. Reduced Microbiome Alpha Diversity in Young Patients with ADHD. PLoS ONE 2018, 13, e0200728. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Yang, C.-Y.; Chou, W.-J.; Lee, M.-J.; Chou, M.-C.; Kuo, H.-C.; Yeh, Y.-M.; Lee, S.-Y.; Huang, L.-H.; Li, S.-C. Gut Microbiota and Dietary Patterns in Children with Attention-Deficit/Hyperactivity Disorder. Eur. Child. Adolesc. Psychiatry 2020, 29, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhou, Y.; Zhou, G.; Li, Y.; Yuan, J.; Li, X.; Ruan, B. Gut Microbiota Profiles in Treatment-Naïve Children with Attention Deficit Hyperactivity Disorder. Behav. Brain Res. 2018, 347, 408–413. [Google Scholar] [CrossRef]
- Szopinska-Tokov, J.; Dam, S.; Naaijen, J.; Konstanti, P.; Rommelse, N.; Belzer, C.; Buitelaar, J.; Franke, B.; Aarts, E.; Arias Vasquez, A. Investigating the Gut Microbiota Composition of Individuals with Attention-Deficit/Hyperactivity Disorder and Association with Symptoms. Microorganisms 2020, 8, 406. [Google Scholar] [CrossRef]
- Wan, L.; Ge, W.-R.; Zhang, S.; Sun, Y.-L.; Wang, B.; Yang, G. Case-Control Study of the Effects of Gut Microbiota Composition on Neurotransmitter Metabolic Pathways in Children with Attention Deficit Hyperactivity Disorder. Front. Neurosci. 2020, 14, 127. [Google Scholar] [CrossRef]
- Stevens, A.J.; Purcell, R.V.; Darling, K.A.; Eggleston, M.J.F.; Kennedy, M.A.; Rucklidge, J.J. Human Gut Microbiome Changes during a 10 Week Randomised Control Trial for Micronutrient Supplementation in Children with Attention Deficit Hyperactivity Disorder. Sci. Rep. 2019, 9, 10128. [Google Scholar] [CrossRef]
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer Disease. Nat. Rev. Dis. Primers 2021, 7, 33. [Google Scholar] [CrossRef]
- Rajan, K.B.; Weuve, J.; Barnes, L.L.; McAninch, E.A.; Wilson, R.S.; Evans, D.A. Population Estimate of People with Clinical Alzheimer’s Disease and Mild Cognitive Impairment in the United States (2020–2060). Alzheimer’s Dement. 2021, 17, 1966–1975. [Google Scholar] [CrossRef]
- Migliore, L.; Coppedè, F. Gene–Environment Interactions in Alzheimer Disease: The Emerging Role of Epigenetics. Nat. Rev. Neurol. 2022, 18, 643–660. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, H.; Li, P.; Peng, Y.; Cui, P.; Miao, F.; Zhang, Y.; Zhang, A.; Zhang, J. MHC Class I Molecules and PirB Shape Neuronal Morphology by Affecting the Dendritic Arborization of Cortical Neurons. Neurochem. Res. 2019, 44, 312–322. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Xiayu, X.; Shi, C.; Chen, W.; Song, N.; Fu, X.; Zhou, R.; Xu, Y.-F.; Huang, L.; et al. Altered Gut Microbiota in a Mouse Model of Alzheimer’s Disease. JAD 2017, 60, 1241–1257. [Google Scholar] [CrossRef]
- Kim, M.-S.; Kim, Y.; Choi, H.; Kim, W.; Park, S.; Lee, D.; Kim, D.K.; Kim, H.J.; Choi, H.; Hyun, D.-W.; et al. Transfer of a Healthy Microbiota Reduces Amyloid and Tau Pathology in an Alzheimer’s Disease Animal Model. Gut 2020, 69, 283–294. [Google Scholar] [CrossRef]
- D’Amato, A.; Di Cesare Mannelli, L.; Lucarini, E.; Man, A.L.; Le Gall, G.; Branca, J.J.V.; Ghelardini, C.; Amedei, A.; Bertelli, E.; Regoli, M.; et al. Faecal Microbiota Transplant from Aged Donor Mice Affects Spatial Learning and Memory via Modulating Hippocampal Synaptic Plasticity- and Neurotransmission-Related Proteins in Young Recipients. Microbiome 2020, 8, 140. [Google Scholar] [CrossRef]
- Wang, M.; Cao, J.; Gong, C.; Amakye, W.K.; Yao, M.; Ren, J. Exploring the Microbiota-Alzheimer’s Disease Linkage Using Short-Term Antibiotic Treatment Followed by Fecal Microbiota Transplantation. Brain Behav. Immun. 2021, 96, 227–238. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. APOE and Alzheimer’s Disease: Advances in Genetics, Pathophysiology, and Therapeutic Approaches. Lancet Neurol. 2021, 20, 68–80. [Google Scholar] [CrossRef]
- Koutsodendris, N.; Nelson, M.R.; Rao, A.; Huang, Y. Apolipoprotein E and Alzheimer’s Disease: Findings, Hypotheses, and Potential Mechanisms. Annu. Rev. Pathol. 2022, 17, 73–99. [Google Scholar] [CrossRef]
- Raulin, A.-C.; Doss, S.V.; Trottier, Z.A.; Ikezu, T.C.; Bu, G.; Liu, C.-C. ApoE in Alzheimer’s Disease: Pathophysiology and Therapeutic Strategies. Mol. Neurodegener. 2022, 17, 72. [Google Scholar] [CrossRef]
- Luo, Y.-X.; Yang, L.-L.; Yao, X.-Q. Gut Microbiota-Host Lipid Crosstalk in Alzheimer’s Disease: Implications for Disease Progression and Therapeutics. Mol. Neurodegener. 2024, 19, 35. [Google Scholar] [CrossRef]
- Tran, T.T.T.; Corsini, S.; Kellingray, L.; Hegarty, C.; Le Gall, G.; Narbad, A.; Müller, M.; Tejera, N.; O’Toole, P.W.; Minihane, A.-M.; et al. APOE Genotype Influences the Gut Microbiome Structure and Function in Humans and Mice: Relevance for Alzheimer’s Disease Pathophysiology. FASEB J. 2019, 33, 8221–8231. [Google Scholar] [CrossRef]
- Demirci, M.; Kadirhan, O.A. What We Have Learned to Date from the Omics Approach to Non-Alzheimer’s Dementias. J. Integr. Neurosci. 2022, 21, 76. [Google Scholar] [CrossRef] [PubMed]
- Grochowska, M.; Laskus, T.; Radkowski, M. Gut Microbiota in Neurological Disorders. Arch. Immunol. Et Ther. Exp. 2019, 67, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Tyler Patterson, T.; Grandhi, R. Gut Microbiota and Neurologic Diseases and Injuries. In Gut Microbiota and Pathogenesis of Organ Injury; Springer: Berlin/Heidelberg, Germany, 2020; pp. 73–91. [Google Scholar]
- Chunchai, T.; Thunapong, W.; Yasom, S.; Wanchai, K.; Eaimworawuthikul, S.; Metzler, G.; Lungkaphin, A.; Pongchaidecha, A.; Sirilun, S.; Chaiyasut, C.; et al. Decreased Microglial Activation through Gut-Brain Axis by Prebiotics, Probiotics, or Synbiotics Effectively Restored Cognitive Function in Obese-Insulin Resistant Rats. J. Neuroinflamm. 2018, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Tremlett, H.; Bauer, K.C.; Appel-Cresswell, S.; Finlay, B.B.; Waubant, E. The Gut Microbiome in Human Neurological Disease: A Review. Ann. Neurol. 2017, 81, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Suganya, K.; Koo, B.-S. Gut–Brain Axis: Role of Gut Microbiota on Neurological Disorders and How Probiotics/Prebiotics Beneficially Modulate Microbial and Immune Pathways to Improve Brain Functions. Int. J. Mol. Sci. 2020, 21, 7551. [Google Scholar] [CrossRef]
- Chen, C.-H.; Lin, C.-L.; Kao, C.-H. Irritable Bowel Syndrome Is Associated with an Increased Risk of Dementia: A Nationwide Population-Based Study. PLoS ONE 2016, 11, e0144589. [Google Scholar] [CrossRef]
- Saji, N.; Murotani, K.; Hisada, T.; Kunihiro, T.; Tsuduki, T.; Sugimoto, T.; Kimura, A.; Niida, S.; Toba, K.; Sakurai, T. Relationship between Dementia and Gut Microbiome-Associated Metabolites: A Cross-Sectional Study in Japan. Sci. Rep. 2020, 10, 8088. [Google Scholar] [CrossRef]
- Roth, S.C. What Is Genomic Medicine? J. Med. Libr. Assoc. JMLA 2019, 107, 442. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Z.; Shi, J.; An, Y.; Zhang, K.; Wang, Y.; Li, S.; Jin, L.; Ye, W.; Cui, M.; et al. Metabolomics in the Development and Progression of Dementia: A Systematic Review. Front. Neurosci. 2019, 13, 343. [Google Scholar] [CrossRef]
- Westfall, S.; Lomis, N.; Kahouli, I.; Dia, S.Y.; Singh, S.P.; Prakash, S. Microbiome, Probiotics and Neurodegenerative Diseases: Deciphering the Gut Brain Axis. Cell. Mol. Life Sci. 2017, 74, 3769–3787. [Google Scholar] [CrossRef]
- Rosario, D.; Boren, J.; Uhlen, M.; Proctor, G.; Aarsland, D.; Mardinoglu, A.; Shoaie, S. Systems Biology Approaches to Understand the Host–Microbiome Interactions in Neurodegenerative Diseases. Front. Neurosci. 2020, 14, 716. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Alam, M.T.; Dey, J.; Sasidharan, B.C.P.; Ray, U.; Srivastava, A.K.; Gandhi, S.; Tripathi, P.P. Healthy Gut, Healthy Brain: The Gut Microbiome in Neurodegenerative Disorders. Curr. Top. Med. Chem. 2020, 20, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, S.; Kim, J.Y.; Roh, J.H.; Kee, B.S.; An, H.; Choi, B. [P3–430]: Gastrointestinal Microbiome Status of Early Onset Alzheimer’s Disease (EOAD) and Frontotemporal Lobar Degeneration (FTLD) Subjects. Alzheimer’s Dement. 2017, 13, P1132–P1133. [Google Scholar] [CrossRef]
- Ji, D.; Chen, W.-Z.; Zhang, L.; Zhang, Z.-H.; Chen, L.-J. Gut Microbiota, Circulating Cytokines and Dementia: A Mendelian Randomization Study. J. Neuroinflamm. 2024, 21, 2. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-Y.; Kim, J.-K.; Yun, S.-W.; Han, M.J.; Kim, D.-H. DW2009 Elevates the Efficacy of Donepezil against Cognitive Impairment in Mice. Nutrients 2021, 13, 3273. [Google Scholar] [CrossRef] [PubMed]
- Cammann, D.; Lu, Y.; Cummings, M.J.; Zhang, M.L.; Cue, J.M.; Do, J.; Ebersole, J.; Chen, X.; Oh, E.C.; Cummings, J.L.; et al. Genetic Correlations between Alzheimer’s Disease and Gut Microbiome Genera. Sci. Rep. 2023, 13, 5258. [Google Scholar] [CrossRef]
- Amir, I.; Bouvet, P.; Legeay, C.; Gophna, U.; Weinberger, A. Eisenbergiella tayi gen. nov., sp. nov., isolated from human blood. Int. J. Syst. Evol. Microbiol. 2014, 64, 907–914. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef]
- Sikorska, B.; Knight, R.; Ironside, J.W.; Liberski, P.P. Creutzfeldt-Jakob Disease. Adv. Exp. Med. Biol. 2012, 724, 76–90. [Google Scholar] [CrossRef]
- D’Argenio, V.; Sarnataro, D. Microbiome Influence in the Pathogenesis of Prion and Alzheimer’s Diseases. Int. J. Mol. Sci. 2019, 20, 4704. [Google Scholar] [CrossRef]
- Mossad, O.; Erny, D. The Microbiota–Microglia Axis in Central Nervous System Disorders. Brain Pathol. 2020, 30, 1159–1177. [Google Scholar] [CrossRef] [PubMed]
- Doroszkiewicz, J.; Groblewska, M.; Mroczko, B. The Role of Gut Microbiota and Gut–Brain Interplay in Selected Diseases of the Central Nervous System. Int. J. Mol. Sci. 2021, 22, 10028. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, S.; Kalam, N.; Rashid, S.; Bano, G. Effects of Gut Microbiota on Neurodegenerative Diseases. Front. Aging Neurosci. 2023, 15, 1145241. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Huo, Y.-J.; Li, Y.; Han, Y.; Zhou, D. Gut-Brain Axis: Focus on Gut Metabolites Short-Chain Fatty Acids. World J. Clin. Cases 2022, 10, 1754. [Google Scholar] [CrossRef] [PubMed]
- Salim, S.; Ahmad, F.; Banu, A.; Mohammad, F. Gut Microbiome and Parkinson’s Disease: Perspective on Pathogenesis and Treatment. J. Adv. Res. 2023, 50, 83–105. [Google Scholar] [CrossRef]
- Stein, P.S.; Steffen, M.J.; Smith, C.; Jicha, G.; Ebersole, J.L.; Abner, E.; Dawson, D., III. Serum Antibodies to Periodontal Pathogens Are a Risk Factor for Alzheimer’s Disease. Alzheimer’s Dement. 2012, 8, 196–203. [Google Scholar] [CrossRef]
- Bairamian, D.; Sha, S.; Rolhion, N.; Sokol, H.; Dorothée, G.; Lemere, C.A.; Krantic, S. Microbiota in Neuroinflammation and Synaptic Dysfunction: A Focus on Alzheimer’s Disease. Mol. Neurodegener. 2022, 17, 19. [Google Scholar] [CrossRef]
- O’Riordan, K.J.; Collins, M.K.; Moloney, G.M.; Knox, E.G.; Aburto, M.R.; Fülling, C.; Morley, S.J.; Clarke, G.; Schellekens, H.; Cryan, J.F. Short Chain Fatty Acids: Microbial Metabolites for Gut-Brain Axis Signalling. Mol. Cell. Endocrinol. 2022, 546, 111572. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Ghosh, S.K.; Shokeen, B.; Eapan, B.; Lux, R.; Kiselar, J.; Nithianantham, S.; Young, A.; Pandiyan, P.; McCormick, T.S.; et al. FAD-I, a Fusobacterium Nucleatum Cell Wall-Associated Diacylated Lipoprotein That Mediates Human Beta Defensin 2 Induction through Toll-like Receptor-1/2 (TLR-1/2) and TLR-2/6. Infect. Immun. 2016, 84, 1446–1456. [Google Scholar] [CrossRef]
- Donaldson, D.S.; Sehgal, A.; Rios, D.; Williams, I.R.; Mabbott, N.A. Increased Abundance of M Cells in the Gut Epithelium Dramatically Enhances Oral Prion Disease Susceptibility. PloS Pathog. 2016, 12, e1006075. [Google Scholar] [CrossRef]
- Prinz, M.; Priller, J. Microglia and Brain Macrophages in the Molecular Age: From Origin to Neuropsychiatric Disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Ticinesi, A.; Parise, A.; Nouvenne, A.; Cerundolo, N.; Prati, B.; Meschi, T. The Possible Role of Gut Microbiota Dysbiosis in the Pathophysiology of Delirium in Older Persons. Microbiome Res. Rep. 2023, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Medici, V.; Weiss, K.-H. Genetic and Environmental Modifiers of Wilson Disease. Handb. Clin. Neurol. 2017, 142, 35–41. [Google Scholar] [PubMed]
- Geng, H.; Shu, S.; Dong, J.; Li, H.; Xu, C.; Han, Y.; Hu, J.; Han, Y.; Yang, R.; Cheng, N. Association Study of Gut Flora in Wilson’s Disease through High-Throughput Sequencing. Medicine 2018, 97, e11743. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Deng, L.; Ma, X.; Guo, Y.; Feng, Z.; Liu, M.; Guan, Y.; Huang, Y.; Deng, J.; Li, H.; et al. Altered Diversity and Composition of Gut Microbiota in Wilson’s Disease. Sci. Rep. 2020, 10, 21825. [Google Scholar] [CrossRef]
- Anderson, J.R.; Carroll, I.; Azcarate-Peril, M.A.; Rochette, A.D.; Heinberg, L.J.; Peat, C.; Steffen, K.; Manderino, L.M.; Mitchell, J.; Gunstad, J. A Preliminary Examination of Gut Microbiota, Sleep, and Cognitive Flexibility in Healthy Older Adults. Sleep Med. 2017, 38, 104–107. [Google Scholar] [CrossRef]
- Cani, P.D. Human Gut Microbiome: Hopes, Threats and Promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef]
- Park, S.-N.; Lim, Y.K.; Shin, J.H.; Kim, H.-S.; Jo, E.; Lee, W.-P.; Shin, Y.; Paek, J.; Chang, Y.-H.; Kim, H.; et al. Fusobacterium Pseudoperiodonticum Sp. Nov., Isolated from the Human Oral Cavity. Curr. Microbiol. 2019, 76, 659–665. [Google Scholar] [CrossRef]
- Mobeen, F.; Sharma, V.; Tulika, P. Enterotype Variations of the Healthy Human Gut Microbiome in Different Geographical Regions. Bioinformation 2018, 14, 560. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut Microbiome Structure and Metabolic Activity in Inflammatory Bowel Disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Gai, W.; Power, J.; Blumbergs, P.; Blessing, W. Multiple-System Atrophy: A New α-Synuclein Disease? Lancet 1998, 352, 547–548. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Chen, Y.; Zhou, Q.; Zhang, L.; Ou, R.; Wei, Q.; Wu, Y.; Shang, H.-F. Functional Variant Rs3135500 in NOD2 Increases the Risk of Multiple System Atrophy in a Chinese Population. Front. Aging Neurosci. 2018, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Engen, P.A.; Dodiya, H.B.; Naqib, A.; Forsyth, C.B.; Green, S.J.; Voigt, R.M.; Kordower, J.H.; Mutlu, E.A.; Shannon, K.M.; Keshavarzian, A. The Potential Role of Gut-Derived Inflammation in Multiple System Atrophy. J. Park. Dis. 2017, 7, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.H.; Chong, C.W.; Song, S.L.; Teh, C.S.J.; Yap, I.K.S.; Loke, M.F.; Tan, Y.Q.; Yong, H.S.; Mahadeva, S.; Lang, A.E.; et al. Altered Gut Microbiome and Metabolome in Patients with Multiple System Atrophy. J. Neurol. Sci. 2017, 381, 96–97. [Google Scholar] [CrossRef]
- Wan, L.; Zhou, X.; Wang, C.; Chen, Z.; Peng, H.; Hou, X.; Peng, Y.; Wang, P.; Li, T.; Yuan, H.; et al. Alterations of the Gut Microbiota in Multiple System Atrophy Patients. Front. Neurosci. 2019, 13, 1102. [Google Scholar] [CrossRef]
- Derrien, M.; Van Baarlen, P.; Hooiveld, G.; Norin, E.; Müller, M.; de Vos, W.M. Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia Muciniphila. Front. Microbiol. 2011, 2, 166. [Google Scholar] [CrossRef]
- Ganesh, B.P.; Klopfleisch, R.; Loh, G.; Blaut, M. Commensal Akkermansia Muciniphila Exacerbates Gut Inflammation in Salmonella Typhimurium-Infected Gnotobiotic Mice. PLoS ONE 2013, 8, e74963. [Google Scholar] [CrossRef]
- Freitas, R.G.B.d.O.N.; Vasques, A.C.J.; Fernandes, G.d.R.; Ribeiro, F.B.; Solar, I.; Barbosa, M.G.; Pititto, B.d.A.; Geloneze, B.; Ferreira, S.R.G. Associations of Blautia Genus with Early-Life Events and Later Phenotype in the NutriHS. Front. Cell. Infect. Microbiol. 2022, 12, 838750. [Google Scholar] [CrossRef]
- Ewaschuk, J.B.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Looijer-van Langen, M.; Madsen, K.L. Secreted Bioactive Factors from Bifidobacterium Infantis Enhance Epithelial Cell Barrier Function. Am. J. Physiol.-Gastrointest. Liver Physiol. 2008, 295, G1025–G1034. [Google Scholar] [CrossRef]
- Kong, G.; Lê Cao, K.-A.; Judd, L.M.; Li, S.; Renoir, T.; Hannan, A.J. Microbiome Profiling Reveals Gut Dysbiosis in a Transgenic Mouse Model of Huntington’s Disease. Neurobiol. Dis. 2020, 135, 104268. [Google Scholar] [CrossRef]
- Radulescu, C.I.; Garcia-Miralles, M.; Sidik, H.; Bardile, C.F.; Yusof, N.A.B.M.; Lee, H.U.; Ho, E.X.P.; Chu, C.W.; Layton, E.; Low, D.; et al. Manipulation of Microbiota Reveals Altered Callosal Myelination and White Matter Plasticity in a Model of Huntington Disease. Neurobiol. Dis. 2019, 127, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Benito, M.; Granado, N.; García-Sanz, P.; Michel, A.; Dumoulin, M.; Moratalla, R. Modeling Parkinson’s Disease with the Alpha-Synuclein Protein. Front. Pharmacol. 2020, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Koeglsperger, T.; Rumpf, S.-L.; Schließer, P.; Struebing, F.L.; Brendel, M.; Levin, J.; Trenkwalder, C.; Höglinger, G.U.; Herms, J. Neuropathology of Incidental Lewy Body & Prodromal Parkinson’s Disease. Mol. Neurodegener. 2023, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Sulzer, D.; Surmeier, D.J. Neuronal Vulnerability, Pathogenesis, and Parkinson’s Disease. Mov. Disord. 2013, 28, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Beach, T.G.; Adler, C.H.; Serrano, G.; Sue, L.I.; Walker, D.G.; Dugger, B.N.; Shill, H.A.; Driver-Dunckley, E.; Caviness, J.N.; Intorcia, A.; et al. Prevalence of Submandibular Gland Synucleinopathy in Parkinson’s Disease, Dementia with Lewy Bodies and Other Lewy Body Disorders. J. Park. Dis. 2016, 6, 153–163. [Google Scholar] [CrossRef]
- Liddle, R.A. Parkinson’s Disease from the Gut. Brain Res. 2018, 1693, 201–206. [Google Scholar] [CrossRef]
- Ganguly, U.; Singh, S.; Pal, S.; Prasad, S.; Agrawal, B.K.; Saini, R.V.; Chakrabarti, S. Alpha-Synuclein as a Biomarker of Parkinson’s Disease: Good, but Not Good Enough. Front. Aging Neurosci. 2021, 13, 702639. [Google Scholar] [CrossRef]
- Unger, M.M.; Spiegel, J.; Dillmann, K.-U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.-H. Short Chain Fatty Acids and Gut Microbiota Differ between Patients with Parkinson’s Disease and Age-Matched Controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-Analysis of the Parkinson’s Disease Gut Microbiome Suggests Alterations Linked to Intestinal Inflammation. NPJ Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Zapała, B.; Stefura, T.; Wójcik-Pędziwiatr, M.; Kabut, R.; Bałajewicz-Nowak, M.; Milewicz, T.; Dudek, A.; Stój, A.; Rudzińska-Bar, M. Differences in the Composition of Gut Microbiota between Patients with Parkinson’s Disease and Healthy Controls: A Cohort Study. J. Clin. Med. 2021, 10, 5698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qu, Y.; Yang, J.; Liu, J.; Li, S.; He, X. A Pilot Study to Investigate the Alteration of Gut Microbial Profile in Dip2a Knockout Mice. Int. Microbiol. 2022, 25, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Forero-Rodríguez, J.; Zimmermann, J.; Taubenheim, J.; Arias-Rodríguez, N.; Caicedo-Narvaez, J.D.; Best, L.; Mendieta, C.V.; López-Castiblanco, J.; Gómez-Muñoz, L.A.; Gonzalez-Santos, J.; et al. Changes in Bacterial Gut Composition in Parkinson’s Disease and Their Metabolic Contribution to Disease Development: A Gut Community Reconstruction Approach. Microorganisms 2024, 12, 325. [Google Scholar] [CrossRef] [PubMed]
- Heravi, F.S.; Naseri, K.; Hu, H. Gut Microbiota Composition in Patients with Neurodegenerative Disorders (Parkinson’s and Alzheimer’s) and Healthy Controls: A Systematic Review. Nutrients 2023, 15, 4365. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic Bacterial Composition in Parkinson’s Disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef]
- Zhang, K.; Paul, K.C.; Jacobs, J.P.; Chou, H.-C.L.; Duarte Folle, A.; Del Rosario, I.; Yu, Y.; Bronstein, J.M.; Keener, A.M.; Ritz, B. Parkinson’s Disease and the Gut Microbiome in Rural California. J. Park. Dis. 2022, 12, 2441–2452. [Google Scholar] [CrossRef]
- Hillion, S.; Arleevskaya, M.I.; Blanco, P.; Bordron, A.; Brooks, W.H.; Cesbron, J.Y.; Kaveri, S.; Vivier, E.; Renaudineau, Y. The Innate Part of the Adaptive Immune System. Clinic Rev. Allerg. Immunol. 2020, 58, 151–154. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Oettgen, H.C. Adaptive Immunity. J. Allergy Clin. Immunol. 2010, 125, S33–S40. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal Epithelial Cells: Regulators of Barrier Function and Immune Homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; Van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef]
- Francino, M. Early Development of the Gut Microbiota and Immune Health. Pathogens 2014, 3, 769–790. [Google Scholar] [CrossRef] [PubMed]
- Owaga, E.; Hsieh, R.-H.; Mugendi, B.; Masuku, S.; Shih, C.-K.; Chang, J.-S. Th17 Cells as Potential Probiotic Therapeutic Targets in Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2015, 16, 20841–20858. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, L.; van Hooft, J.A.; Chameau, P. Altered Dendritic Complexity Affects Firing Properties of Cortical Layer 2/3 Pyramidal Neurons in Mice Lacking the 5-HT3A Receptor. J. Neurophysiol. 2012, 108, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.A.; Ryu, J.H.; Jo, Y.; Hong, C. The Role of Gut Microbiota in T Cell Immunity and Immune Mediated Disorders. Int. J. Biol. Sci. 2023, 19, 1178–1191. [Google Scholar] [CrossRef]
- Schepici, G.; Silvestro, S.; Bramanti, P.; Mazzon, E. The Gut Microbiota in Multiple Sclerosis: An Overview of Clinical Trials. Cell Transplant. 2019, 28, 1507–1527. [Google Scholar] [CrossRef]
- Walton, C.; King, R.; Rechtman, L.; Kaye, W.; Leray, E.; Marrie, R.A.; Robertson, N.; La Rocca, N.; Uitdehaag, B.; van der Mei, I.; et al. Rising Prevalence of Multiple Sclerosis Worldwide: Insights from the Atlas of MS, Third Edition. Mult. Scler. 2020, 26, 1816–1821. [Google Scholar] [CrossRef]
- Huang, W.-J.; Chen, W.-W.; Zhang, X. Multiple Sclerosis: Pathology, Diagnosis and Treatments. Exp. Ther. Med. 2017, 13, 3163–3166. [Google Scholar] [CrossRef]
- Frohman, E.M.; Racke, M.K.; Raine, C.S. Multiple Sclerosis—The Plaque and Its Pathogenesis. N. Engl. J. Med. 2006, 354, 942–955. [Google Scholar] [CrossRef]
- Baecher-Allan, C.; Kaskow, B.J.; Weiner, H.L. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 2018, 97, 742–768. [Google Scholar] [CrossRef]
- Janeway, C.A. How the Immune System Protects the Host from Infection. Microbes Infect. 2001, 3, 1167–1171. [Google Scholar] [CrossRef]
- Reith, W.; LeibundGut-Landmann, S.; Waldburger, J.-M. Regulation of MHC Class II Gene Expression by the Class II Transactivator. Nat. Rev. Immunol. 2005, 5, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Arellano, G.; Ottum, P.A.; Reyes, L.I.; Burgos, P.I.; Naves, R. Stage-Specific Role of Interferon-Gamma in Experimental Autoimmune Encephalomyelitis and Multiple Sclerosis. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Legroux, L.; Arbour, N. Multiple Sclerosis and T Lymphocytes: An Entangled Story. J. Neuroimmune Pharmacol. 2015, 10, 528–546. [Google Scholar] [CrossRef] [PubMed]
- Hjorth, M.; Dandu, N.; Mellergård, J. Treatment Effects of Fingolimod in Multiple Sclerosis: Selective Changes in Peripheral Blood Lymphocyte Subsets. PLoS ONE 2020, 15, e0228380. [Google Scholar] [CrossRef]
- Correale, J.; Farez, M.F. The Role of Astrocytes in Multiple Sclerosis Progression. Front. Neurol. 2015, 6, 180. [Google Scholar] [CrossRef]
- Bosworth, A.P.; Allen, N.J. The Diverse Actions of Astrocytes during Synaptic Development. Curr. Opin. Neurobiol. 2017, 47, 38–43. [Google Scholar] [CrossRef]
- Purushotham, S.S.; Buskila, Y. Astrocytic Modulation of Neuronal Signalling. Front. Netw. Physiol. 2023, 3, 1205544. [Google Scholar] [CrossRef]
- Bonvento, G.; Bolaños, J.P. Astrocyte-Neuron Metabolic Cooperation Shapes Brain Activity. Cell Metab. 2021, 33, 1546–1564. [Google Scholar] [CrossRef]
- Heithoff, B.P.; George, K.K.; Phares, A.N.; Zuidhoek, I.A.; Munoz-Ballester, C.; Robel, S. Astrocytes Are Necessary for Blood–Brain Barrier Maintenance in the Adult Mouse Brain. Glia 2021, 69, 436–472. [Google Scholar] [CrossRef]
- Mahmoud, S.; Gharagozloo, M.; Simard, C.; Gris, D. Astrocytes Maintain Glutamate Homeostasis in the CNS by Controlling the Balance between Glutamate Uptake and Release. Cells 2019, 8, 184. [Google Scholar] [CrossRef]
- Brambilla, R. The Contribution of Astrocytes to the Neuroinflammatory Response in Multiple Sclerosis and Experimental Autoimmune Encephalomyelitis. Acta Neuropathol. 2019, 137, 757–783. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.; Ghorpade, A. Tissue Inhibitor of Metalloproteinase (TIMP)-1: The TIMPed Balance of Matrix Metalloproteinases in the Central Nervous System. J. Neurosci. Res. 2003, 74, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Yiu, G.; He, Z. Glial Inhibition of CNS Axon Regeneration. Nat. Rev. Neurosci. 2006, 7, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.J.M.; ffrench-Constant, C. Remyelination in the CNS: From Biology to Therapy. Nat. Rev. Neurosci. 2008, 9, 839–855. [Google Scholar] [CrossRef]
- Perry, V.H.; Teeling, J. Microglia and Macrophages of the Central Nervous System: The Contribution of Microglia Priming and Systemic Inflammation to Chronic Neurodegeneration. Semin. Immunopathol. 2013, 35, 601–612. [Google Scholar] [CrossRef]
- Norden, D.M.; Fenn, A.M.; Dugan, A.; Godbout, J.P. TGFβ Produced by IL-10 Redirected Astrocytes Attenuates Microglial Activation. Glia 2014, 62, 881–895. [Google Scholar] [CrossRef]
- Wang, K.; Song, F.; Fernandez-Escobar, A.; Luo, G.; Wang, J.-H.; Sun, Y. The Properties of Cytokines in Multiple Sclerosis: Pros and Cons. Am. J. Med. Sci. 2018, 356, 552–560. [Google Scholar] [CrossRef]
- Molina-Gonzalez, I.; Miron, V.E. Astrocytes in Myelination and Remyelination. Neurosci. Lett. 2019, 713, 134532. [Google Scholar] [CrossRef]
- Albini, M.; Krawczun-Rygmaczewska, A.; Cesca, F. Astrocytes and Brain-Derived Neurotrophic Factor (BDNF). Neurosci. Res. 2023, 197, 42–51. [Google Scholar] [CrossRef]
- Wan, M.; Ding, L.; Wang, D.; Han, J.; Gao, P. Serotonin: A Potent Immune Cell Modulator in Autoimmune Diseases. Front. Immunol. 2020, 11, 186. [Google Scholar] [CrossRef]
- Mandolesi, G.; Gentile, A.; Musella, A.; Fresegna, D.; De Vito, F.; Bullitta, S.; Sepman, H.; Marfia, G.A.; Centonze, D. Synaptopathy Connects Inflammation and Neurodegeneration in Multiple Sclerosis. Nat. Rev. Neurol. 2015, 11, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, E.; Miles, R. The CA3 Region of the Hippocampus: How Is It? What Is It for? How Does It Do It? Front. Cell Neurosci. 2015, 9, 19. [Google Scholar] [CrossRef]
- Krapić, M.; Kavazović, I.; Wensveen, F.M. Immunological Mechanisms of Sickness Behavior in Viral Infection. Viruses 2021, 13, 2245. [Google Scholar] [CrossRef] [PubMed]
- Altieri, C.; Speranza, B.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Gut-Microbiota, and Multiple Sclerosis: Background, Evidence, and Perspectives. Nutrients 2023, 15, 942. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Waubant, E.; Chehoud, C.; Kuczynski, J.; DeSantis, T.Z.; Warrington, J.; Venkatesan, A.; Fraser, C.M.; Mowry, E.M. Gut Microbiota in Multiple Sclerosis: Possible Influence of Immunomodulators. J. Investig. Med. 2015, 63, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut Microbiota from Multiple Sclerosis Patients Enables Spontaneous Autoimmune Encephalomyelitis in Mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Pröbstel, A.-K.; Thomann, A.; Runia, T.F.; Casaccia, P.; Katz Sand, I.; Crabtree, E.; Singh, S.; Morrissey, J.; Barba, P.; et al. Multiple Sclerosis-Associated Changes in the Composition and Immune Functions of Spore-Forming Bacteria. mSystems 2018, 3, e00083-18. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the Gut Microbiota in Disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Zhu, F.; Hart, J.; Roalstad, S.; Graves, J.; Lynch, S.; Waubant, E. US Network of Pediatric MS Centers Gut Microbiota in Early Pediatric Multiple Sclerosis: A Case-Control Study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef]
- Toepfer, M.C.; Folwaczny, A.; Klauser, R.L.; Riepl, W.; Muller-Felber, D. Pongratz Gastrointestinal Dysfunction in Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 15–19. [Google Scholar] [CrossRef]
- Fang, X.; Wang, X.; Yang, S.; Meng, F.; Wang, X.; Wei, H.; Chen, T. Evaluation of the Microbial Diversity in Amyotrophic Lateral Sclerosis Using High-Throughput Sequencing. Front. Microbiol. 2016, 7, 1479. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Yilmaz, R.; Müller, K.; Grehl, T.; Petri, S.; Meyer, T.; Grosskreutz, J.; Weydt, P.; Ruf, W.; Neuwirth, C.; et al. Hot-Spot KIF5A Mutations Cause Familial ALS. Brain 2018, 141, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Mazzini, L.; Ferrari, D.; Andjus, P.R.; Buzanska, L.; Cantello, R.; De Marchi, F.; Gelati, M.; Giniatullin, R.; Glover, J.C.; Grilli, M.; et al. Advances in Stem Cell Therapy for Amyotrophic Lateral Sclerosis. Expert Opin. Biol. Ther. 2018, 18, 865–881. [Google Scholar] [CrossRef] [PubMed]
- Rowin, J.; Xia, Y.; Jung, B.; Sun, J. Gut Inflammation and Dysbiosis in Human Motor Neuron Disease. Physiol. Rep. 2017, 5, e13443. [Google Scholar] [CrossRef] [PubMed]
- Dohm-Hansen, S.; Donoso, F.; Lucassen, P.J.; Clarke, G.; Nolan, Y.M. The Gut Microbiome and Adult Hippocampal Neurogenesis: A New Focal Point for Epilepsy? Neurobiol. Dis. 2022, 170, 105746. [Google Scholar] [CrossRef]
- Cho, K.-O.; Lybrand, Z.R.; Ito, N.; Brulet, R.; Tafacory, F.; Zhang, L.; Good, L.; Ure, K.; Kernie, S.G.; Birnbaum, S.G.; et al. Aberrant Hippocampal Neurogenesis Contributes to Epilepsy and Associated Cognitive Decline. Nat. Commun. 2015, 6, 6606. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, Z.; Guo, Z.; Chen, L.; Ma, Y.; Wang, Z.; Xiao, W.; Wang, Y. Systems-Level Analysis Identifies Key Regulators Driving Epileptogenesis in Temporal Lobe Epilepsy. Genomics 2020, 112, 1768–1780. [Google Scholar] [CrossRef]
- Hoogland, G.; Spierenburg, H.A.; Van Veelen, C.W.M.; Van Rijen, P.C.; Van Huffelen, A.C.; De Graan, P.N.E. Synaptosomal Glutamate and GABA Transport in Patients with Temporal Lobe Epilepsy. J. Neurosci. Res. 2004, 76, 881–890. [Google Scholar] [CrossRef]
- Wei, S.; Mai, Y.; Hu, L.; Zheng, R.; Zheng, D.; Chen, W.; Cai, Y.; Wang, J. Altered Gut Microbiota in Temporal Lobe Epilepsy with Anxiety Disorders. Front. Microbiol. 2023, 14, 1165787. [Google Scholar] [CrossRef]
- Seid, J.; Mebrahtu, K.; Andualem, F. Prevalence and Associated Factors of Anxiety Disorder Symptoms among People with Epilepsy in Mekelle, Ethiopia, 2019: Institutional-based Cross-sectional Study. Nurs. Open 2022, 9, 1731–1743. [Google Scholar] [CrossRef]
- Beghi, E. The Epidemiology of Epilepsy. Neuroepidemiology 2020, 54, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Buchin, A.; De Frates, R.; Nandi, A.; Mann, R.; Chong, P.; Ng, L.; Miller, J.; Hodge, R.; Kalmbach, B.; Bose, S.; et al. Multi-Modal Characterization and Simulation of Human Epileptic Circuitry. Cell Rep. 2022, 41, 111873. [Google Scholar] [CrossRef] [PubMed]
- Munger Clary, H.M. Epilepsy-Specific Anxiety: A Potential Clue to the Seizure Onset Zone. Epilepsy Curr. 2022, 22, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Nutt, D.J.; Tyacke, R.J.; Spriggs, M.; Jacoby, V.; Borthwick, A.D.; Belelli, D. Functional Alternatives to Alcohol. Nutrients 2022, 14, 3761. [Google Scholar] [CrossRef]
- Xu, Z.; Tian, X.; Wang, X.; Chen, Y.; Xu, Q.; Assenza, G. Editorial: The Comorbid Anxiety and Depression Disorder in Patients with Epilepsy: Diagnosis, Prevention and Treatment. Front. Neurol. 2022, 13, 974462. [Google Scholar] [CrossRef]
- Mattson, M.P. Glutamate and Neurotrophic Factors in Neuronal Plasticity and Disease. Ann. N. Y. Acad. Sci. 2008, 1144, 97–112. [Google Scholar] [CrossRef]
- Picciotto, M.R.; Higley, M.J.; Mineur, Y.S. Acetylcholine as a Neuromodulator: Cholinergic Signaling Shapes Nervous System Function and Behavior. Neuron 2012, 76, 116–129. [Google Scholar] [CrossRef]
- Speranza, L.; di Porzio, U.; Viggiano, D.; de Donato, A.; Volpicelli, F. Dopamine: The Neuromodulator of Long-Term Synaptic Plasticity, Reward and Movement Control. Cells 2021, 10, 735. [Google Scholar] [CrossRef]
- Crispino, M.; Volpicelli, F.; Perrone-Capano, C. Role of the Serotonin Receptor 7 in Brain Plasticity: From Development to Disease. Int. J. Mol. Sci. 2020, 21, 505. [Google Scholar] [CrossRef]
- Maffei, A.; Charrier, C.; Caiati, M.D.; Barberis, A.; Mahadevan, V.; Woodin, M.A.; Tyagarajan, S.K. Emerging Mechanisms Underlying Dynamics of GABAergic Synapses. J. Neurosci. 2017, 37, 10792–10799. [Google Scholar] [CrossRef]
- Xu, N.-J.; Sun, S.; Gibson, J.R.; Henkemeyer, M. A Dual Shaping Mechanism for Postsynaptic Ephrin-B3 as a Receptor That Sculpts Dendrites and Synapses. Nat. Neurosci. 2011, 14, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Brierley, S.M.; Linden, D.R. Neuroplasticity and Dysfunction after Gastrointestinal Inflammation. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Y.; Chen, Z. Adult Neurogenesis in Epileptogenesis: An Update for Preclinical Finding and Potential Clinical Translation. Curr. Neuropharmacol. 2020, 18, 464–484. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Shen, X.; Hao, Y.; Li, J.; Li, H.; Xu, H.; Yin, L.; Kuang, W. Gut Microbiome: A Potential Indicator for Differential Diagnosis of Major Depressive Disorder and General Anxiety Disorder. Front. Psychiatry 2021, 12, 651536. [Google Scholar] [CrossRef] [PubMed]
- Wlaź, P.; Wiater, A.; Majewska, M.; Wyska, E.; Grąz, M.; Śliwa-Dominiak, J.; Gapińska, N.; Socała, K. Effect of Dietary Supplementation with Lactobacillus Helveticus R0052 on Seizure Thresholds and Antiseizure Potency of Sodium Valproate in Mice. Psychopharmacology 2024, 241, 327–340. [Google Scholar] [CrossRef]
- Reinoso Webb, C.; Koboziev, I.; Furr, K.L.; Grisham, M.B. Protective and Pro-Inflammatory Roles of Intestinal Bacteria. Pathophysiology 2016, 23, 67–80. [Google Scholar] [CrossRef]
- Ceccarani, C.; Bassanini, G.; Montanari, C.; Casiraghi, M.C.; Ottaviano, E.; Morace, G.; Biasucci, G.; Paci, S.; Borghi, E.; Verduci, E. Proteobacteria Overgrowth and Butyrate-Producing Taxa Depletion in the Gut Microbiota of Glycogen Storage Disease Type 1 Patients. Metabolites 2020, 10, 133. [Google Scholar] [CrossRef]
- Trifu, S.C.; Trifu, A.C.; Aluaş, E.; Tătaru, M.A.; Costea, R.V. Brain Changes in Depression. Rom. J. Morphol. Embryol. 2020, 61, 361–370. [Google Scholar] [CrossRef]
- WHO Depressive Disorder (Depression). Available online: https://www.who.int/news-room/fact-sheets/detail/depression (accessed on 19 June 2024).
- Limbana, T.; Khan, F.; Eskander, N. Gut Microbiome and Depression: How Microbes Affect the Way We Think. Cureus 2020, 12, e9966. [Google Scholar] [CrossRef]
- Carlessi, A.S.; Borba, L.A.; Zugno, A.I.; Quevedo, J.; Réus, G.Z. Gut Microbiota–Brain Axis in Depression: The Role of Neuroinflammation. Eur. J. Neurosci. 2021, 53, 222–235. [Google Scholar] [CrossRef]
- Chinna Meyyappan, A.; Forth, E.; Wallace, C.J.K.; Milev, R. Effect of Fecal Microbiota Transplant on Symptoms of Psychiatric Disorders: A Systematic Review. BMC Psychiatry 2020, 20, 299. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, J.K.; Michaelsen, T.Y.; Bundgaard-Nielsen, C.; Nielsen, R.E.; Hjerrild, S.; Leutscher, P.; Wegener, G.; Sørensen, S. Faecal Microbiota Transplantation from Patients with Depression or Healthy Individuals into Rats Modulates Mood-Related Behaviour. Sci. Rep. 2021, 11, 21869. [Google Scholar] [CrossRef] [PubMed]
- Van De Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain Fatty Acids: Microbial Metabolites That Alleviate Stress-induced Brain–Gut Axis Alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Gui, S.; Zeng, B.; Pu, J.; Zheng, P.; Zeng, L.; Luo, Y.; Wu, Y.; Zhou, C.; et al. Proteomics Analysis of the Gut–Brain Axis in a Gut Microbiota-Dysbiosis Model of Depression. Transl. Psychiatry 2021, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered Fecal Microbiota Composition in Patients with Major Depressive Disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef]
- Naseribafrouei, A.; Hestad, K.; Avershina, E.; Sekelja, M.; Linløkken, A.; Wilson, R.; Rudi, K. Correlation between the Human Fecal Microbiota and Depression. Neurogastroenterol. Motil. 2014, 26, 1155–1162. [Google Scholar] [CrossRef]
- Tillmann, S.; Abildgaard, A.; Winther, G.; Wegener, G. Altered Fecal Microbiota Composition in the Flinders Sensitive Line Rat Model of Depression. Psychopharmacology 2019, 236, 1445–1457. [Google Scholar] [CrossRef]
- Yang, J.; Zheng, P.; Li, Y.; Wu, J.; Tan, X.; Zhou, J.; Sun, Z.; Chen, X.; Zhang, G.; Zhang, H.; et al. Landscapes of Bacterial and Metabolic Signatures and Their Interaction in Major Depressive Disorders. Sci. Adv. 2020, 6, eaba8555. [Google Scholar] [CrossRef]
- Zheng, P.; Yang, J.; Li, Y.; Wu, J.; Liang, W.; Yin, B.; Tan, X.; Huang, Y.; Chai, T.; Zhang, H.; et al. Gut Microbial Signatures Can Discriminate Unipolar from Bipolar Depression. Adv. Sci. 2020, 7, 1902862. [Google Scholar] [CrossRef]
- Kahn, R.S.; Sommer, I.E.; Murray, R.M.; Meyer-Lindenberg, A.; Weinberger, D.R.; Cannon, T.D.; O’Donovan, M.; Correll, C.U.; Kane, J.M.; Van Os, J.; et al. Schizophrenia. Nat. Rev. Dis. Primers 2015, 1, 15067. [Google Scholar] [CrossRef]
- Luvsannyam, E.; Jain, M.S.; Pormento, M.K.L.; Siddiqui, H.; Balagtas, A.R.A.; Emuze, B.O.; Poprawski, T. Neurobiology of Schizophrenia: A Comprehensive Review. Cureus 2022, 14, e23959. [Google Scholar] [CrossRef] [PubMed]
- Brisch, R.; Saniotis, A.; Wolf, R.; Bielau, H.; Bernstein, H.-G.; Steiner, J.; Bogerts, B.; Braun, K.; Jankowski, Z.; Kumaratilake, J.; et al. The Role of Dopamine in Schizophrenia from a Neurobiological and Evolutionary Perspective: Old Fashioned, but Still in Vogue. Front. Psychiatry 2014, 5, 47. [Google Scholar] [CrossRef]
- Severance, E.G.; Prandovszky, E.; Castiglione, J.; Yolken, R.H. Gastroenterology Issues in Schizophrenia: Why the Gut Matters. Curr. Psychiatry Rep. 2015, 17, 27. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Kosciolek, T.; Maldonado, Y.; Daly, R.E.; Martin, A.S.; McDonald, D.; Knight, R.; Jeste, D.V. Differences in Gut Microbiome Composition between Persons with Chronic Schizophrenia and Healthy Comparison Subjects. Schizophr. Res. 2019, 204, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Thirion, F.; Speyer, H.; Hansen, T.H.; Nielsen, T.; Fan, Y.; Le Chatelier, E.; Fromentin, S.; Berland, M.; Plaza Oñate, F.; Pons, N.; et al. Alteration of Gut Microbiome in Patients with Schizophrenia Indicates Links between Bacterial Tyrosine Biosynthesis and Cognitive Dysfunction. Biol. Psychiatry Glob. Open Sci. 2023, 3, 283–291. [Google Scholar] [CrossRef]
- Macedo E Cordeiro, T.; Zhang, X.; Graubics, K.; Colwell, R.; Teixeira, A.L. Microbiome and Schizophrenia: Current Evidence and Future Challenges. Curr. Behav. Neurosci. Rep. 2020, 7, 51–61. [Google Scholar] [CrossRef]
- Schwarz, E.; Maukonen, J.; Hyytiäinen, T.; Kieseppä, T.; Orešič, M.; Sabunciyan, S.; Mantere, O.; Saarela, M.; Yolken, R.; Suvisaari, J. Analysis of Microbiota in First Episode Psychosis Identifies Preliminary Associations with Symptom Severity and Treatment Response. Schizophr. Res. 2018, 192, 398–403. [Google Scholar] [CrossRef]
- Mándi, Y.; Vécsei, L. The Kynurenine System and Immunoregulation. J. Neural. Transm. 2012, 119, 197–209. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Jarosz, Ł.S.; Socała, K.; Michalak, K.; Wiater, A.; Ciszewski, A.; Majewska, M.; Marek, A.; Grądzki, Z.; Wlaź, P. The Effect of Psychoactive Bacteria, Bifidobacterium Longum Rosell®-175 and Lactobacillus Rhamnosus JB-1, on Brain Proteome Profiles in Mice. Psychopharmacology 2024, 241, 925–945. [Google Scholar] [CrossRef]
- Sjöstedt, P.; Enander, J.; Isung, J. Serotonin Reuptake Inhibitors and the Gut Microbiome: Significance of the Gut Microbiome in Relation to Mechanism of Action, Treatment Response, Side Effects, and Tachyphylaxis. Front. Psychiatry 2021, 12, 682868. [Google Scholar] [CrossRef] [PubMed]
- Ait Chait, Y.; Mottawea, W.; Tompkins, T.A.; Hammami, R. Unravelling the Antimicrobial Action of Antidepressants on Gut Commensal Microbes. Sci. Rep. 2020, 10, 17878. [Google Scholar] [CrossRef]
- Lukić, I.; Getselter, D.; Ziv, O.; Oron, O.; Reuveni, E.; Koren, O.; Elliott, E. Antidepressants Affect Gut Microbiota and Ruminococcus Flavefaciens Is Able to Abolish Their Effects on Depressive-like Behavior. Transl. Psychiatry 2019, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Chen, X.; Zhang, Y.; Zhang, H.; Xie, P. Gut Microbiota and Its Metabolites in Depression: From Pathogenesis to Treatment. EBioMedicine 2023, 90, 104527. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Dong, T.S.; Krause-Sorio, B.; Siddarth, P.; Milillo, M.M.; Lagishetty, V.; Datta, T.; Aguilar-Faustino, Y.; Jacobs, J.P.; Lavretsky, H. The Intestinal Microbiota as a Predictor for Antidepressant Treatment Outcome in Geriatric Depression: A Prospective Pilot Study. Int. Psychogeriatr. 2022, 34, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Yang, X.; Li, G.; Gao, J.; Liang, Y. The Change of Gut Microbiota in MDD Patients under SSRIs Treatment. Sci. Rep. 2021, 11, 14918. [Google Scholar] [CrossRef]
- Gao, M.; Tu, H.; Liu, P.; Zhang, Y.; Zhang, R.; Jing, L.; Zhang, K. Association Analysis of Gut Microbiota and Efficacy of SSRIs Antidepressants in Patients with Major Depressive Disorder. J. Affect. Disord. 2023, 330, 40–47. [Google Scholar] [CrossRef]
- Lin, S.-K.K.; Chen, H.-C.; Chen, C.-H.; Chen, I.-M.; Lu, M.-L.; Hsu, C.-D.; Chiu, Y.-H.; Wang, T.-Y.; Chen, H.-M.; Chung, Y.-C.E.; et al. Exploring the Human Gut Microbiota Targets in Relation to the Use of Contemporary Antidepressants. J. Affect. Disord. 2024, 344, 473–484. [Google Scholar] [CrossRef]
- Edinoff, A.N.; Akuly, H.A.; Hanna, T.A.; Ochoa, C.O.; Patti, S.J.; Ghaffar, Y.A.; Kaye, A.D.; Viswanath, O.; Urits, I.; Boyer, A.G.; et al. Selective Serotonin Reuptake Inhibitors and Adverse Effects: A Narrative Review. Neurol. Int. 2021, 13, 387–401. [Google Scholar] [CrossRef]
- Skonieczna-Żydecka, K.; Łoniewski, I.; Misera, A.; Stachowska, E.; Maciejewska, D.; Marlicz, W.; Galling, B. Second-Generation Antipsychotics and Metabolism Alterations: A Systematic Review of the Role of the Gut Microbiome. Psychopharmacology 2019, 236, 1491–1512. [Google Scholar] [CrossRef]
- Davey, K.J.; O’Mahony, S.M.; Schellekens, H.; O’Sullivan, O.; Bienenstock, J.; Cotter, P.D.; Dinan, T.G.; Cryan, J.F. Gender-Dependent Consequences of Chronic Olanzapine in the Rat: Effects on Body Weight, Inflammatory, Metabolic and Microbiota Parameters. Psychopharmacology 2012, 221, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Davey, K.J.; Cotter, P.D.; O’Sullivan, O.; Crispie, F.; Dinan, T.G.; Cryan, J.F.; O’Mahony, S.M. Antipsychotics and the Gut Microbiome: Olanzapine-Induced Metabolic Dysfunction Is Attenuated by Antibiotic Administration in the Rat. Transl. Psychiatry 2013, 3, e309. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.P.; Crowley, J.J.; Nonneman, R.J.; Quackenbush, C.R.; Miller, C.N.; Ryan, A.K.; Bogue, M.A.; Paredes, S.H.; Yourstone, S.; Carroll, I.M.; et al. The Antipsychotic Olanzapine Interacts with the Gut Microbiome to Cause Weight Gain in Mouse. PLoS ONE 2014, 9, e115225. [Google Scholar] [CrossRef] [PubMed]
- Bahr, S.M.; Weidemann, B.J.; Castro, A.N.; Walsh, J.W.; deLeon, O.; Burnett, C.M.L.; Pearson, N.A.; Murry, D.J.; Grobe, J.L.; Kirby, J.R. Risperidone-Induced Weight Gain Is Mediated through Shifts in the Gut Microbiome and Suppression of Energy Expenditure. EBioMedicine 2015, 2, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Grobe, J.; Bahr, S.; Weidemann, B.; Burnett, C.; Pearson, N.; DeLeon, O.; Murry, D.; Kirby, J. Transfer of Obesity via the Gut Microbiome Is Mediated Specifically through Suppression of Non-Aerobic Resting Metabolism. FASEB J. 2015, 29, 857.2. [Google Scholar] [CrossRef]
- Riedl, R.A.; Burnett, C.M.; Pearson, N.A.; Atkinson, S.N.; Ollinger, T.L.; Edwards, R.A.; Mokadem, M.; Kirby, J.R.; Grobe, J.L. The Biomass and Composition of the Gut Microbiota Modify Anaerobic Metabolism. FASEB J. 2017, 31, 890.2. [Google Scholar] [CrossRef]
- Minichino, A.; Preston, T.; Fanshawe, J.B.; Fusar-Poli, P.; McGuire, P.; Burnet, P.W.J.; Lennox, B.R. Psycho-Pharmacomicrobiomics: A Systematic Review and Meta-Analysis. Biol. Psychiatry 2024, 95, 611–628. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, P.; Wang, Y.; Liu, Y.; Li, X.; Kumar, B.U.; Hei, G.; Lv, L.; Huang, X.-F.; Fan, X.; et al. Changes in Metabolism and Microbiota after 24-Week Risperidone Treatment in Drug Naïve, Normal Weight Patients with First Episode Schizophrenia. Schizophr. Res. 2018, 201, 299–306. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, Y.; Li, X.; Jiang, J.; Kang, Y.; Pang, L.; Zhang, P.; Li, A.; Lv, L.; Andreassen, O.A.; et al. Gut Microbial Biomarkers for the Treatment Response in First-Episode, Drug-Naïve Schizophrenia: A 24-Week Follow-up Study. Transl. Psychiatry 2021, 11, 422. [Google Scholar] [CrossRef]
- Bahr, S.M.; Tyler, B.C.; Wooldridge, N.; Butcher, B.D.; Burns, T.L.; Teesch, L.M.; Oltman, C.L.; Azcarate-Peril, M.A.; Kirby, J.R.; Calarge, C.A. Use of the Second-Generation Antipsychotic, Risperidone, and Secondary Weight Gain Are Associated with an Altered Gut Microbiota in Children. Transl. Psychiatry 2015, 5, e652. [Google Scholar] [CrossRef]
- Hu, S.; Li, A.; Huang, T.; Lai, J.; Li, J.; Sublette, M.E.; Lu, H.; Lu, Q.; Du, Y.; Hu, Z.; et al. Gut Microbiota Changes in Patients with Bipolar Depression. Adv. Sci. 2019, 6, 1900752. [Google Scholar] [CrossRef] [PubMed]
- Pełka-Wysiecka, J.; Kaczmarczyk, M.; Bąba-Kubiś, A.; Liśkiewicz, P.; Wroński, M.; Skonieczna-Żydecka, K.; Marlicz, W.; Misiak, B.; Starzyńska, T.; Kucharska-Mazur, J.; et al. Analysis of Gut Microbiota and Their Metabolic Potential in Patients with Schizophrenia Treated with Olanzapine: Results from a Six-Week Observational Prospective Cohort Study. JCM 2019, 8, 1605. [Google Scholar] [CrossRef]
- Zheng, J.; Lin, Z.; Ko, C.-Y.; Xu, J.-H.; Lin, Y.; Wang, J. Analysis of Gut Microbiota in Patients with Exacerbated Symptoms of Schizophrenia Following Therapy with Amisulpride: A Pilot Study. Behav. Neurol. 2022, 2022, 4262094. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, G.D.; Luca, A.; Rusu, R.N.; Bild, V.; Beschea Chiriac, S.I.; Solcan, C.; Bild, W.; Ababei, D.C. Alzheimer’s Disease Pharmacotherapy in Relation to Cholinergic System Involvement. Biomolecules 2019, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.-K.; Lee, G.; Nguyen, C.D.; Park, S.-E.; Kim, E.-J.; Kim, H.-W.; Seo, S.-H.; Cho, K.-M.; Kwon, S.J.; Kim, J.-H.; et al. Effects of Donepezil Treatment on Brain Metabolites, Gut Microbiota, and Gut Metabolites in an Amyloid Beta-Induced Cognitive Impairment Mouse Pilot Model. Molecules 2022, 27, 6591. [Google Scholar] [CrossRef] [PubMed]
- Valiukas, Z.; Ephraim, R.; Tangalakis, K.; Davidson, M.; Apostolopoulos, V.; Feehan, J. Immunotherapies for Alzheimer’s Disease—A Review. Vaccines 2022, 10, 1527. [Google Scholar] [CrossRef]
- Dai, C.-L.; Liu, F.; Iqbal, K.; Gong, C.-X. Gut Microbiota and Immunotherapy for Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 15230. [Google Scholar] [CrossRef]
- Koch, J.M.; Dashtipour, K.; Chen, J.J. Parkinson Disease. In DiPiro’s Pharmacotherapy: A Pathophysiologic Approach, 12th ed.; DiPiro, J.T., Yee, G.C., Haines, S.T., Nolin, T.D., Ellingrod, V.L., Posey, L.M., Eds.; McGraw Hill: New York, NY, USA, 2023. [Google Scholar]
- Radisavljevic, N.; Metcalfe-Roach, A.; Cirstea, M.; Tabusi, M.M.; Bozorgmehr, T.; Bar-Yoseph, H.; Finlay, B.B. Microbiota-Mediated Effects of Parkinson’s Disease Medications on Parkinsonian Non-Motor Symptoms in Male Transgenic Mice. mSphere 2024, 9, e00379-23. [Google Scholar] [CrossRef]
- Van Kessel, S.P.; Bullock, A.; Van Dijk, G.; El Aidy, S. Parkinson’s Disease Medication Alters Small Intestinal Motility and Microbiota Composition in Healthy Rats. mSystems 2022, 7, e01191-21. [Google Scholar] [CrossRef]
- Weis, S.; Schwiertz, A.; Unger, M.M.; Becker, A.; Faßbender, K.; Ratering, S.; Kohl, M.; Schnell, S.; Schäfer, K.-H.; Egert, M. Effect of Parkinson’s Disease and Related Medications on the Composition of the Fecal Bacterial Microbiota. NPJ Park. Dis. 2019, 5, 28. [Google Scholar] [CrossRef]
- Jameson, K.G.; Hsiao, E.Y. A Novel Pathway for Microbial Metabolism of Levodopa. Nat. Med. 2019, 25, 1195–1197. [Google Scholar] [CrossRef]
- Maini Rekdal, V.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and Inhibition of an Interspecies Gut Bacterial Pathway for Levodopa Metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef]
- Van Kessel, S.P.; Frye, A.K.; El-Gendy, A.O.; Castejon, M.; Keshavarzian, A.; Van Dijk, G.; El Aidy, S. Gut Bacterial Tyrosine Decarboxylases Restrict Levels of Levodopa in the Treatment of Parkinson’s Disease. Nat. Commun. 2019, 10, 310. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, Z.E.; Brochard, V.; Lapaque, N.; Auvin, S.; Lepage, P. Exposure to Anti-Seizure Medications Impact Growth of Gut Bacterial Species and Subsequent Host Response. Neurobiol. Dis. 2022, 167, 105664. [Google Scholar] [CrossRef] [PubMed]
- Thai, K.; Taylor, M.W.; Fernandes, T.; Akinade, E.A.; Campbell, S.L. Topiramate Alters the Gut Microbiome to Aid in Its Anti-Seizure Effect. Front. Microbiol. 2023, 14, 1242856. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Latif, A.; Shehzad, A.; Niazi, S.; Zahid, A.; Ashraf, W.; Iqbal, M.W.; Rehman, A.; Riaz, T.; Aadil, R.M.; Khan, I.M.; et al. Probiotics: Mechanism of Action, Health Benefits and Their Application in Food Industries. Front. Microbiol. 2023, 14, 1216674. [Google Scholar] [CrossRef]
- Mousavi Khaneghah, A.; Abhari, K.; Eş, I.; Soares, M.B.; Oliveira, R.B.A.; Hosseini, H.; Rezaei, M.; Balthazar, C.F.; Silva, R.; Cruz, A.G.; et al. Interactions between Probiotics and Pathogenic Microorganisms in Hosts and Foods: A Review. Trends Food Sci. Technol. 2020, 95, 205–218. [Google Scholar] [CrossRef]
- Neri-Numa, I.A.; Arruda, H.S.; Geraldi, M.V.; Maróstica Júnior, M.R.; Pastore, G.M. Natural Prebiotic Carbohydrates, Carotenoids and Flavonoids as Ingredients in Food Systems. Curr. Opin. Food Sci. 2020, 33, 98–107. [Google Scholar] [CrossRef]
- Aghamohammad, S.; Hafezi, A.; Rohani, M. Probiotics as Functional Foods: How Probiotics Can Alleviate the Symptoms of Neurological Disabilities. Biomed. Pharmacother. 2023, 163, 114816. [Google Scholar] [CrossRef]
- Shamsipour, S.; Sharifi, G.; Taghian, F. An 8-Week Administration of Bifidobacterium Bifidum and Lactobacillus Plantarum Combined with Exercise Training Alleviates Neurotoxicity of Aβ and Spatial Learning via Acetylcholine in Alzheimer Rat Model. J. Mol. Neurosci. 2021, 71, 1495–1505. [Google Scholar] [CrossRef]
- Athari Nik Azm, S.; Djazayeri, A.; Safa, M.; Azami, K.; Ahmadvand, B.; Sabbaghziarani, F.; Sharifzadeh, M.; Vafa, M. Lactobacilli and Bifidobacteria Ameliorate Memory and Learning Deficits and Oxidative Stress in β-Amyloid (1-42) Injected Rats. Appl. Physiol. Nutr. Metab. 2018, 43, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, S.; Sadr, S.S. Assessment of Probiotics Mixture on Memory Function, Inflammation Markers, and Oxidative Stress in an Alzheimer’s Disease Model of Rats. Iran. Biomed. J. 2020, 24, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Bonfili, L.; Cecarini, V.; Cuccioloni, M.; Angeletti, M.; Berardi, S.; Scarpona, S.; Rossi, G.; Eleuteri, A.M. SLAB51 Probiotic Formulation Activates SIRT1 Pathway Promoting Antioxidant and Neuroprotective Effects in an AD Mouse Model. Mol. Neurobiol. 2018, 55, 7987–8000. [Google Scholar] [CrossRef] [PubMed]
- Akbari, E.; Asemi, Z.; Daneshvar Kakhaki, R.; Bahmani, F.; Kouchaki, E.; Tamtaji, O.R.; Hamidi, G.A.; Salami, M. Effect of Probiotic Supplementation on Cognitive Function and Metabolic Status in Alzheimer’s Disease: A Randomized, Double-Blind and Controlled Trial. Front. Aging Neurosci. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Katsumata, N.; Bernier, F.; Ohno, K.; Yamauchi, Y.; Odamaki, T.; Yoshikawa, K.; Ito, K.; Kaneko, T. Probiotic Bifidobacterium Breve in Improving Cognitive Functions of Older Adults with Suspected Mild Cognitive Impairment: A Randomized, Double-Blind, Placebo-Controlled Trial. JAD 2020, 77, 139–147. [Google Scholar] [CrossRef]
- Purdel, C.; Ungurianu, A.; Adam-Dima, I.; Margină, D. Exploring the Potential Impact of Probiotic Use on Drug Metabolism and Efficacy. Biomed. Pharmacother. 2023, 161, 114468. [Google Scholar] [CrossRef]
- Shepilov, D.; Osadchenko, I.; Kovalenko, T.; Yamada, C.; Chereshynska, A.; Smozhanyk, K.; Ostrovska, G.; Groppa, S.; Movila, A.; Skibo, G. Maternal Antibiotic Administration during Gestation Can Affect the Memory and Brain Structure in Mouse Offspring. Front. Cell. Neurosci. 2023, 17, 1176676. [Google Scholar] [CrossRef]
- Vorhees, C.V.; Williams, M.T. Morris Water Maze: Procedures for Assessing Spatial and Related Forms of Learning and Memory. Nat. Protoc. 2006, 1, 848–858. [Google Scholar] [CrossRef]
- Lueptow, L.M. Novel Object Recognition Test for the Investigation of Learning and Memory in Mice. JoVE 2017, 55718. [Google Scholar] [CrossRef]
- Graves, A.R.; Moore, S.J.; Bloss, E.B.; Mensh, B.D.; Kath, W.L.; Spruston, N. Hippocampal Pyramidal Neurons Comprise Two Distinct Cell Types That Are Countermodulated by Metabotropic Receptors. Neuron 2012, 76, 776–789. [Google Scholar] [CrossRef]
- Parker, D.C.; Kraus, W.E.; Whitson, H.E.; Kraus, V.B.; Smith, P.J.; Cohen, H.J.; Pieper, C.F.; Faldowski, R.A.; Hall, K.S.; Huebner, J.L.; et al. Tryptophan Metabolism and Neurodegeneration: Longitudinal Associations of Kynurenine Pathway Metabolites with Cognitive Performance and Plasma Alzheimer’s Disease and Related Dementias Biomarkers in the Duke Physical Performance Across the LifeSpan Study. J. Alzheimer’s Dis. 2023, 91, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Woodburn, S.C.; Asrat, H.S.; Flurer, J.K.; Schwierling, H.C.; Bollinger, J.L.; Vollmer, L.L.; Wohleb, E.S. Depletion of Microglial BDNF Increases Susceptibility to the Behavioral and Synaptic Effects of Chronic Unpredictable Stress. Brain Behav. Immun. 2023, 109, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Abramova, O.; Zorkina, Y.; Ushakova, V.; Zubkov, E.; Morozova, A.; Chekhonin, V. The Role of Oxytocin and Vasopressin Dysfunction in Cognitive Impairment and Mental Disorders. Neuropeptides 2020, 83, 102079. [Google Scholar] [CrossRef] [PubMed]
- Ceylani, T.; Jakubowska-Doğru, E.; Gurbanov, R.; Teker, H.T.; Gozen, A.G. The Effects of Repeated Antibiotic Administration to Juvenile BALB/c Mice on the Microbiota Status and Animal Behavior at the Adult Age. Heliyon 2018, 4, e00644. [Google Scholar] [CrossRef] [PubMed]
- Seibenhener, M.L.; Wooten, M.C. Use of the Open Field Maze to Measure Locomotor and Anxiety-like Behavior in Mice. J. Vis. Exp. 2015, e52434. [Google Scholar] [CrossRef]
- Walf, A.A.; Frye, C.A. The Use of the Elevated plus Maze as an Assay of Anxiety-Related Behavior in Rodents. Nat. Protoc. 2007, 2, 322–328. [Google Scholar] [CrossRef]
- Yankelevitch-Yahav, R.; Franko, M.; Huly, A.; Doron, R. The Forced Swim Test as a Model of Depressive-like Behavior. J. Vis. Exp. 2015, 52587. [Google Scholar] [CrossRef]
- Pham, K.; Nacher, J.; Hof, P.R.; McEwen, B.S. Repeated Restraint Stress Suppresses Neurogenesis and Induces Biphasic PSA-NCAM Expression in the Adult Rat Dentate Gyrus. Eur. J. Neurosci. 2003, 17, 879–886. [Google Scholar] [CrossRef]
- Lu, L.; Bao, G.; Chen, H.; Xia, P.; Fan, X.; Zhang, J.; Pei, G.; Ma, L. Modification of Hippocampal Neurogenesis and Neuroplasticity by Social Environments. Exp. Neurol. 2003, 183, 600–609. [Google Scholar] [CrossRef]
- Kempermann, G.; Song, H.; Gage, F.H. Neurogenesis in the Adult Hippocampus. Cold Spring Harb. Perspect. Biol. 2015, 7, a018812. [Google Scholar] [CrossRef]
- Appleton, J. The Gut-Brain Axis: Influence of Microbiota on Mood and Mental Health. Integr. Med 2018, 17, 28–32. [Google Scholar]
- Wolf, S.A.; Steiner, B.; Akpinarli, A.; Kammertoens, T.; Nassenstein, C.; Braun, A.; Blankenstein, T.; Kempermann, G. CD4-Positive T Lymphocytes Provide a Neuroimmunological Link in the Control of Adult Hippocampal Neurogenesis. J. Immunol. 2009, 182, 3979–3984. [Google Scholar] [CrossRef] [PubMed]
- Hoban, A.E.; Stilling, R.M.; Ryan, F.J.; Shanahan, F.; Dinan, T.G.; Claesson, M.J.; Clarke, G.; Cryan, J.F. Regulation of Prefrontal Cortex Myelination by the Microbiota. Transl. Psychiatry 2016, 6, e774. [Google Scholar] [CrossRef] [PubMed]
- Švob Štrac, D.; Pivac, N.; Mück-Šeler, D. The Serotonergic System and Cognitive Function. Transl. Neurosci. 2016, 7, 35–49. [Google Scholar] [CrossRef]
- Jeanneteau, F.D.; Lambert, W.M.; Ismaili, N.; Bath, K.G.; Lee, F.S.; Garabedian, M.J.; Chao, M.V. BDNF and Glucocorticoids Regulate Corticotrophin-Releasing Hormone (CRH) Homeostasis in the Hypothalamus. Proc. Natl. Acad. Sci. USA 2012, 109, 1305–1310. [Google Scholar] [CrossRef]
- Widodo, A.D.; Juffrie, M. Caesarean Section and Gut Microbiota in Children. World Nutr. J. 2020, 4, 8–16. [Google Scholar] [CrossRef]
- Chavoya-Guardado, M.A.; Vasquez-Garibay, E.M.; Ruiz-Quezada, S.L.; Ramírez-Cordero, M.I.; Larrosa-Haro, A.; Castro-Albarran, J. Firmicutes, Bacteroidetes and Actinobacteria in Human Milk and Maternal Adiposity. Nutrients 2022, 14, 2887. [Google Scholar] [CrossRef]
- Baud, A.; Hillion, K.-H.; Plainvert, C.; Tessier, V.; Tazi, A.; Mandelbrot, L.; Poyart, C.; Kennedy, S.P. Microbial Diversity in the Vaginal Microbiota and Its Link to Pregnancy Outcomes. Sci. Rep. 2023, 13, 9061. [Google Scholar] [CrossRef]
- Barcan, A.S.; Barcan, R.A.; Vamanu, E. Therapeutic Potential of Fungal Polysaccharides in Gut Microbiota Regulation: Implications for Diabetes, Neurodegeneration, and Oncology. JoF 2024, 10, 394. [Google Scholar] [CrossRef]
- Wong, C.B.; Huang, H.; Ning, Y.; Xiao, J. Probiotics in the New Era of Human Milk Oligosaccharides (HMOs): HMO Utilization and Beneficial Effects of Bifidobacterium Longum Subsp. Infantis M-63 on Infant Health. Microorganisms 2024, 12, 1014. [Google Scholar] [CrossRef]
- Shin, H.; Pei, Z.; Martinez, K.A.; Rivera-Vinas, J.I.; Mendez, K.; Cavallin, H.; Dominguez-Bello, M.G. The First Microbial Environment of Infants Born by C-Section: The Operating Room Microbes. Microbiome 2015, 3, 59. [Google Scholar] [CrossRef]
- Murata, C.; Gutiérrez-Castrellón, P.; Pérez-Villatoro, F.; García-Torres, I.; Enríquez-Flores, S.; De La Mora-de La Mora, I.; Fernández-Lainez, C.; Werner, J.; López-Velázquez, G. Delivery Mode-Associated Gut Microbiota in the First 3 Months of Life in a Country with High Obesity Rates: A Descriptive Study. Medicine 2020, 99, e22442. [Google Scholar] [CrossRef] [PubMed]
- Čoklo, M.; Maslov, D.R.; Kraljević Pavelić, S. Modulation of Gut Microbiota in Healthy Rats after Exposure to Nutritional Supplements. Gut Microbes 2020, 12, 1779002. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Cabré, S.; Ratsika, A.; Rea, K.; Stanton, C.; Cryan, J.F. Animal Models for Assessing Impact of C-Section Delivery on Biological Systems. Neurosci. Biobehav. Rev. 2022, 135, 104555. [Google Scholar] [CrossRef]
- Greenberg, J.M.; Romero, R.; Winters, A.D.; Galaz, J.; Garcia-Flores, V.; Arenas-Hernandez, M.; Panzer, J.; Shaffer, Z.; Kracht, D.J.; Gomez-Lopez, N.; et al. Microbiota of the Pregnant Mouse: Characterization of the Bacterial Communities in the Oral Cavity, Lung, Intestine, and Vagina through Culture and DNA Sequencing. Microbiol. Spectr. 2022, 10, e01286-22. [Google Scholar] [CrossRef]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-Induced Inflammatory Bowel Disease Mice Model. eBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef]
- Bobba, P.S. Early Brain Microstructural Development among Preterm Infants Requiring Caesarean Section versus Those Delivered Vaginally. Sci. Rep. 2023, 13, 21514. [Google Scholar] [CrossRef]
- Feng, K.; Rowell, A.C.; Andres, A.; Bellando, B.J.; Lou, X.; Glasier, C.M.; Ramakrishnaiah, R.H.; Badger, T.M.; Ou, X. Diffusion Tensor MRI of White Matter of Healthy Full-Term Newborns: Relationship to Neurodevelopmental Outcomes. Radiology 2019, 292, 179–187. [Google Scholar] [CrossRef]
- Anand, N.; Gorantla, V.R.; Chidambaram, S.B. The Role of Gut Dysbiosis in the Pathophysiology of Neuropsychiatric Disorders. Cells 2022, 12, 54. [Google Scholar] [CrossRef]
- Zhang, T.; Sidorchuk, A.; Sevilla-Cermeño, L.; Vilaplana-Pérez, A.; Chang, Z.; Larsson, H.; Mataix-Cols, D.; Fernández De La Cruz, L. Association of Cesarean Delivery with Risk of Neurodevelopmental and Psychiatric Disorders in the Offspring: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2019, 2, e1910236. [Google Scholar] [CrossRef] [PubMed]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxidative Med. Cell. Longev. 2017, 2017, 3831972. [Google Scholar] [CrossRef] [PubMed]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Bressa, C.; Bailén-Andrino, M.; Pérez-Santiago, J.; González-Soltero, R.; Pérez, M.; Montalvo-Lominchar, M.G.; Maté-Muñoz, J.L.; Domínguez, R.; Moreno, D.; Larrosa, M. Differences in Gut Microbiota Profile between Women with Active Lifestyle and Sedentary Women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Stilling, R.M.; Kennedy, P.J.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Minireview: Gut Microbiota: The Neglected Endocrine Organ. Mol. Endocrinol. 2014, 28, 1221–1238. [Google Scholar] [CrossRef]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The Microbiome of Professional Athletes Differs from That of More Sedentary Subjects in Composition and Particularly at the Functional Metabolic Level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Holland, A.M.; Hyatt, H.W.; Smuder, A.J.; Sollanek, K.J.; Morton, A.B.; Roberts, M.D.; Kavazis, A.N. Influence of Endurance Exercise Training on Antioxidant Enzymes, Tight Junction Proteins, and Inflammatory Markers in the Rat Ileum. BMC Res. Notes 2015, 8, 514. [Google Scholar] [CrossRef]
- Cook, M.D.; Allen, J.M.; Pence, B.D.; Wallig, M.A.; Gaskins, H.R.; White, B.A.; Woods, J.A. Exercise and Gut Immune Function: Evidence of Alterations in Colon Immune Cell Homeostasis and Microbiome Characteristics with Exercise Training. Immunol. Cell Biol. 2016, 94, 158–163. [Google Scholar] [CrossRef]
- Zhang, M.; Xiao, B.; Chen, X.; Ou, B.; Wang, S. Physical Exercise Plays a Role in Rebalancing the Bile Acids of Enterohepatic Axis in Non-alcoholic Fatty Liver Disease. Acta Physiol. 2024, 240, e14065. [Google Scholar] [CrossRef]
- Boytar, A.N.; Skinner, T.L.; Wallen, R.E.; Jenkins, D.G.; Dekker Nitert, M. The Effect of Exercise Prescription on the Human Gut Microbiota and Comparison between Clinical and Apparently Healthy Populations: A Systematic Review. Nutrients 2023, 15, 1534. [Google Scholar] [CrossRef]
- Cronin, O.; Barton, W.; Skuse, P.; Penney, N.C.; Garcia-Perez, I.; Murphy, E.F.; Woods, T.; Nugent, H.; Fanning, A.; Melgar, S.; et al. A Prospective Metagenomic and Metabolomic Analysis of the Impact of Exercise and/or Whey Protein Supplementation on the Gut Microbiome of Sedentary Adults. mSystems 2018, 3, e00044-18. [Google Scholar] [CrossRef] [PubMed]
- Torquati, L.; Gajanand, T.; Cox, E.R.; Willis, C.R.G.; Zaugg, J.; Keating, S.E.; Coombes, J.S. Effects of Exercise Intensity on Gut Microbiome Composition and Function in People with Type 2 Diabetes. Eur. J. Sport Sci. 2023, 23, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Resende, A.S.; Leite, G.S.F.; Lancha Junior, A.H. Changes in the Gut Bacteria Composition of Healthy Men with the Same Nutritional Profile Undergoing 10-Week Aerobic Exercise Training: A Randomized Controlled Trial. Nutrients 2021, 13, 2839. [Google Scholar] [CrossRef] [PubMed]
- Warbeck, C.; Dowd, A.J.; Kronlund, L.; Parmar, C.; Daun, J.T.; Wytsma-Fisher, K.; Millet, G.Y.; Schick, A.; Reimer, R.A.; Fung, T.; et al. Feasibility and Effects on the Gut Microbiota of a 12-Week High-Intensity Interval Training plus Lifestyle Education Intervention on Inactive Adults with Celiac Disease. Appl. Physiol. Nutr. Metab. 2021, 46, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Ni, Y.; Cheung, C.K.Y.; Lam, K.S.L.; Wang, Y.; Xia, Z.; Ye, D.; Guo, J.; Tse, M.A.; et al. Gut Microbiome Fermentation Determines the Efficacy of Exercise for Diabetes Prevention. Cell Metab. 2020, 31, 77–91.e5. [Google Scholar] [CrossRef]
- Van Setten, G.-B. Ocular Surface Allostasis—When Homeostasis Is Lost: Challenging Coping Potential, Stress Tolerance, and Resilience. Biomolecules 2023, 13, 1246. [Google Scholar] [CrossRef]
- Chubar, V.; Vaessen, T.; Noortgate, W.V.D.; Lutin, E.; Bosmans, G.; Bekaert, B.; Van Leeuwen, K.; Calders, F.; Weyn, S.; Bijttebier, P.; et al. Mild Daily Stress, in Interaction with NR3C1 DNA Methylation Levels, Is Linked to Alterations in the HPA Axis and ANS Response to Acute Stress in Early Adolescents. Psychoneuroendocrinology 2023, 150, 106045. [Google Scholar] [CrossRef]
- Li, L.-F.; Zou, H.-W.; Song, B.-L.; Wang, Y.; Jiang, Y.; Li, Z.-L.; Niu, Q.-H.; Liu, Y.-J. Increased Lactobacillus Abundance Contributes to Stress Resilience in Mice Exposed to Chronic Social Defeat Stress. Neuroendocrinology 2023, 113, 563–576. [Google Scholar] [CrossRef]
- Mai, N.T.P.; Mai, N.T.T.; Phuong, D.T.M. An Association of Psychosocial Characteristics and Severity of Ulcers in Adolescents with Chronic Peptic Ulcer Disease. Tạp chí Nghiên cứu Y học 2023, 166, 36–43. [Google Scholar] [CrossRef]
- Lotti, S.; Dinu, M.; Colombini, B.; Amedei, A.; Sofi, F. Circadian Rhythms, Gut Microbiota, and Diet: Possible Implications for Health. Nutr. Metab. Cardiovasc. Dis. 2023, 33, 1490–1500. [Google Scholar] [CrossRef]
- Hantsoo, L.; Zemel, B.S. Stress Gets into the Belly: Early Life Stress and the Gut Microbiome. Behav. Brain Res. 2021, 414, 113474. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a Social Stressor Alters the Structure of the Intestinal Microbiota: Implications for Stressor-Induced Immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.D.; Bailey, M.T. Impact of Stressor Exposure on the Interplay between Commensal Microbiota and Host Inflammation. Gut Microbes 2014, 5, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Cao, Q.; Cheng, Y.; Zhao, D.; Wang, Z.; Yang, H.; Wu, Q.; You, L.; Wang, Y.; Lin, Y.; et al. Chronic Stress Promotes Colitis by Disturbing the Gut Microbiota and Triggering Immune System Response. Proc. Natl. Acad. Sci. USA 2018, 115, E2960–E2969. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Q.; Wang, Y.; Sun, A.; Lin, Y.; Jin, Y.; Li, X. Fecal Microbiota Transplantation from Chronic Unpredictable Mild Stress Mice Donors Affects Anxiety-like and Depression-like Behavior in Recipient Mice via the Gut Microbiota-Inflammation-Brain Axis. Stress 2019, 22, 592–602. [Google Scholar] [CrossRef]
- Knowles, S.R.; Nelson, E.A.; Palombo, E.A. Investigating the Role of Perceived Stress on Bacterial Flora Activity and Salivary Cortisol Secretion: A Possible Mechanism Underlying Susceptibility to Illness. Biol. Psychol. 2008, 77, 132–137. [Google Scholar] [CrossRef]
- Nishida, K.; Sawada, D.; Kuwano, Y.; Tanaka, H.; Rokutan, K. Health Benefits of Lactobacillus gasseri CP2305 Tablets in Young Adults Exposed to Chronic Stress: A Randomized, Double-Blind, Placebo-Controlled Study. Nutrients 2019, 11, 1859. [Google Scholar] [CrossRef]
- Michels, N.; Van De Wiele, T.; Fouhy, F.; O’Mahony, S.; Clarke, G.; Keane, J. Gut Microbiome Patterns Depending on Children’s Psychosocial Stress: Reports versus Biomarkers. Brain Behav. Immun. 2019, 80, 751–762. [Google Scholar] [CrossRef]
- Humbel, F.; Rieder, J.H.; Franc, Y.; Juillerat, P.; Scharl, M.; Misselwitz, B.; Schreiner, P.; Begré, S.; Rogler, G.; Von Känel, R.; et al. Association of Alterations in Intestinal Microbiota with Impaired Psychological Function in Patients with Inflammatory Bowel Diseases in Remission. Clin. Gastroenterol. Hepatol. 2020, 18, 2019–2029.e11. [Google Scholar] [CrossRef]
- Mackner, L.M.; Hatzakis, E.; Allen, J.M.; Davies, R.H.; Kim, S.C.; Maltz, R.M.; Bailey, M.T. Fecal Microbiota and Metabolites Are Distinct in a Pilot Study of Pediatric Crohn’s Disease Patients with Higher Levels of Perceived Stress. Psychoneuroendocrinology 2020, 111, 104469. [Google Scholar] [CrossRef]
- Amini-Khoei, H.; Haghani-Samani, E.; Beigi, M.; Soltani, A.; Mobini, G.R.; Balali-Dehkordi, S.; Haj-Mirzaian, A.; Rafieian-Kopaei, M.; Alizadeh, A.; Hojjati, M.R.; et al. On the Role of Corticosterone in Behavioral Disorders, Microbiota Composition Alteration and Neuroimmune Response in Adult Male Mice Subjected to Maternal Separation Stress. Int. Immunopharmacol. 2019, 66, 242–250. [Google Scholar] [CrossRef] [PubMed]
- El Aidy, S.; Ramsteijn, A.S.; Dini-Andreote, F.; Van Eijk, R.; Houwing, D.J.; Salles, J.F.; Olivier, J.D.A. Serotonin Transporter Genotype Modulates the Gut Microbiota Composition in Young Rats, an Effect Augmented by Early Life Stress. Front. Cell. Neurosci. 2017, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Marchesi, J.R.; Scully, P.; Codling, C.; Ceolho, A.-M.; Quigley, E.M.M.; Cryan, J.F.; Dinan, T.G. Early Life Stress Alters Behavior, Immunity, and Microbiota in Rats: Implications for Irritable Bowel Syndrome and Psychiatric Illnesses. Biol. Psychiatry 2009, 65, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, S.A.; Kang, W.S.; Kim, J.W. Early-Life Stress Modulates Gut Microbiota and Peripheral and Central Inflammation in a Sex-Dependent Manner. Int. J. Mol. Sci. 2021, 22, 1899. [Google Scholar] [CrossRef] [PubMed]
- Pusceddu, M.M.; El Aidy, S.; Crispie, F.; O’Sullivan, O.; Cotter, P.; Stanton, C.; Kelly, P.; Cryan, J.F.; Dinan, T.G. N-3 Polyunsaturated Fatty Acids (PUFAs) Reverse the Impact of Early-Life Stress on the Gut Microbiota. PLoS ONE 2015, 10, e0139721. [Google Scholar] [CrossRef]
- Rincel, M.; Olier, M.; Minni, A.; Monchaux De Oliveira, C.; Matime, Y.; Gaultier, E.; Grit, I.; Helbling, J.-C.; Costa, A.M.; Lépinay, A.; et al. Pharmacological Restoration of Gut Barrier Function in Stressed Neonates Partially Reverses Long-Term Alterations Associated with Maternal Separation. Psychopharmacology 2019, 236, 1583–1596. [Google Scholar] [CrossRef]
- Coley, E.J.L.; Mayer, E.A.; Osadchiy, V.; Chen, Z.; Subramanyam, V.; Zhang, Y.; Hsiao, E.Y.; Gao, K.; Bhatt, R.; Dong, T.; et al. Early Life Adversity Predicts Brain-Gut Alterations Associated with Increased Stress and Mood. Neurobiol. Stress 2021, 15, 100348. [Google Scholar] [CrossRef]
- Hemmings, S.M.J.; Malan-Müller, S.; Van Den Heuvel, L.L.; Demmitt, B.A.; Stanislawski, M.A.; Smith, D.G.; Bohr, A.D.; Stamper, C.E.; Hyde, E.R.; Morton, J.T.; et al. The Microbiome in Posttraumatic Stress Disorder and Trauma-Exposed Controls: An Exploratory Study. Psychosom. Med. 2017, 79, 936–946. [Google Scholar] [CrossRef]
- Aatsinki, A.-K.; Keskitalo, A.; Laitinen, V.; Munukka, E.; Uusitupa, H.-M.; Lahti, L.; Kortesluoma, S.; Mustonen, P.; Rodrigues, A.J.; Coimbra, B.; et al. Maternal Prenatal Psychological Distress and Hair Cortisol Levels Associate with Infant Fecal Microbiota Composition at 2.5 Months of Age. Psychoneuroendocrinology 2020, 119, 104754. [Google Scholar] [CrossRef]
- Hu, J.; Ly, J.; Zhang, W.; Huang, Y.; Glover, V.; Peter, I.; Hurd, Y.L.; Nomura, Y. Microbiota of Newborn Meconium Is Associated with Maternal Anxiety Experienced during Pregnancy. Dev. Psychobiol. 2019, 61, 640–649. [Google Scholar] [CrossRef]
- Callaghan, B.L.; Fields, A.; Gee, D.G.; Gabard-Durnam, L.; Caldera, C.; Humphreys, K.L.; Goff, B.; Flannery, J.; Telzer, E.H.; Shapiro, M.; et al. Mind and Gut: Associations between Mood and Gastrointestinal Distress in Children Exposed to Adversity. Dev. Psychopathol. 2020, 32, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Horne, R.; Donzella, B.; Szamosi, J.C.; Coe, C.L.; Foster, J.A.; Gunnar, M.R. Microbiota-immune Alterations in Adolescents Following Early Life Adversity: A Proof of Concept Study. Dev. Psychobiol. 2021, 63, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, H. The Gut-Brain Axis: Literature Overview and Psychiatric Applications. Fed. Pract. 2021, 38, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Eijsbouts, C.; Zheng, T.; Kennedy, N.A.; Bonfiglio, F.; Anderson, C.A.; Moutsianas, L.; Holliday, J.; Shi, J.; Shringarpure, S.; 23andMe Research Team; et al. Genome-Wide Analysis of 53,400 People with Irritable Bowel Syndrome Highlights Shared Genetic Pathways with Mood and Anxiety Disorders. Nat. Genet. 2021, 53, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Tylee, D.S.; Lee, Y.K.; Wendt, F.R.; Pathak, G.A.; Levey, D.F.; De Angelis, F.; Gelernter, J.; Polimanti, R. An Atlas of Genetic Correlations and Genetically Informed Associations Linking Psychiatric and Immune-Related Phenotypes. JAMA Psychiatry 2022, 79, 667. [Google Scholar] [CrossRef]
- Wu, Y.; Murray, G.K.; Byrne, E.M.; Sidorenko, J.; Visscher, P.M.; Wray, N.R. GWAS of Peptic Ulcer Disease Implicates Helicobacter Pylori Infection, Other Gastrointestinal Disorders and Depression. Nat. Commun. 2021, 12, 1146. [Google Scholar] [CrossRef]
- Okbay, A.; Baselmans, B.M.L.; De Neve, J.-E.; Turley, P.; Nivard, M.G.; Fontana, M.A.; Meddens, S.F.W.; Linnér, R.K.; Rietveld, C.A.; Derringer, J.; et al. Genetic Variants Associated with Subjective Well-Being, Depressive Symptoms, and Neuroticism Identified through Genome-Wide Analyses. Nat. Genet. 2016, 48, 624–633. [Google Scholar] [CrossRef]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-Wide Association Analyses Identify 44 Risk Variants and Refine the Genetic Architecture of Major Depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef]
- Demontis, D.; Walters, R.K.; Martin, J.; Mattheisen, M.; Als, T.D.; Agerbo, E.; Baldursson, G.; Belliveau, R.; Bybjerg-Grauholm, J.; Bækvad-Hansen, M.; et al. Discovery of the First Genome-Wide Significant Risk Loci for Attention Deficit/Hyperactivity Disorder. Nat. Genet. 2019, 51, 63–75. [Google Scholar] [CrossRef]
- Hammerschlag, A.R.; Stringer, S.; De Leeuw, C.A.; Sniekers, S.; Taskesen, E.; Watanabe, K.; Blanken, T.F.; Dekker, K.; Te Lindert, B.H.W.; Wassing, R.; et al. Genome-Wide Association Analysis of Insomnia Complaints Identifies Risk Genes and Genetic Overlap with Psychiatric and Metabolic Traits. Nat. Genet. 2017, 49, 1584–1592. [Google Scholar] [CrossRef]
- Lane, J.M.; Liang, J.; Vlasac, I.; Anderson, S.G.; Bechtold, D.A.; Bowden, J.; Emsley, R.; Gill, S.; Little, M.A.; Luik, A.I.; et al. Genome-Wide Association Analyses of Sleep Disturbance Traits Identify New Loci and Highlight Shared Genetics with Neuropsychiatric and Metabolic Traits. Nat. Genet. 2017, 49, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Pouget, J.G.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Han, B.; Wu, Y.; Mignot, E.; Ollila, H.M.; Barker, J.; Spain, S.; Dand, N.; Trembath, R.; et al. Cross-Disorder Analysis of Schizophrenia and 19 Immune-Mediated Diseases Identifies Shared Genetic Risk. Hum. Mol. Genet. 2019, 28, 3498–3513. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Guo, P.; Li, Y.; Liu, L.; Yan, R.; Liu, S.; Wang, S.; Xue, F.; Zhou, X.; Yuan, Z. Role of the Gut-Brain Axis in the Shared Genetic Etiology between Gastrointestinal Tract Diseases and Psychiatric Disorders: A Genome-Wide Pleiotropic Analysis. JAMA Psychiatry 2023, 80, 360. [Google Scholar] [CrossRef] [PubMed]
- Uellendahl-Werth, F.; Maj, C.; Borisov, O.; Juzenas, S.; Wacker, E.M.; Jørgensen, I.F.; Steiert, T.A.; Bej, S.; Krawitz, P.; Hoffmann, P.; et al. Cross-Tissue Transcriptome-Wide Association Studies Identify Susceptibility Genes Shared between Schizophrenia and Inflammatory Bowel Disease. Commun. Biol. 2022, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; He, M.; Liu, Z.; Qin, Z.; Wang, Z.; Duan, L. Shared Genetic Susceptibilities for Irritable Bowel Syndrome and Depressive Disorder in Chinese Patients Uncovered by Pooled Whole-Exome Sequencing. J. Adv. Res. 2020, 23, 113–121. [Google Scholar] [CrossRef]
- Luo, J.; Xu, Z.; Noordam, R.; Van Heemst, D.; Li-Gao, R. Depression and Inflammatory Bowel Disease: A Bidirectional Two-Sample Mendelian Randomization Study. J. Crohn’s Colitis 2022, 16, 633–642. [Google Scholar] [CrossRef]
- Sadik, A.; Dardani, C.; Pagoni, P.; Havdahl, A.; Stergiakouli, E.; The iPSYCH Autism Spectrum Disorder Working Group; Grove, J.; Khandaker, G.M.; Sullivan, S.A.; Zammit, S.; et al. Parental Inflammatory Bowel Disease and Autism in Children. Nat. Med. 2022, 28, 1406–1411. [Google Scholar] [CrossRef]




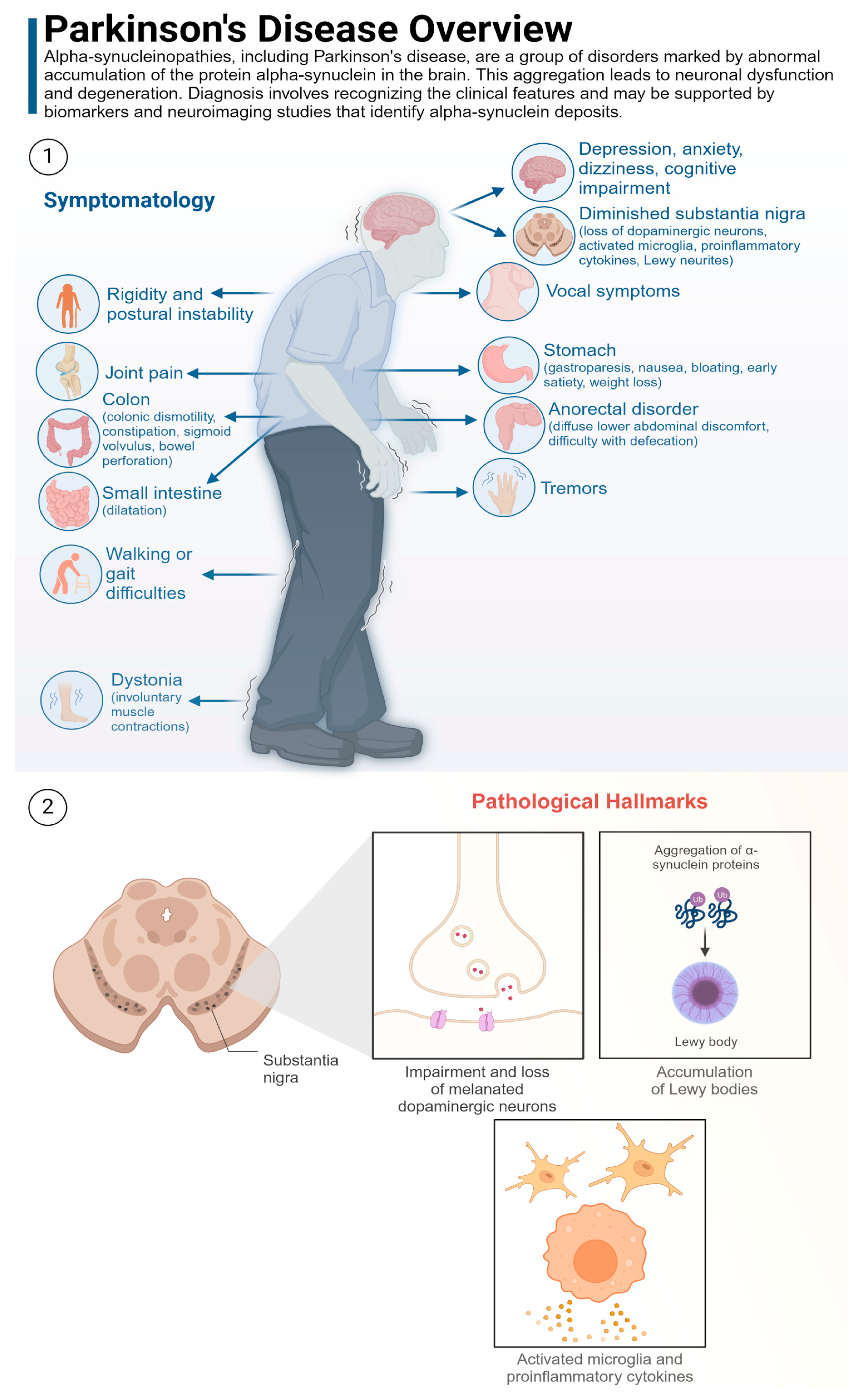
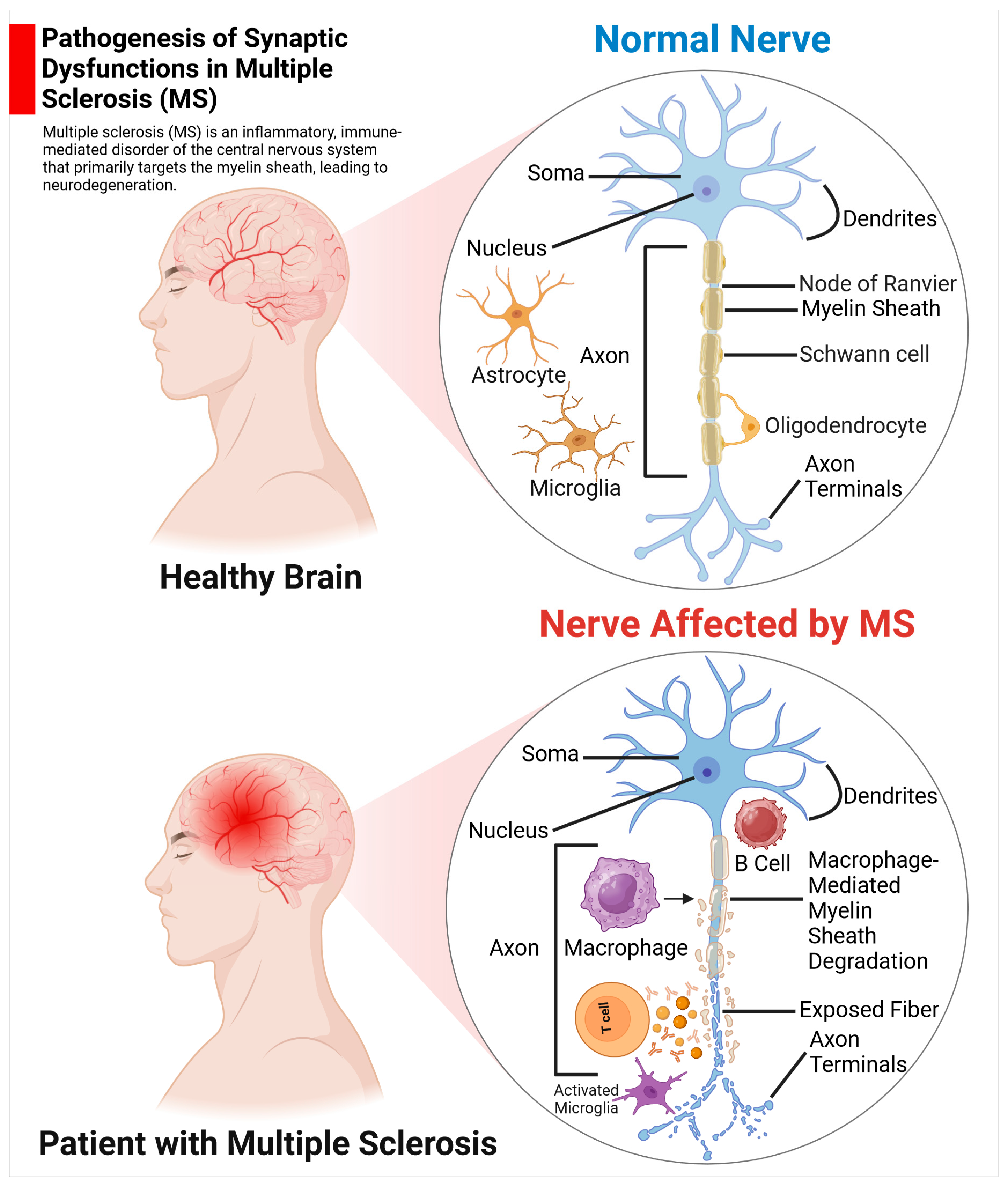
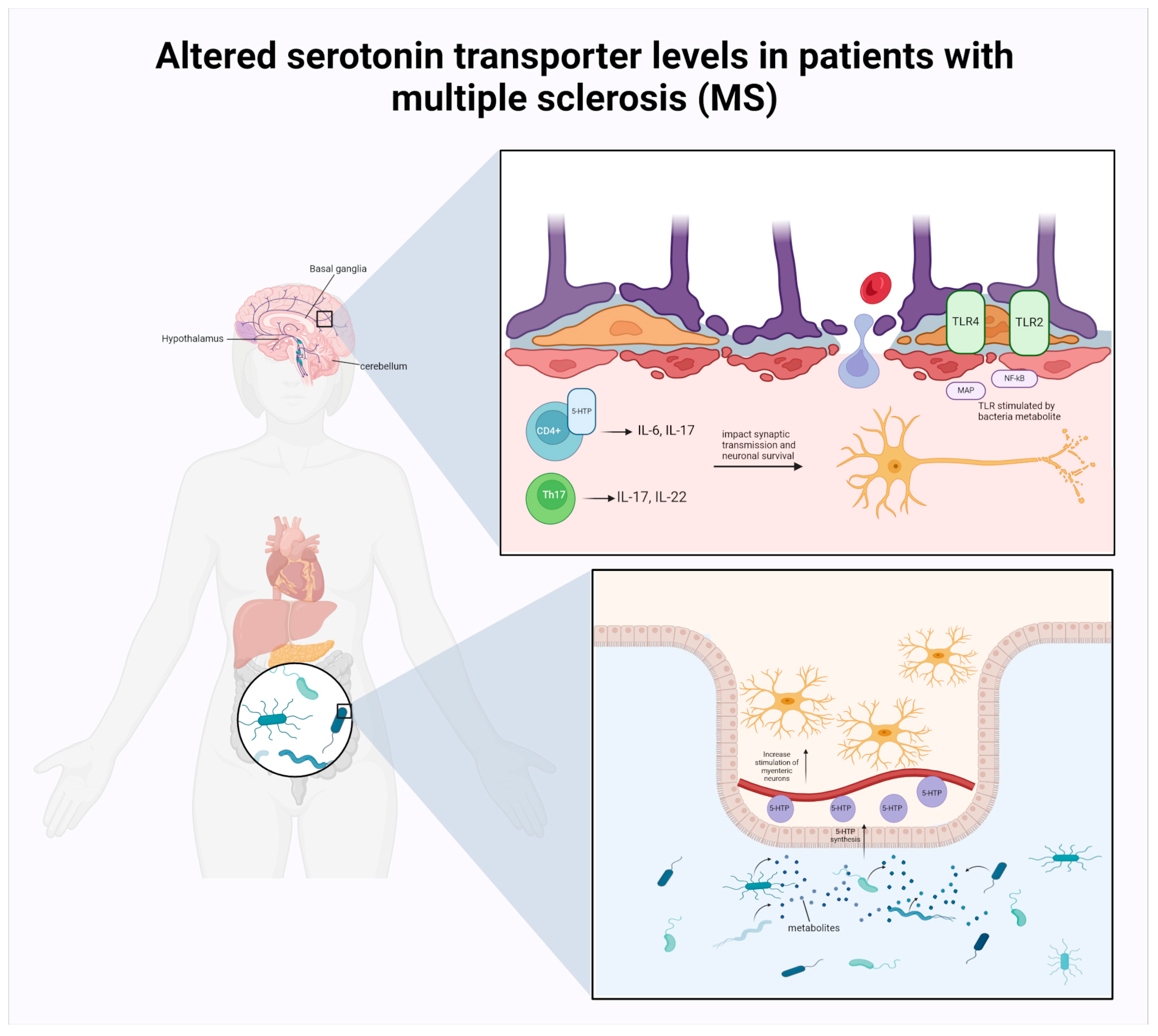
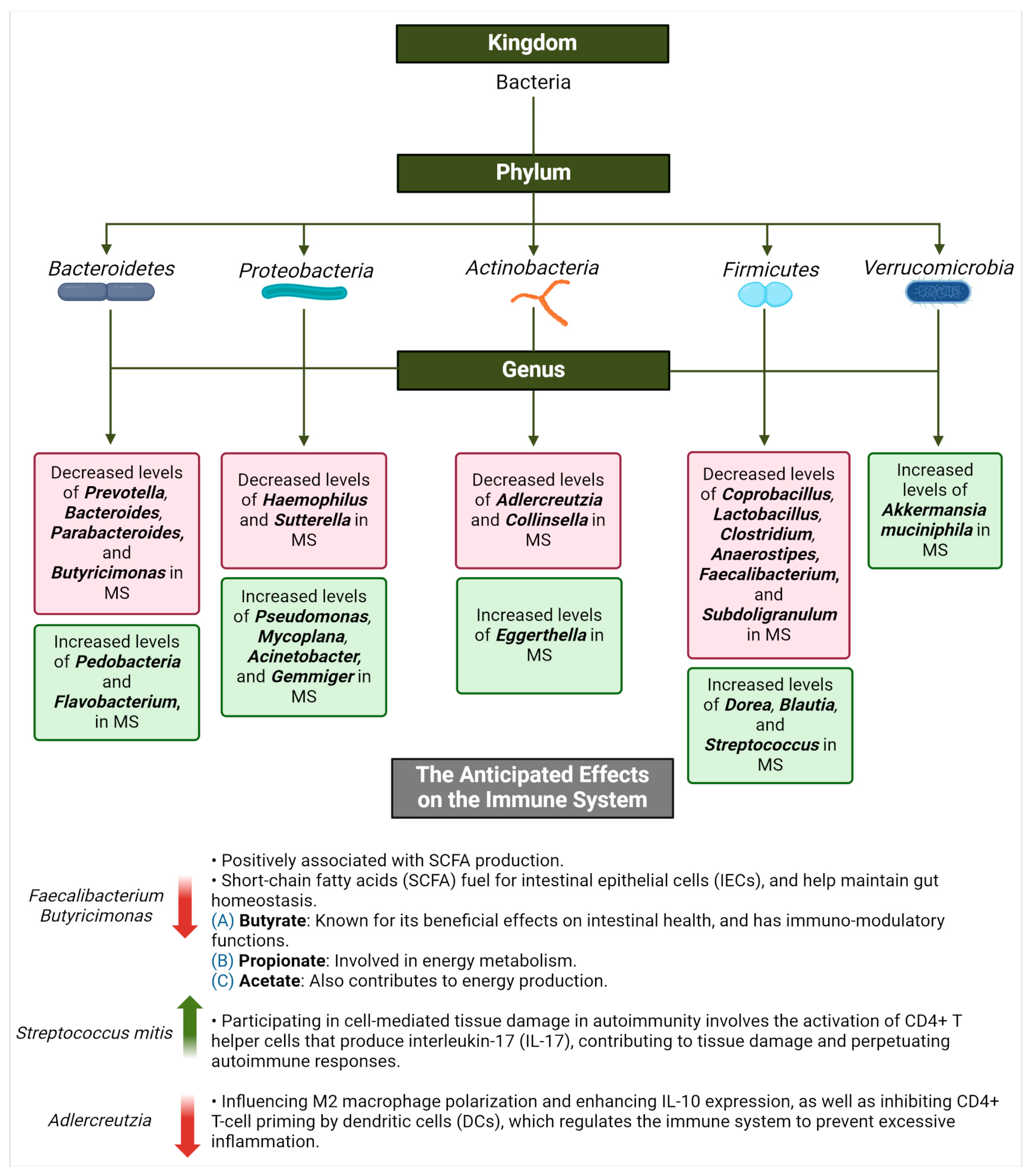
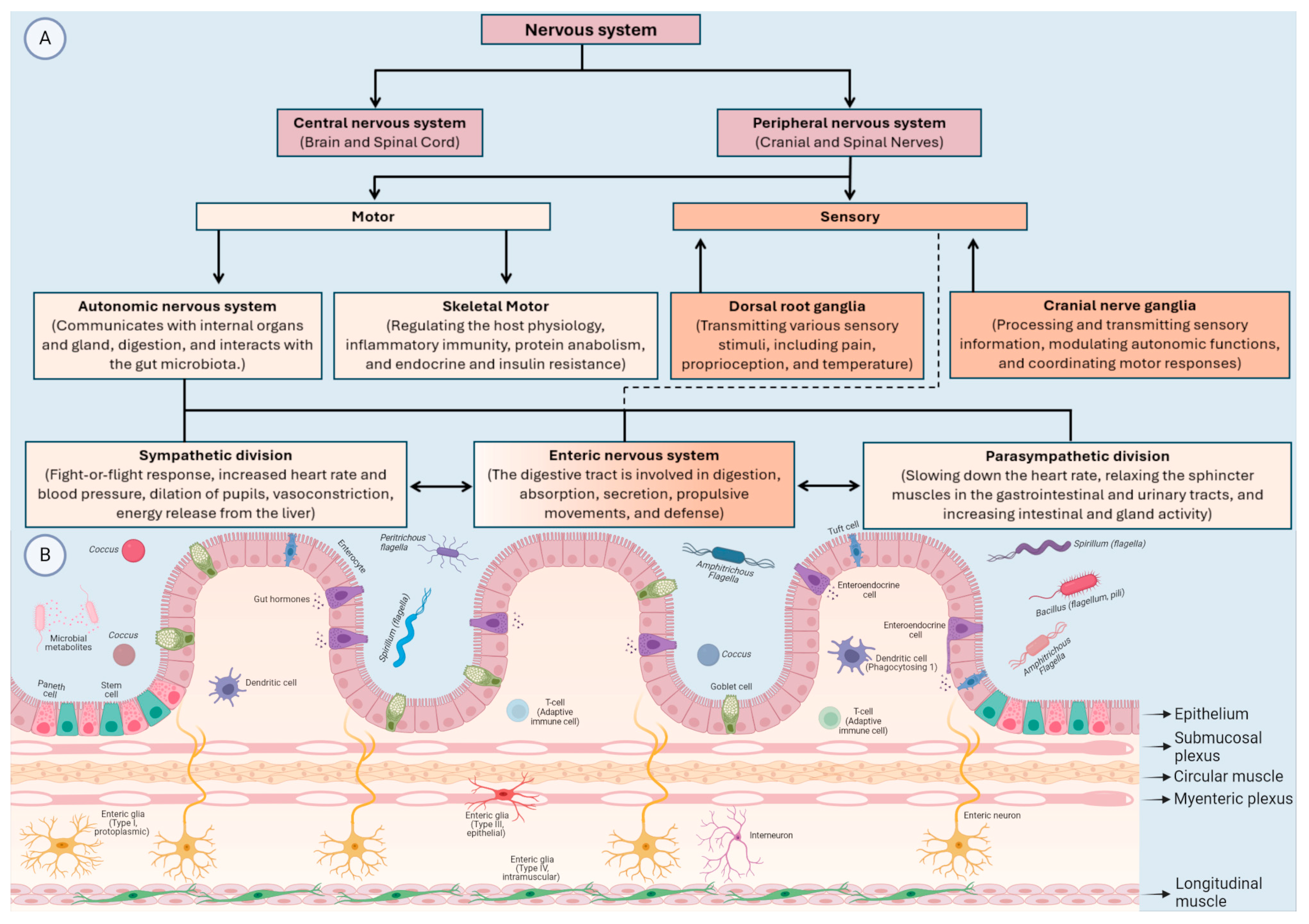
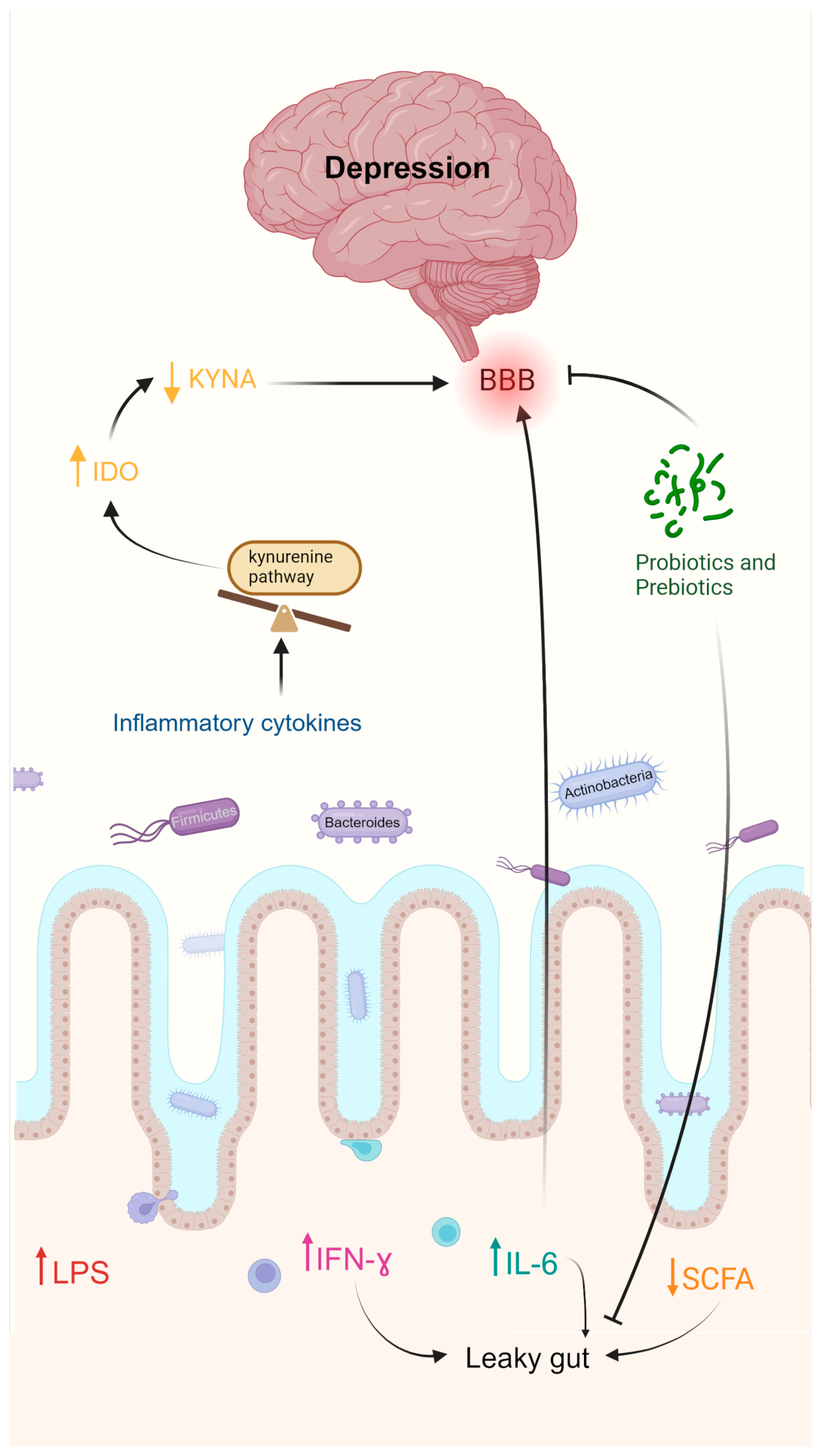
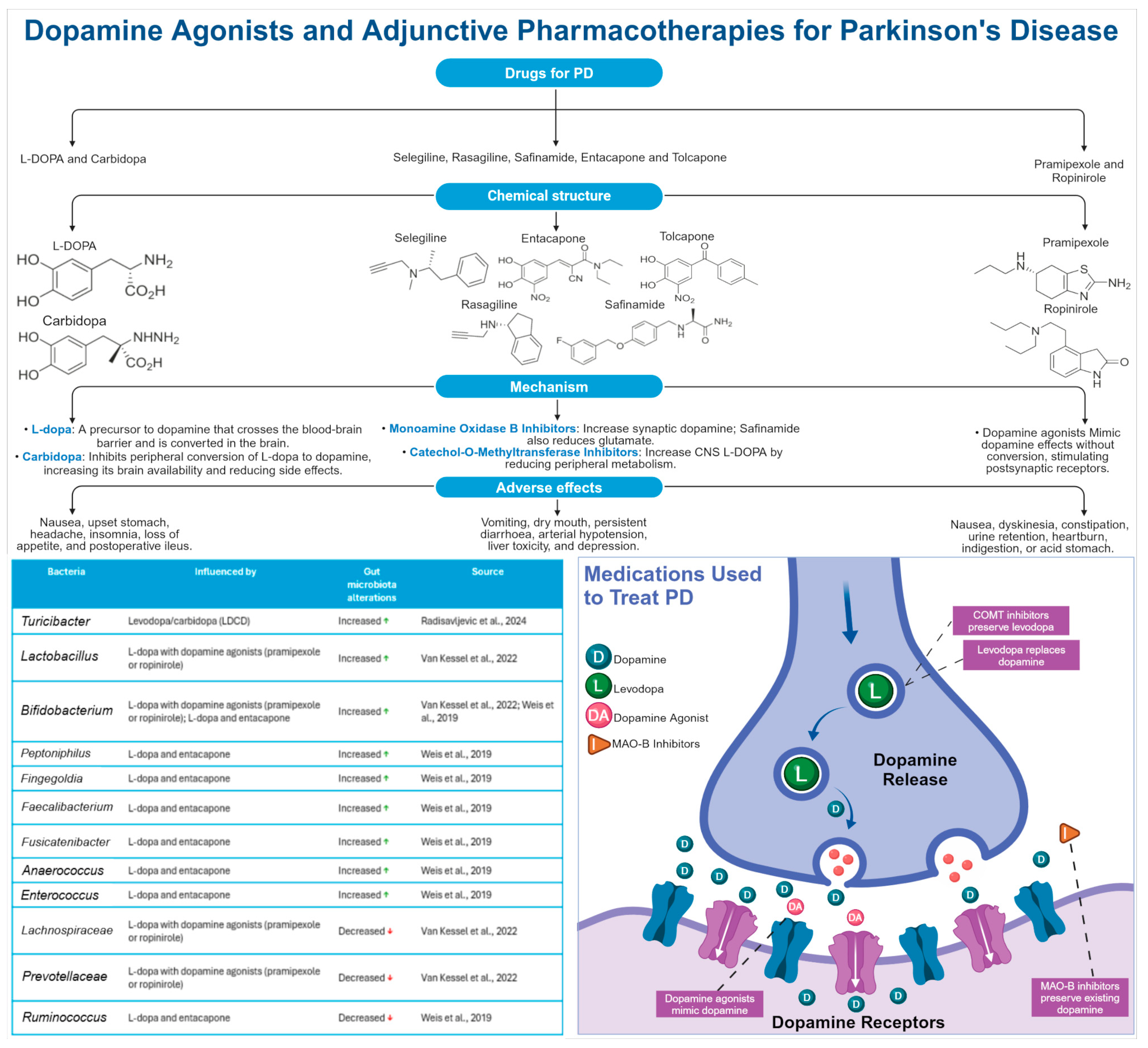
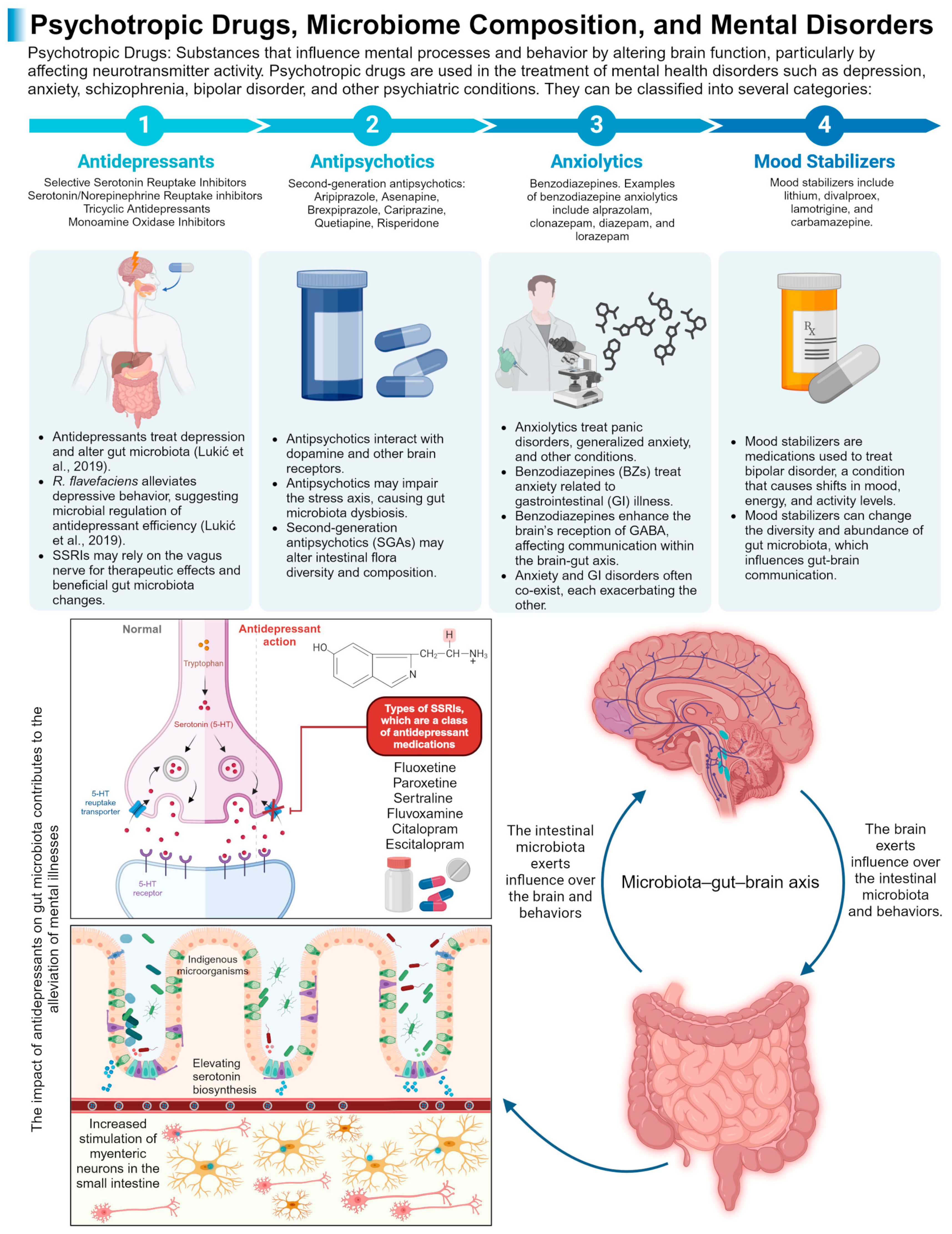
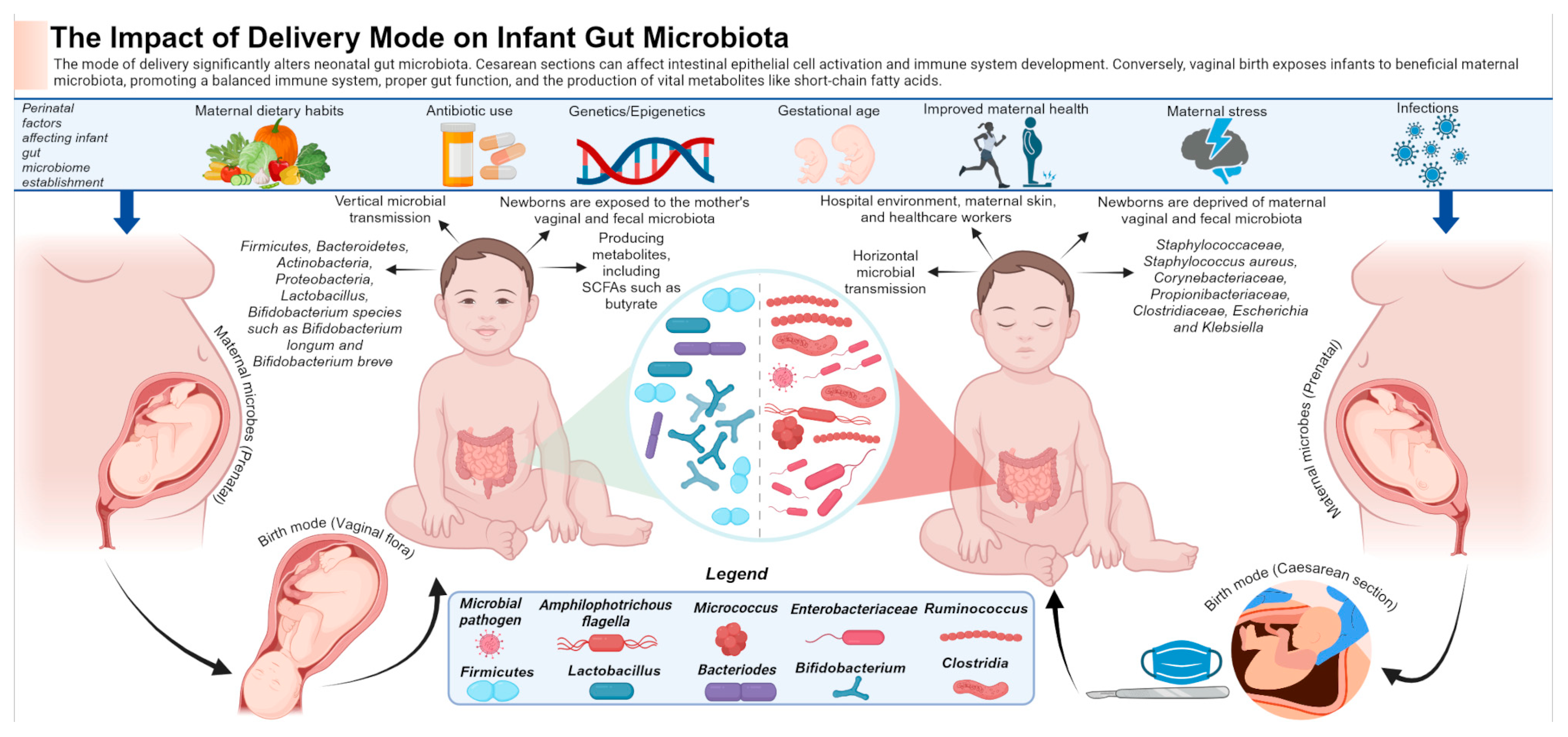

| Condition | Bacteria Phylum | Genus/spp. | Condition Compared to Control Group | References |
|---|---|---|---|---|
| Rett Syndrome | Firmicutes | Ruminococcus spp. | ↓ levels in the intestinal microbiome | [26,29,30] |
| Rett Syndrome | Firmicutes | Faecalibacterium prausnitzii (species) | ↓ levels in the intestinal microbiome | [26,29,30] |
| Rett Syndrome | Firmicutes | Clostridium spp. | ↑ levels in the intestinal microbiome | [26,29,30] |
| Rett Syndrome | Firmicutes | Sutterella spp. | ↑ levels in the intestinal microbiome | [26] |
| Rett Syndrome | Firmicutes | Erysipelatoclostridium | ↑ levels in the intestinal microbiome | [26,29] |
| Rett Syndrome | Firmicutes | Lactobacillaceae spp. | ↑ levels in the intestinal microbiome | [29,30] |
| Rett Syndrome | Firmicutes | Oscillibacter spp. | ↓ levels in the intestinal microbiome | [30] |
| Rett Syndrome | Firmicutes | Sporobacter spp. | ↓ levels in the intestinal microbiome | [30] |
| Rett Syndrome | Firmicutes | Veillonellaceae spp. | ↑ levels in the intestinal microbiome | [31] |
| Rett Syndrome | Firmicutes | Enterococcus spp. | ↑ levels in the intestinal microbiome | [29] |
| Rett Syndrome | Bacteroidetes | Bacteroides spp. | ↑ levels in the intestinal microbiome | [26,29] |
| Rett Syndrome | Bacteroidetes | Prevotella spp. | ↓ levels in the intestinal microbiome | [26,29,30] |
| Rett Syndrome | Bacteroidetes | Barnesiella spp. | ↓ levels in the intestinal microbiome | [30] |
| Rett Syndrome | Bacteroidetes | Alistipes spp. | ↓ levels in the intestinal microbiome | [30] |
| Rett Syndrome | Bacteroidetes | Odoribacter spp. | ↓ levels in the intestinal microbiome | [31] |
| Rett Syndrome | Bacteroidetes | Butyricimonas spp. | ↓ levels in the intestinal microbiome | [31] |
| Rett Syndrome | Bacteroidetes | Rikenellaceae spp. | ↑ levels in the intestinal microbiome | [31] |
| Rett Syndrome | Actinobacteria | Bifidobacterium spp. | ↑ levels in the intestinal microbiome | [26,29,30] |
| Rett Syndrome | Actinobacteria | Actinomyces spp. | ↑ levels in the intestinal microbiome | [26,29] |
| Rett Syndrome | Actinobacteria | Eggerthella spp. | ↑ levels in the intestinal microbiome | [29] |
| Rett Syndrome | Proteobacteria | Escherichia/Shigella spp. | ↑ levels in the intestinal microbiome | [26,29] |
| Rett Syndrome | Verrucomicrobia | Verrucomicrobiaceae spp. | ↓ levels in the intestinal microbiome | [31] |
| Assessment Methods | Potential Confounders | Taxonomic Composition Changes | Potential Changes in Synaptic Plasticity | References |
|---|---|---|---|---|
| ADHD diagnosed using Kiddie-SADS-PL, symptoms severity assessed with CPRS | Dietary habits, gastrointestinal symptoms, depression, ADHD medications | No significant change in α or β-diversity ↓ Faecalibacterium levels | ↑ Systematic inflammation ↑ Gut permeability ↑ Unbalanced neurotransmitters levels in the brain | [47] |
| Diagnosed using DSM-IV via K-SADS, symptom severity assessed with CPRS. | ADHD medications | ↑ β-diversity in ADHD ↑ Ruminococcaceae_UGC_004 | CD74, TNF, cytokine receptors | [48] |
| Wilcoxon tests for species abundance, LEfSe method for taxa differences, symptom severity assessed with CPRS | Gastrointestinal symptoms, depression or anxiety, use of probiotics or antibiotics, obesity, allergy | ↓ Faecalibacterium prausnitzii ↓ Lachnospiraceae spp. ↓ Ruminococcus gnavus ↑ Bacteroides caccae ↑ Odoribacter Splanchnicus ↑ Paraprevotella Xylaniphila ↑ Veillonella parvula | ↑ Inflammatory factors ↓ Neuroplasticity | [49] |
| Children’s Global Assessment Scale, ADHD Rating Scale-IV, 16S rRNA gene sequencing, dietary intake questionnaire | Unspecified | No significant change in α or β-diversity ↓ Bifidobacterium | ↑ Neural signalling modulation ↓ Inflammatory responses ↓ Gut–brain axis dysregulation ↓ Oxidative stress levels | [50] |
| Condition | Bacteria Phylum | Genus/spp. | Condition Compared to Control Group | References |
|---|---|---|---|---|
| Parkinson’s Disease | Verrucomicrobia | Akkermansia spp. | ↑ levels in the intestinal microbiome | [124,127,128,130] |
| Parkinson’s Disease | Firmicutes | Blautia spp. | ↓ levels in the intestinal microbiome | [129] |
| Parkinson’s Disease | Firmicutes | Coprococcus spp. | ↓ levels in the intestinal microbiome | [129] |
| Parkinson’s Disease | Firmicutes | Roseburia spp. | ↓ levels in the intestinal microbiome | [129] |
| Parkinson’s Disease | Firmicutes | Faecalibacterium spp. | ↓ levels in the intestinal microbiome | [123,129] |
| Parkinson’s Disease | Proteobacteria | Ralstonia spp. | ↑ levels in the intestinal microbiome | [129] |
| Parkinson’s Disease | Proteobacteria | Enterobacteriaceae spp. | ↑ levels in the intestinal microbiome | [122] |
| Parkinson’s Disease | Bacteroidetes | Prevotellaceae spp. | ↓ levels in the intestinal microbiome | [122] |
| Neurotransmitters | Precursors | Gut Microbiota | Proposed Roles within the Gut–Brain Axis | References |
|---|---|---|---|---|
| Glutamate | Acetate | Lactobacillus plantarum Bacteroides vulgatus Campylobacter jejuni | Transfer intestinal sensory signals to the brain through the vagus nerve; regulate neurogenesis; synaptogenesis; neuron survival | [190] |
| Acetylcholine | Choline | Lactobacillus plantarum Bacillus acetylcholine Bacillus subtilis Escherichia coli Staphylococcus aureus | Regulate intestinal motility and secretion and enteric neurotransmission; retain brain plasticity | [191] |
| Dopamine | Tyrosine L-DOPA | Staphylococcus | Promote intestinal motility and modulates STDP | [192] |
| Serotonin | 5-HTP Tryptophan | Staphylococcus Clostridial spp. | Remodels neuronal cytoarchitecture | [193] |
| GABA | Acetate | Bifidobacterium Bacteroides fragilis Parabacteroides Eubacterium | Modulates synaptic transmission in the ENS; regulates inhibitory–excitatory balance | [194,195] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakhal, M.M.; Yassin, L.K.; Alyaqoubi, R.; Saeed, S.; Alderei, A.; Alhammadi, A.; Alshehhi, M.; Almehairbi, A.; Al Houqani, S.; BaniYas, S.; et al. The Microbiota–Gut–Brain Axis and Neurological Disorders: A Comprehensive Review. Life 2024, 14, 1234. https://doi.org/10.3390/life14101234
Nakhal MM, Yassin LK, Alyaqoubi R, Saeed S, Alderei A, Alhammadi A, Alshehhi M, Almehairbi A, Al Houqani S, BaniYas S, et al. The Microbiota–Gut–Brain Axis and Neurological Disorders: A Comprehensive Review. Life. 2024; 14(10):1234. https://doi.org/10.3390/life14101234
Chicago/Turabian StyleNakhal, Mohammed M., Lidya K. Yassin, Rana Alyaqoubi, Sara Saeed, Alreem Alderei, Alya Alhammadi, Mirah Alshehhi, Afra Almehairbi, Shaikha Al Houqani, Shamsa BaniYas, and et al. 2024. "The Microbiota–Gut–Brain Axis and Neurological Disorders: A Comprehensive Review" Life 14, no. 10: 1234. https://doi.org/10.3390/life14101234








