Viability and Motility of Escherichia coli Under Elevated Martian Salt Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms Used
2.2. Incubation of Microorganisms
2.3. Plating
2.4. Tracking and Analysis of Microbial Motility
3. Results
3.1. Effects of Sodium Chloride on Viability
3.2. Effects of Sodium Chlorate on Viability
3.3. Effects of Sodium Perchlorate on Viability
3.4. Effects of Sodium Chloride on Motility
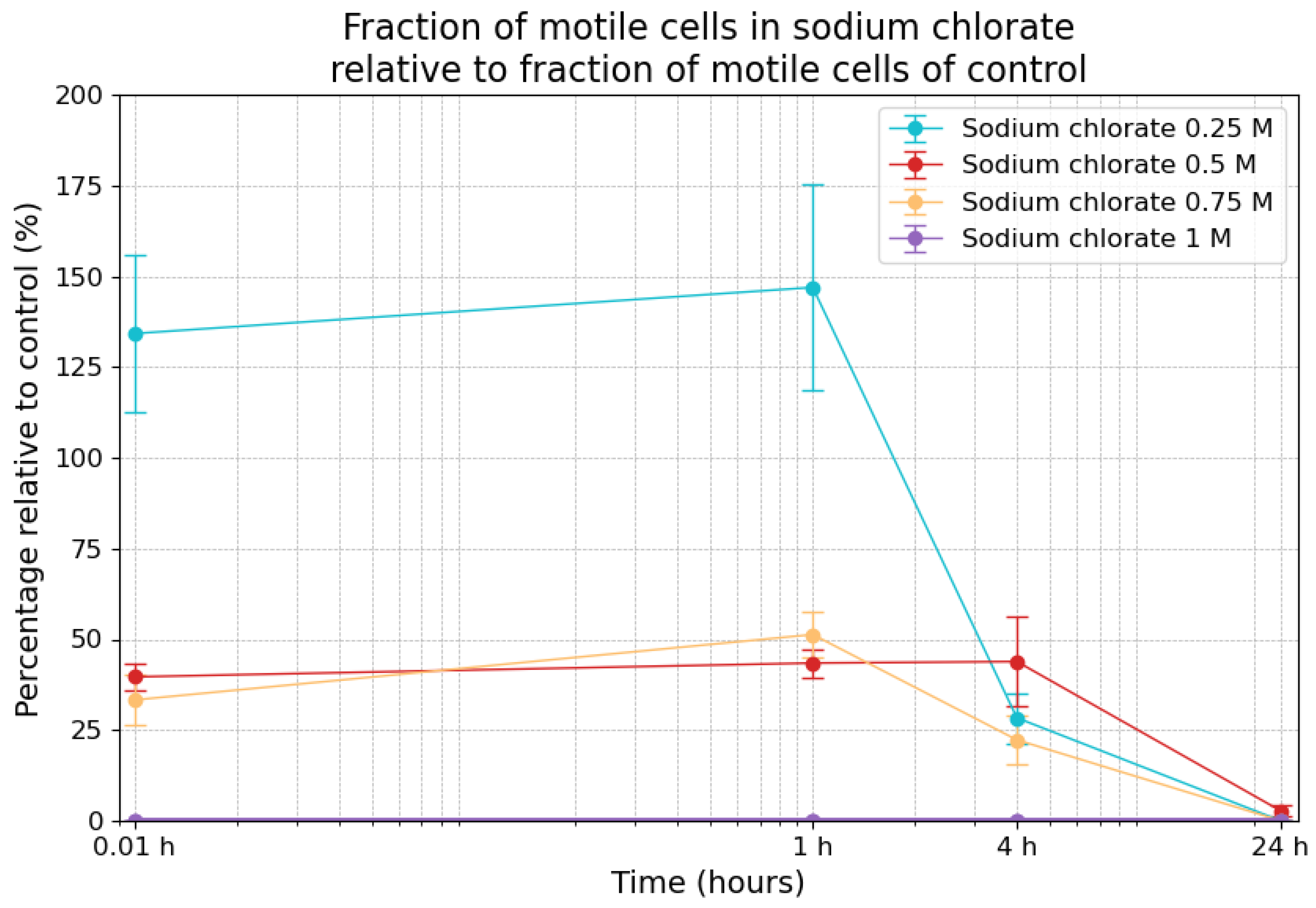
3.5. Effects of Sodium Chlorate on Motility
3.6. Effects of Sodium Perchlorate on Motility
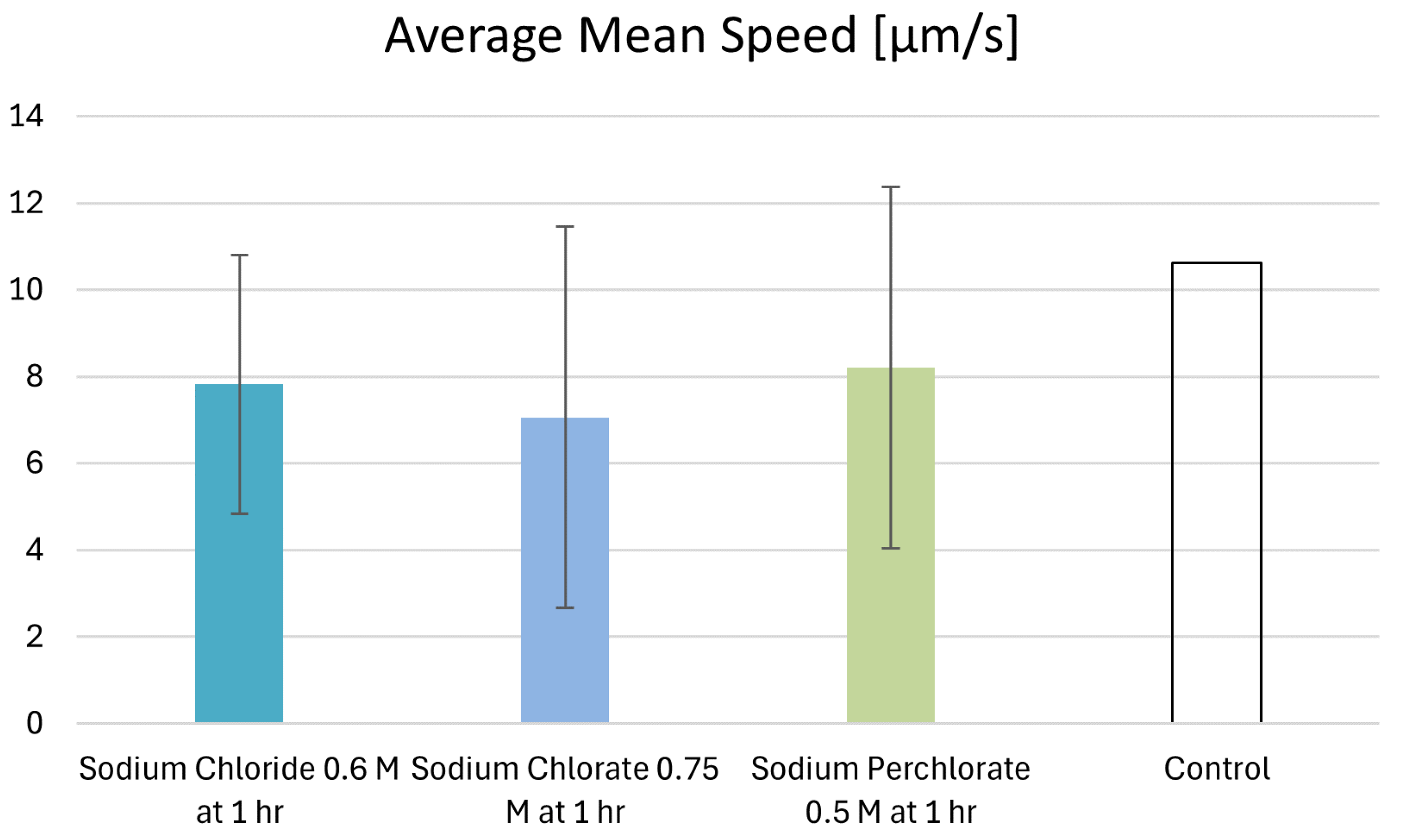
3.7. Motility Behavior of Individual Cells
4. Discussion
5. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cockell, C.S. Trajectories of martian habitability. Astrobiology 2014, 14, 182–203. [Google Scholar] [CrossRef] [PubMed]
- Haberle, R.M.; McKay, C.P.; Schaeffer, J.; Cabrol, N.A.; Grin, E.A.; Zent, A.P.; Quinn, R. On the possibility of liquid water on present-day Mars. J. Geophys. Res. 2001, 106, 23317–23326. [Google Scholar] [CrossRef]
- Masson, P.; Carr, M.H.; Costard, F.; Greeley, R.; Hauber, E.; Jaumann, R. Geomorphologic Evidence for Liquid Water. Space Sci. Rev. 2001, 96, 333–364. [Google Scholar] [CrossRef]
- Nair, C.P.R.; Unnikrishnan, V. Stability of the Liquid Water Phase on Mars: A Thermodynamic Analysis Considering Martian Atmospheric Conditions and Perchlorate Brine Solutions. ACS Omega 2020, 5, 9391–9397. [Google Scholar] [CrossRef]
- Renno, N.O.; Fischer, E.; Martínez, G.; Hanley, J. Complex Brines and Their Implications for Habitability. Life 2021, 11, 847. [Google Scholar] [CrossRef]
- Zorzano, M.-P.; Mateo-Martí, E.; Prieto-Ballesteros, O.; Osuna, S.; Renno, N. Stability of liquid saline water on present day Mars. Geophys. Res. Lett. 2009, 36, L20201. [Google Scholar] [CrossRef]
- Gough, R.V.; Chevrier, V.F.; Baustian, K.J.; Wise, M.E.; Tolbert, M.A. Laboratory studies of perchlorate phase transitions: Support for metastable aqueous perchlorate solutions on Mars. Earth Planet. Sci. Lett. 2011, 312, 371–377. [Google Scholar] [CrossRef]
- Martínez, G.M.; Renno, N.O. Water and Brines on Mars: Current Evidence and Implications for MSL. Space Sci. Rev. 2013, 175, 29–51. [Google Scholar] [CrossRef]
- Fischer, E.; Martínez, G.M.; Elliott, H.M.; Rennó, N.O. Experimental evidence for the formation of liquid saline water on Mars. Geophys. Res. Lett. 2014, 41, 4456–4462. [Google Scholar] [CrossRef]
- Gough, R.V.; Chevrier, V.F.; Tolbert, M.A. Formation of aqueous solutions on Mars via deliquescence of chloride–perchlorate binary mixtures. Earth Planet. Sci. Lett. 2014, 393, 73–82. [Google Scholar] [CrossRef]
- Nuding, D.L.; Rivera-Valentin, E.G.; Davis, R.D.; Gough, R.V.; Chevrier, V.F.; Tolbert, M.A. Deliquescence and efflorescence of calcium perchlorate: An investigation of stable aqueous solutions relevant to Mars. Icarus 2014, 243, 420–428. [Google Scholar] [CrossRef]
- Nikolakakos, G.; Whiteway, J.A. Laboratory investigation of perchlorate deliquescence at the surface of Mars with a Raman scattering lidar. Geophys. Res. Lett. 2015, 42, 7899–7906. [Google Scholar] [CrossRef]
- Nuding, D.L.; Davis, R.D.; Gough, R.V.; Tolbert, M.A. The aqueous stability of a Mars salt analog: Instant Mars. J. Geophys. Res. 2015, 120, 588–598. [Google Scholar] [CrossRef]
- Heinz, J.; Schulze-Makuch, D.; Kounaves, S.P. Deliquescence-induced wetting and RSL-like darkening of a Mars analogue soil containing various perchlorate and chloride salts. Geophys. Res. Lett. 2016, 43, 4880–4884. [Google Scholar] [CrossRef] [PubMed]
- Maus, D.; Heinz, J.; Schirmack, J.; Airo, A.; Kounaves, S.P.; Wagner, D.; Schulze-Makuch, D. Methanogenic Archaea Can Produce Methane in Deliquescence-Driven Mars Analog Environments. Sci. Rep. 2020, 10, 6. [Google Scholar] [CrossRef]
- Rivera-Valentín, E.G.; Chevrier, V.F.; Soto, A.; Martínez, G. Distribution and habitability of (meta)stable brines on present-day Mars. Nat. Astron. 2020, 4, 756–761. [Google Scholar] [CrossRef]
- Fischer, F.C.; Schulze-Makuch, D.; Heinz, J. Microbial preference for chlorate over perchlorate under simulated shallow subsurface Mars-like conditions. Sci. Rep. 2024, 14, 11537. [Google Scholar] [CrossRef]
- Rennó, N.O.; Bos, B.J.; Catling, D.; Clark, B.C.; Drube, L.; Fisher, D.; Goetz, W.; Hviid, S.F.; Keller, H.U.; Kok, J.F.; et al. Possible physical and thermodynamical evidence for liquid water at the Phoenix landing site. J. Geophys. Res. 2009, 114, E00E03. [Google Scholar] [CrossRef]
- Davila, A.F.; Willson, D.; Coates, J.D.; McKay, C.P. Perchlorate on Mars: A chemical hazard and a resource for humans. Int. J. Astrobiol. 2013, 12, 321–325. [Google Scholar] [CrossRef]
- Ming, D.W.; Archer, P.D.; Glavin, D.P.; Eigenbrode, J.L.; Franz, H.B.; Sutter, B.; Brunner, A.E.; Stern, J.C.; Freissinet, C.; McAdam, A.C.; et al. Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale crater, Mars. Science 2014, 343, 1245267. [Google Scholar] [CrossRef]
- Stern, J.C.; Sutter, B.; Jackson, W.A.; Navarro-González, R.; McKay, C.P.; Ming, D.W.; Archer, P.D.; Mahaffy, P.R. The nitrate/(per)chlorate relationship on Mars. Geophys. Res. Lett. 2017, 44, 2643–2651. [Google Scholar] [CrossRef]
- Kounaves, S.P.; Carrier, B.L.; O’Neil, G.D.; Stroble, S.T.; Claire, M.W. Evidence of martian perchlorate, chlorate, and nitrate in Mars meteorite EETA79001: Implications for oxidants and organics. Icarus 2014, 229, 206–213. [Google Scholar] [CrossRef]
- Schulze-Makuch, D. We may be looking for Martian life in the wrong place. Nat. Astron. 2024, 8, 1208–1210. [Google Scholar] [CrossRef]
- Bridges, J. Evaporite mineral assemblages in the nakhlite (martian) meteorites. Earth Planet. Sci. Lett. 2000, 176, 267–279. [Google Scholar] [CrossRef]
- Osterloo, M.M.; Anderson, F.S.; Hamilton, V.E.; Hynek, B.M. Geologic context of proposed chloride-bearing materials on Mars. J. Geophys. Res. 2010, 115, E10012. [Google Scholar] [CrossRef]
- Leask, E.K.; Ehlmann, B.L. Evidence for deposition of chlorideon Mars from small-volume surfacewater events into the Late Hesperian-Early Amazonian. AGU Adv. 2022, 3, e2021AV000534. [Google Scholar] [CrossRef]
- Bramble, M.S.; Hand, K.P. Spectral evidence for irradiated halite on Mars. Sci. Rep. 2024, 14, 5503. [Google Scholar] [CrossRef] [PubMed]
- Heinz, J.; Waajen, A.C.; Airo, A.; Alibrandi, A.; Schirmack, J.; Schulze-Makuch, D. Bacterial Growth in Chloride and Perchlorate Brines: Halotolerances and Salt Stress Responses of Planococcus halocryophilus. Astrobiology 2019, 19, 1377–1387. [Google Scholar] [CrossRef]
- Davila, A.F.; Schulze-Makuch, D. The Last Possible Outposts for Life on Mars. Astrobiology 2016, 16, 159–168. [Google Scholar] [CrossRef]
- Miyata, M.; Robinson, R.C.; Uyeda, T.Q.P.; Fukumori, Y.; Fukushima, S.-I.; Haruta, S.; Homma, M.; Inaba, K.; Ito, M.; Kaito, C.; et al. Tree of motility—A proposed history of motility systems in the tree of life. Genes Cells 2020, 25, 6–21. [Google Scholar] [CrossRef]
- Thévenet, D.; D’Ari, R.; Bouloc, P. The SIGNAL experiment in BIORACK: Escherichia coli in microgravity. J. Biotechnol. 1996, 47, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Klaus, D.M.; Benoit, M.R.; Nelson, E.S.; Hammond, T.G. Extracellular mass transport considerations for space flight research concerning suspended and adherent in vitro cell cultures. J. Gravit. Physiol. 2004, 11, 17–27. [Google Scholar] [PubMed]
- Horneck, G.; Klaus, D.M.; Mancinelli, R.L. Space microbiology. Microbiol. Mol. Biol. Rev. 2010, 74, 121–156. [Google Scholar] [CrossRef] [PubMed]
- Hirt, C.; Claessens, S.J.; Kuhn, M.; Featherstone, W.E. Kilometer-resolution gravity field of Mars: MGM2011. Planet. Space Sci. 2012, 67, 147–154. [Google Scholar] [CrossRef]
- Berg, H.C. How to track bacteria. Rev. Sci. Instrum. 1971, 42, 868–871. [Google Scholar] [CrossRef]
- Berg, H.C.; Brown, D.A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 1972, 239, 500–504. [Google Scholar] [CrossRef]
- Berg, H.C.; Anderson, R.A. Bacteria swim by rotating their flagellar filaments. Nature 1973, 245, 380–382. [Google Scholar] [CrossRef]
- Berg, H.C.; Brown, D.A. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. In Antibiotics and Chemotherapy; S.Karger AG: Basel, Switzerland, 1974; Volume 19. [Google Scholar]
- Watari, N.; Larson, R.G. The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella. Biophys. J. 2010, 98, 12–17. [Google Scholar] [CrossRef]
- Patteson, A.E.; Gopinath, A.; Goulian, M.; Arratia, P.E. Running and tumbling with E. coli in polymeric solutions. Sci. Rep. 2015, 5, 15761. [Google Scholar] [CrossRef]
- Lemelle, L.; Cajgfinger, T.; Nguyen, C.C.; Dominjon, A.; Place, C.; Chatre, E.; Barbier, R.; Palierne, J.-F.; Vaillant, C. Tumble Kinematics of Escherichia coli near a Solid Surface. Biophys. J. 2020, 118, 2400–2410. [Google Scholar] [CrossRef]
- Kurzthaler, C.; Zhao, Y.; Zhou, N.; Schwarz-Linek, J.; Devailly, C.; Arlt, J.; Huang, J.-D.; Poon, W.C.K.; Franosch, T.; Tailleur, J.; et al. Characterization and Control of the Run-and-Tumble Dynamics of Escherichia Coli. Phys. Rev. Lett. 2024, 132, 38302. [Google Scholar] [CrossRef] [PubMed]
- Acres, J.M.; Youngapelian, M.J.; Nadeau, J. The influence of spaceflight and simulated microgravity on bacterial motility and chemotaxis. NPJ Microgravity 2021, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.C. E. Coli in Motion; Springer: New York, NY, USA, 2004; ISBN 978-0-387-21638-6. [Google Scholar]
- Mitchell, J.G.; Martinez-Alonso, M.; Lalucat, J.; Esteve, I.; Brown, S. Velocity changes, long runs, and reversals in the Chromatium minus swimming response. J. Bacteriol. 1991, 173, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.G.; Pearson, L.; Bonazinga, A.; Dillon, S.; Khouri, H.; Paxinos, R. Long lag times and high velocities in the motility of natural assemblages of marine bacteria. Appl. Environ. Microbiol. 1995, 61, 877–882. [Google Scholar] [CrossRef]
- Riekeles, M.; Schirmack, J.; Schulze-Makuch, D. Machine Learning Algorithms Applied to Identify Microbial Species by Their Motility. Life 2021, 11, 44. [Google Scholar] [CrossRef]
- Khan, S.; Trentham, D.R. Biphasic excitation by leucine in Escherichia coli chemotaxis. J. Bacteriol. 2004, 186, 588–592. [Google Scholar] [CrossRef]
- Mitchell, J.G.; Kogure, K. Bacterial motility: Links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 2006, 55, 3–16. [Google Scholar] [CrossRef]
- Qi, Y.L.; Adler, J. Salt taxis in Escherichia coli bacteria and its lack in mutants. Proc. Natl. Acad. Sci. USA 1989, 86, 8358–8362. [Google Scholar] [CrossRef]
- Li, F.; Xiong, X.-S.; Yang, Y.-Y.; Wang, J.-J.; Wang, M.-M.; Tang, J.-W.; Liu, Q.-H.; Wang, L.; Gu, B. Effects of NaCl Concentrations on Growth Patterns, Phenotypes Associated with Virulence, and Energy Metabolism in Escherichia coli BW25113. Front. Microbiol. 2021, 12, 705326. [Google Scholar] [CrossRef]
- Pfeffer, W. Uber chemotaktische Bewegungen von Bacterien, Flagellaten und Volvocineen. Untersuch. Bot. Inst. Tub. 1888, 2, 582–661. [Google Scholar]
- Nadeau, J.L.; Bedrossian, M.; Lindensmith, C.A. Imaging technologies and strategies for detection of extant extraterrestrial microorganisms. Adv. Phys. X 2018, 3, 1424032. [Google Scholar] [CrossRef]
- Thomas-Keprta, K.L.; Clemett, S.J.; Bazylinski, D.A.; Kirschvink, J.L.; McKay, D.S.; Wentworth, S.J.; Vali, H.; Gibson, E.K.; Romanek, C.S. Magnetofossils from ancient Mars: A robust biosignature in the martian meteorite ALH84001. Appl. Environ. Microbiol. 2002, 68, 3663–3672. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, J.L.; Cho, Y.B.; Kühn, J.; Liewer, K. Improved Tracking and Resolution of Bacteria in Holographic Microscopy Using Dye and Fluorescent Protein Labeling. Front. Chem. 2016, 4, 17. [Google Scholar] [CrossRef]
- Nadeau, J.; Lindensmith, C.; Deming, J.W.; Fernandez, V.I.; Stocker, R. Microbial Morphology and Motility as Biosignatures for Outer Planet Missions. Astrobiology 2016, 16, 755–774. [Google Scholar] [CrossRef]
- Lindensmith, C.A.; Rider, S.; Bedrossian, M.; Wallace, J.K.; Serabyn, E.; Showalter, G.M.; Deming, J.W.; Nadeau, J.L. A Submersible, Off-Axis Holographic Microscope for Detection of Microbial Motility and Morphology in Aqueous and Icy Environments. PLoS ONE 2016, 11, e0147700. [Google Scholar] [CrossRef] [PubMed]
- Lingam, M.; Loeb, A. Life in the Cosmos: From Biosignatures to Technosignatures; Harvard University Press: Cambridge, MA, USA, 2021; ISBN 9780674987579. [Google Scholar]
- Riekeles, M.; Albalkhi, H.; Dubay, M.M.; Nadeau, J.; Lindensmith, C.A. Motion history images: A new method for tracking microswimmers in 3D. Front. Imaging 2024, 3, 1393314. [Google Scholar] [CrossRef]
- Madigan, M.T.; Brock, T.D.; Martinko, J.M. Brock Biology of Microorganisms, 14th ed.; Pearson: London, UK, 2015; ISBN 9780321897398. [Google Scholar]
- Sim, M.; Koirala, S.; Picton, D.; Strahl, H.; Hoskisson, P.A.; Rao, C.V.; Gillespie, C.S.; Aldridge, P.D. Growth rate control of flagellar assembly in Escherichia coli strain RP437. Sci. Rep. 2017, 7, 41189. [Google Scholar] [CrossRef]
- Wronkiewicz, M.; Lee, J.; Mandrake, L.; Lightholder, J.; Doran, G.; Mauceri, S.; Kim, T.; Oborny, N.; Schibler, T.; Nadeau, J.; et al. Onboard Science Instrument Autonomy for the Detection of Microscopy Biosignatures on the Ocean Worlds Life Surveyor. Planet. Sci. J. 2024, 5, 19. [Google Scholar] [CrossRef]
- Ali, S.N.; Ahmad, M.K.; Mahmood, R. Sodium chlorate, a herbicide and major water disinfectant byproduct, generates reactive oxygen species and induces oxidative damage in human erythrocytes. Environ. Sci. Pollut. Res. Int. 2017, 24, 1898–1909. [Google Scholar] [CrossRef]
- Srinivasan, A.; Viraraghavan, T. Perchlorate: Health effects and technologies for its removal from water resources. Int. J. Environ. Res. Public Health 2009, 6, 1418–1442. [Google Scholar] [CrossRef]
- Kestin, J.; Khalifa, H.E.; Correia, R.J. Tables of the dynamic and kinematic viscosity of aqueous NaCl solutions in the temperature range 20–150 °C and the pressure range 0.1–35 MPa. J. Phys. Chem. Ref. Data 1981, 10, 71–88. [Google Scholar] [CrossRef]
- Bera, K.; Kiepas, A.; Godet, I.; Li, Y.; Mehta, P.; Ifemembi, B.; Paul, C.D.; Sen, A.; Serra, S.A.; Stoletov, K.; et al. Extracellular fluid viscosity enhances cell migration and cancer dissemination. Nature 2022, 611, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Ni, B.; Colin, R.; Link, H.; Endres, R.G.; Sourjik, V. Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc. Natl. Acad. Sci. USA 2020, 117, 595–601. [Google Scholar] [CrossRef]
- Mellon, M.T.; Sizemore, H.G.; Heldmann, J.L.; McKay, C.P.; Stoker, C.R. The habitability conditions of possible Mars landing sites for life exploration. Icarus 2024, 408, 115836. [Google Scholar] [CrossRef]
- Mitrofanov, I.; Malakhov, A.; Djachkova, M.; Golovin, D.; Litvak, M.; Mokrousov, M.; Sanin, A.; Svedhem, H.; Zelenyi, L. The evidence for unusually high hydrogen abundances in the central part of Valles Marineris on Mars. Icarus 2022, 374, 114805. [Google Scholar] [CrossRef]
- Wallace, J.K.; Rider, S.; Serabyn, E.; Kühn, J.; Liewer, K.; Deming, J.; Showalter, G.; Lindensmith, C.; Nadeau, J. Robust, compact implementation of an off-axis digital holographic microscope. Opt. Express 2015, 23, 17367–17378. [Google Scholar] [CrossRef]
- Snyder, C.; Centlivre, J.P.; Bhute, S.; Shipman, G.; Friel, A.D.; Viver, T.; Palmer, M.; Konstantinidis, K.T.; Sun, H.J.; Rossello-Mora, R.; et al. Microbial Motility at the Bottom of North America: Digital Holographic Microscopy and Genomic Motility Signatures in Badwater Spring, Death Valley National Park. Astrobiology 2023, 23, 295–307. [Google Scholar] [CrossRef]

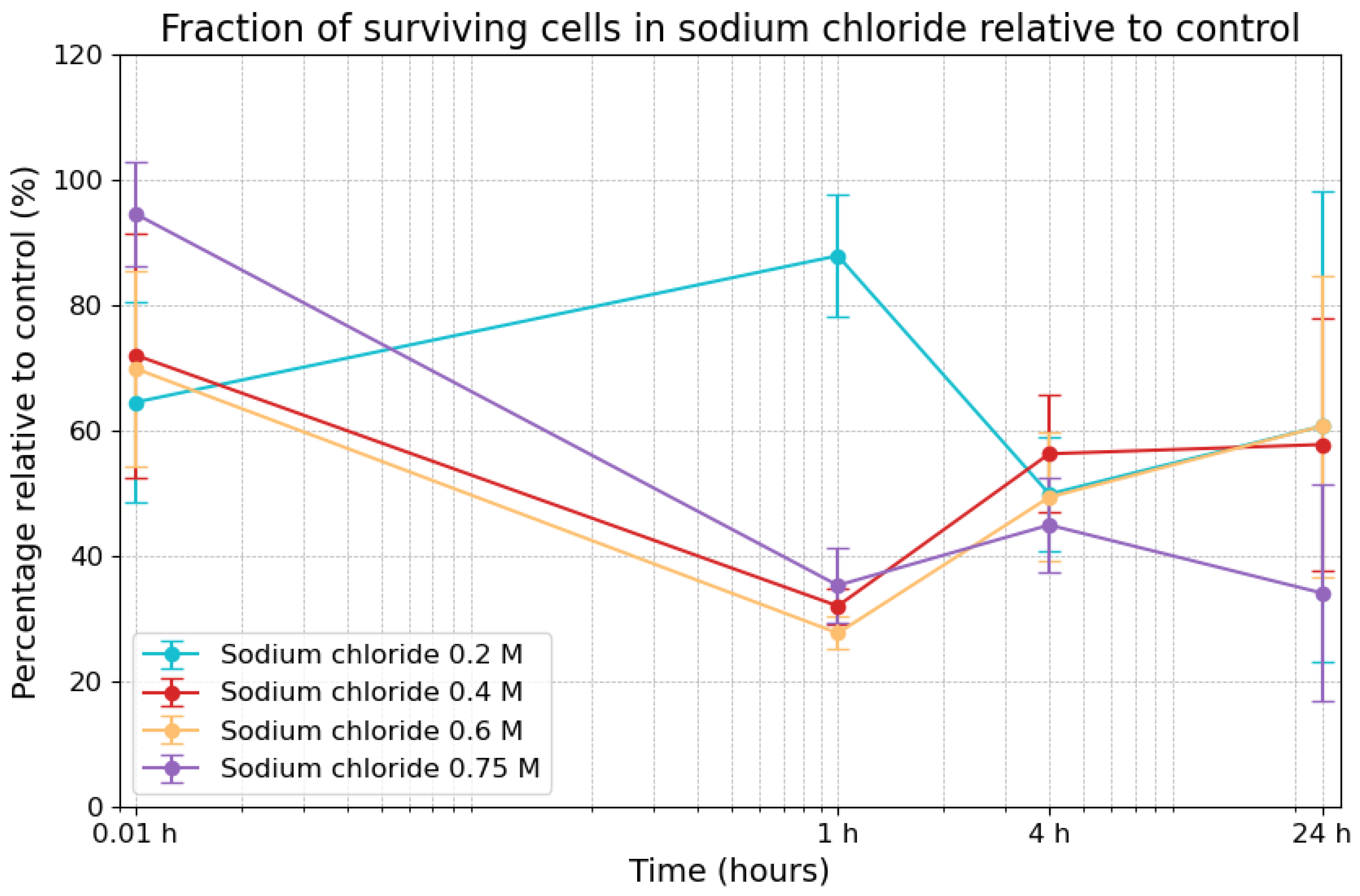
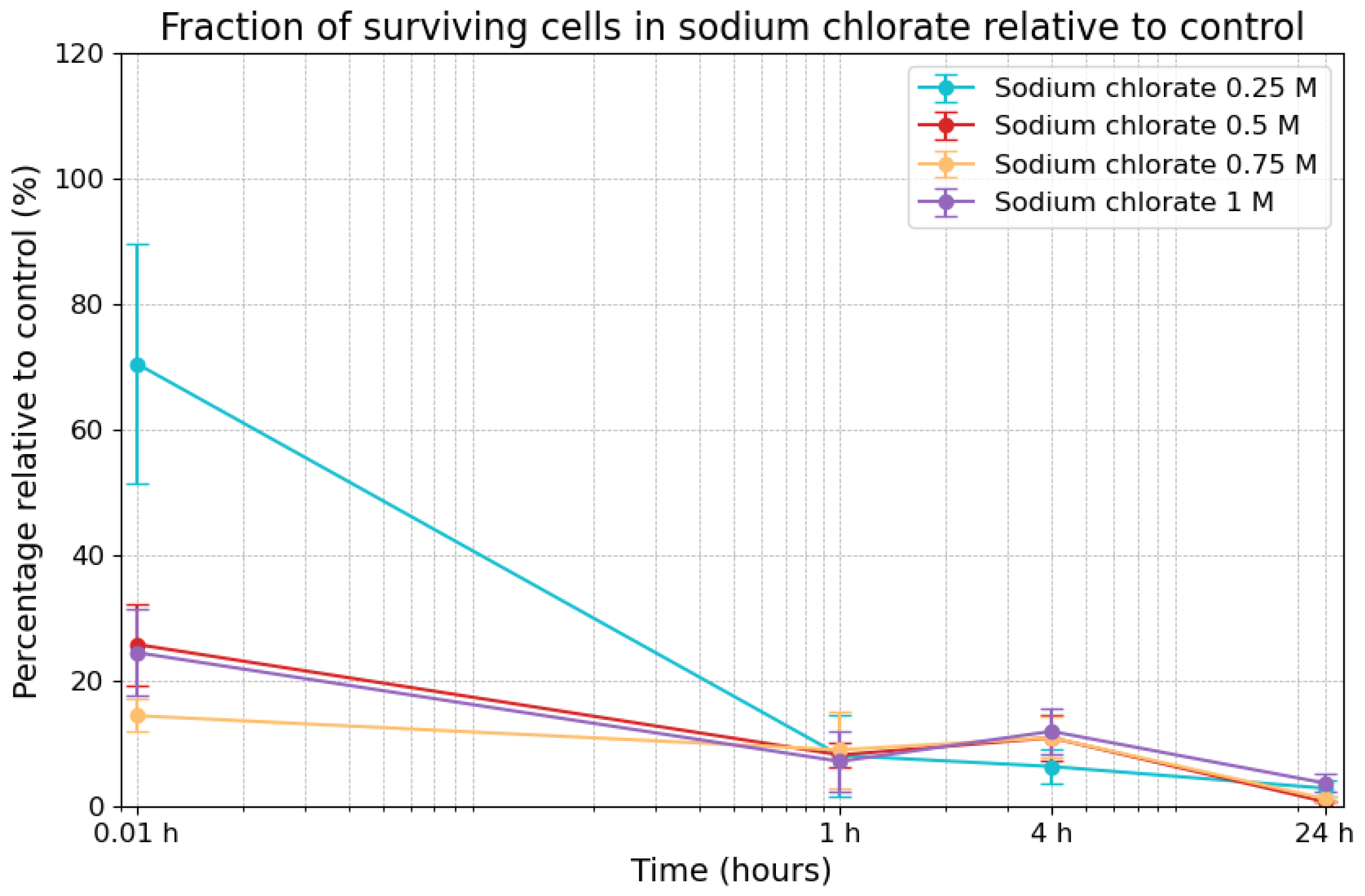
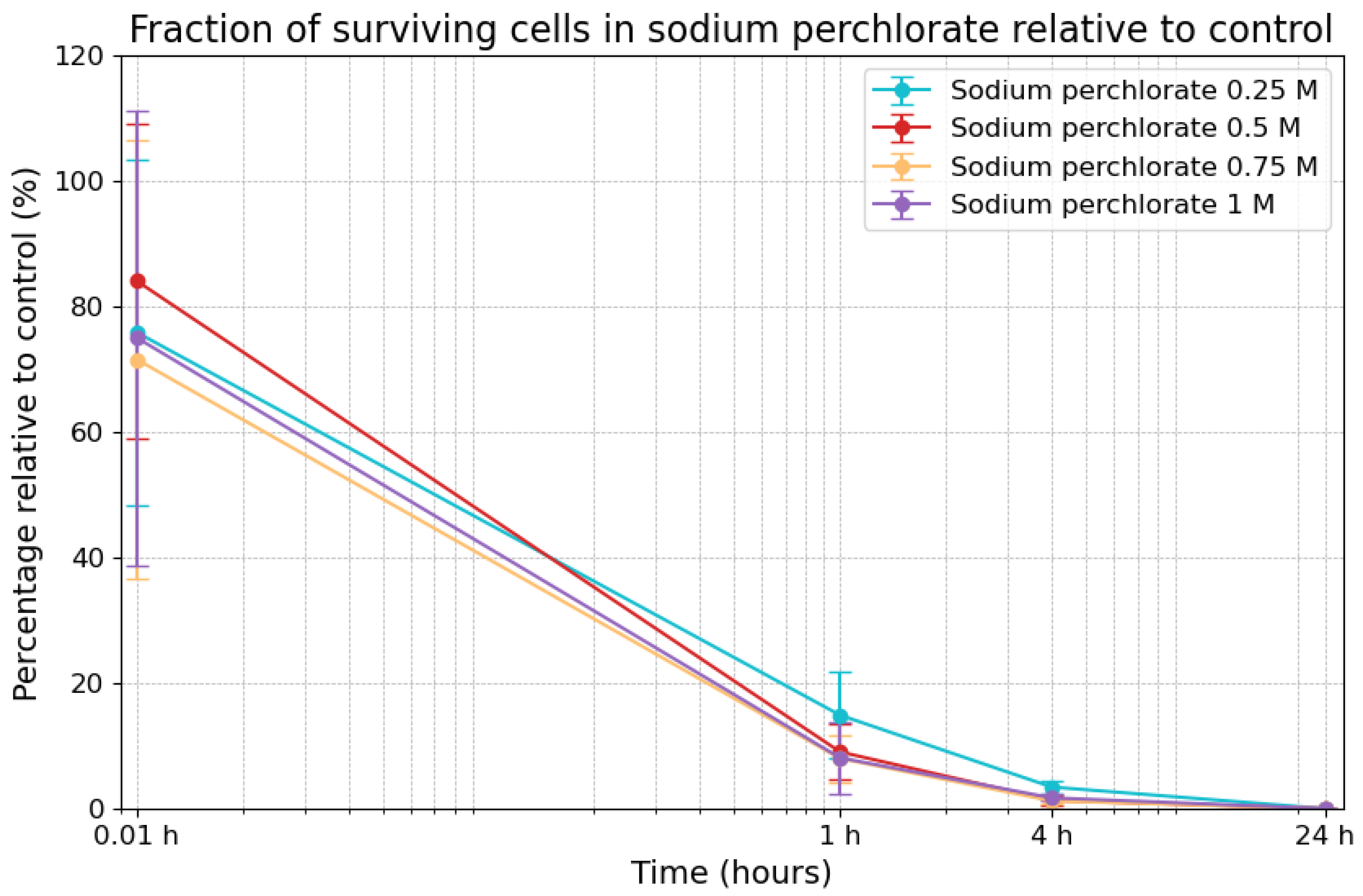
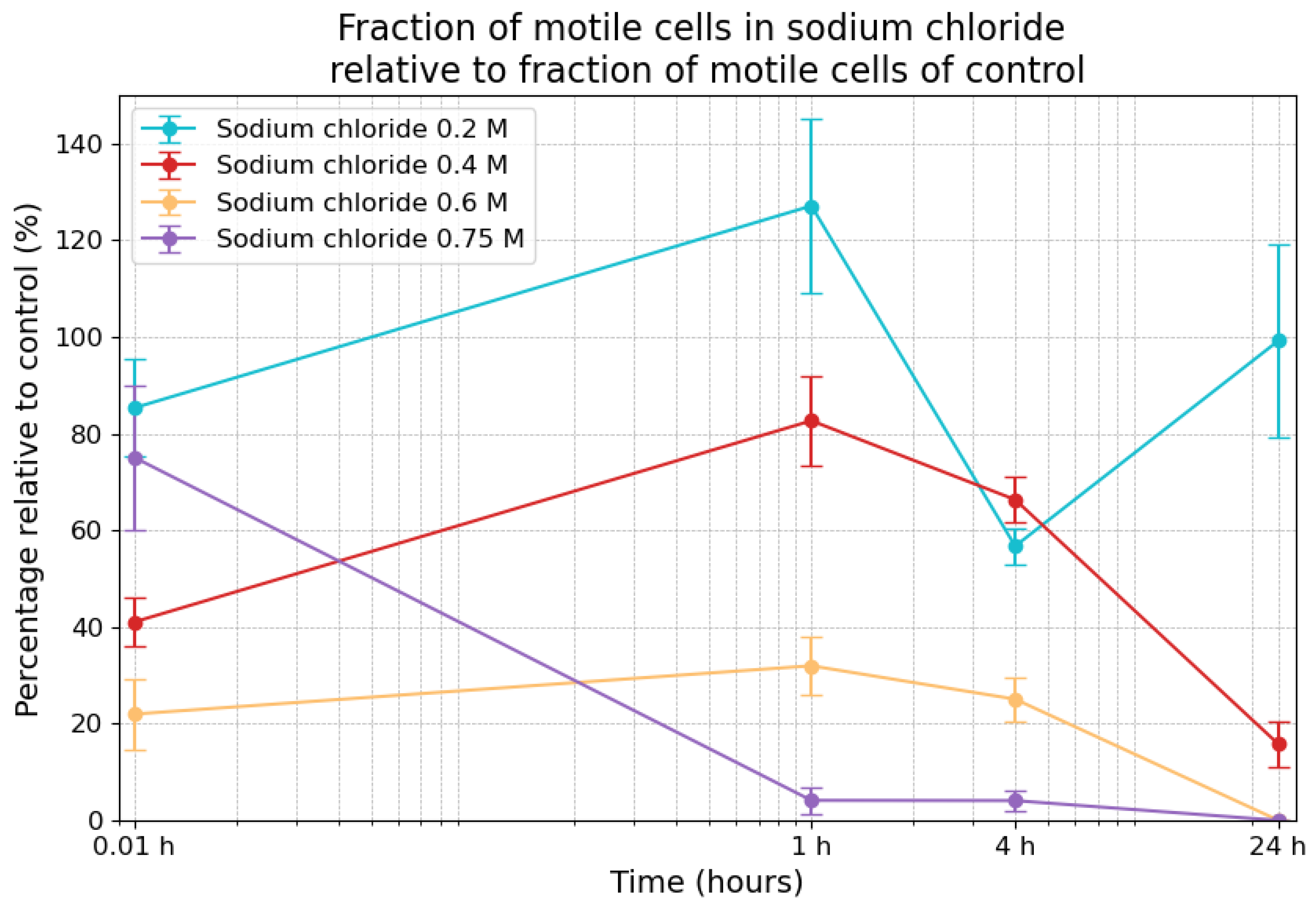


| Sodium Chloride Concentrations | 0.01 h | 1 h | 4 h | 24 h |
|---|---|---|---|---|
| 0.2 M NaCl | 8.4% (±2.4%) | 9.2% (±1.8%) | 9.0% (±1.8%) | 5.9% (±3.9%) |
| 0.4 M NaCl | 3.2% (±1.0%) | 14.5% (±2.3%) | 9.6% (±1.8%) | 1.5% (±0.7%) |
| 0.6 M NaCl | 2.0% (±0.8%) | 7.5% (±1.7%) | 4.2% (±1.2%) | 0.0% (±0.0%) |
| 0.75 M NaCl | 5.0% (±1.2%) | 0.7% (±0.5%) | 0.7% (±0.4%) | 0.0% (±0.0%) |
| Sodium Chlorate Concentrations | 0.01 h | 1 h | 4 h | 24 h |
|---|---|---|---|---|
| 0.25 M NaClO3 | 9.2% (±3.0%) | 87.6% (±72.5%) | 26.6% (±13.8%) | 0.0% |
| 0.5 M NaClO3 | 17.6% (±4.9%) | 60.9% (±16.6%) | 24.6% (±10.9%) | 43.5% (±22.3%) |
| 0.75 M NaClO3 | 22.1% (±6.4%) | 54.6% (±38.2%) | 13.5% (±5.9%) | 0.0% |
| 1 M NaClO3 | 0.0% | 0.0% | 0.0% | 0.0% |
| Sodium Perchlorate Concentrations | 0.01 h | 1 h | 4 h | 24 h |
|---|---|---|---|---|
| 0.25 M NaClO4 | 14.5% (±5.6%) | 61.6% (±28.7%) | 100.0% (±38.7%) | 0.0% |
| 0.5 M NaClO4 | 2.0% (±0.8%) | 33.3% (±16.7%) | 17.3% (±15.6%) | 0.0% |
| 0.75 M NaClO4 | 0.2% (±0.2%) | 1.0% (±1.1%) | 0.0% | 0.0% |
| 1 M NaClO4 | 0.0% | 0.0% | 0.0% | 0.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riekeles, M.; Santos, B.; Youssef, S.A.-M.; Schulze-Makuch, D. Viability and Motility of Escherichia coli Under Elevated Martian Salt Stresses. Life 2024, 14, 1526. https://doi.org/10.3390/life14121526
Riekeles M, Santos B, Youssef SA-M, Schulze-Makuch D. Viability and Motility of Escherichia coli Under Elevated Martian Salt Stresses. Life. 2024; 14(12):1526. https://doi.org/10.3390/life14121526
Chicago/Turabian StyleRiekeles, Max, Berke Santos, Sherif Al-Morssy Youssef, and Dirk Schulze-Makuch. 2024. "Viability and Motility of Escherichia coli Under Elevated Martian Salt Stresses" Life 14, no. 12: 1526. https://doi.org/10.3390/life14121526
APA StyleRiekeles, M., Santos, B., Youssef, S. A.-M., & Schulze-Makuch, D. (2024). Viability and Motility of Escherichia coli Under Elevated Martian Salt Stresses. Life, 14(12), 1526. https://doi.org/10.3390/life14121526







