Evaluation of an Antisense Oligonucleotide Targeting CAG Repeats: A Patient-Customized Therapy Study for Huntington’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Sample Collection and Separation of Leukocytes by Density Gradient
- -
- Group 1: Leukocytes of patient with Huntington’s Disease (H).
- -
- Group 2: Leukocytes of patient with Huntington’s Disease + Transfection Vehicle (Lipofectamine (Thermo Fisher Scientific, Waltham, MA, USA)) (H + L).
- -
- Group 3: Leukocytes of patients with Huntington’s Disease + Transfection Vehicle (Lipofectamine (Thermo Fisher Scientific, Waltham, MA, USA)) + Long antisense oligonucleotide HTT 90-5 (H + L + A)
2.3. Design and Synthesis of the Antisense Targeting the HTT Cluster
2.4. Transfection of Sequences to Lymphocytes
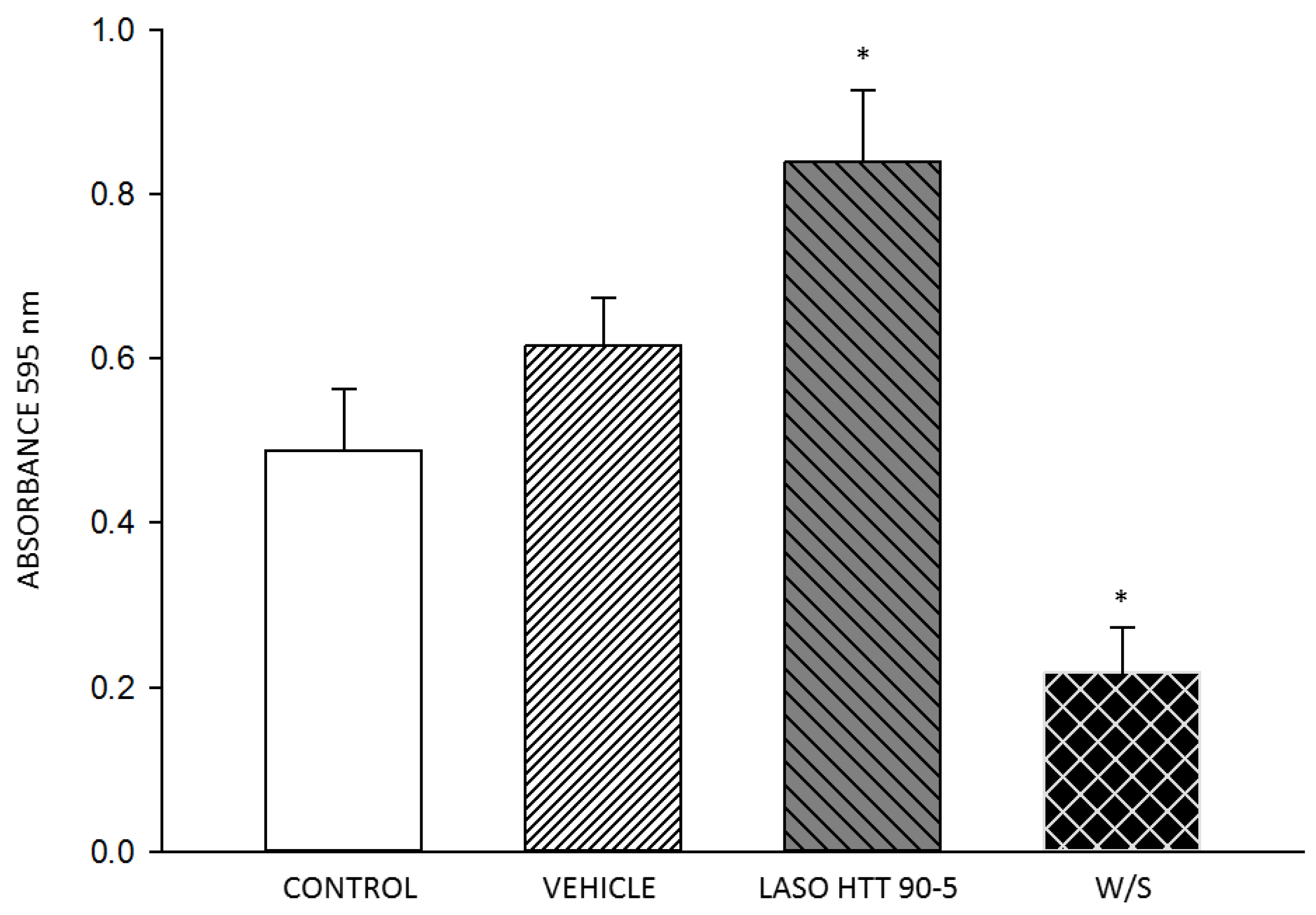
2.5. MTT Assay Cell Viability
2.6. Real-Time PCR Measuring of Huntingtin Expression
2.7. Statistical Analysis
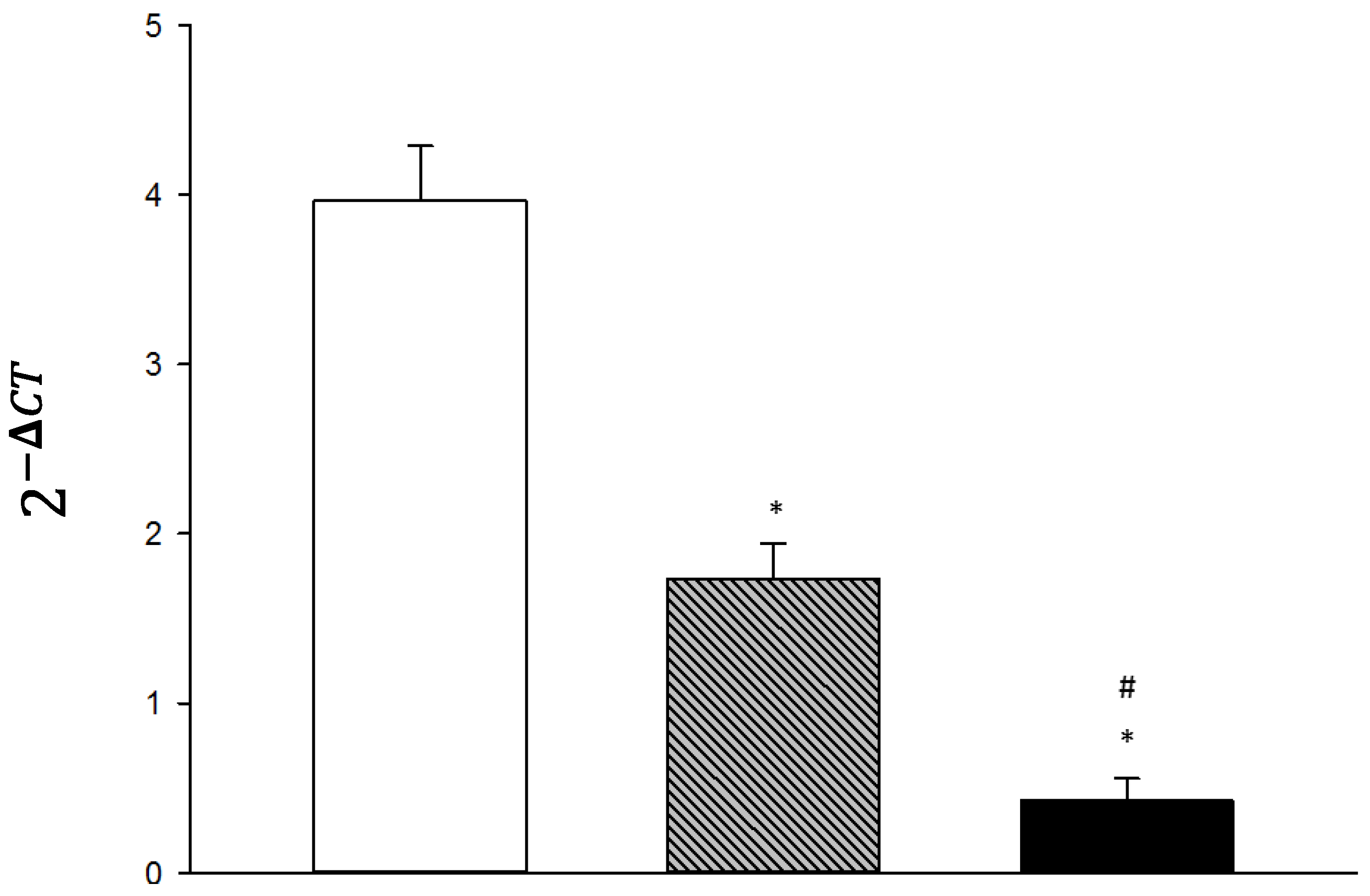
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Andhale, R.; Shrivastava, D.; Disease, H. Huntington’s Disease: A Clinical Review. Cureus 2022, 14, e28484. [Google Scholar] [CrossRef] [PubMed]
- Majerová, V.; Kalinčík, T.; Laczó, J.; Vyhnálek, M.; Hort, J.; Bojar, M.; Růžička, E.; Roth, J. Disturbance of real space navigation in moderately advanced but not in early Huntington’s disease. J. Neurol. Sci. 2012, 312, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.; Handley, R.R.; Lehnert, K.; Snell, R.G. From Pathogenesis to Therapeutics: A Review of 150 Years of Huntington’s Disease Research. Int. J. Mol. Sci. 2023, 24, 13021. [Google Scholar] [CrossRef]
- Vonsattel, J.-P.; Myers, R.H.; Stevens, T.J.; Ferrante, R.J.; Bird, E.D.; Richardson, E.P. Neuropathological Classification of Huntington’s Disease. J. Neuropathol. Exp. Neurol. 1985, 44, 559–577. [Google Scholar] [CrossRef]
- Kwa, L.; Larson, D.; Yeh, C.; Bega, D. Influence of Age of Onset on Huntington’s Disease Phenotype. Tremor Other Hyperkinet. Mov. 2020, 10, 21. [Google Scholar] [CrossRef]
- Squitieri, F.; Maffi, S.; Al Harasi, S.; Al Salmi, Q.; D’Alessio, B.; Capelli, G.; Mazza, T. Incidence and prevalence of Huntington disease (HD) in the Sultanate of Oman: The first Middle East post-HTT service-based study. J. Neurol. Neurosurg. Psychiatry 2020, 91, 1359–1360. [Google Scholar] [CrossRef]
- Rawlins, M. Huntington’s disease out of the closet? Lancet 2010, 376, 1372–1373. [Google Scholar] [CrossRef]
- Evans, S.J.; Douglas, I.; Rawlins, M.D.; Wexler, N.S.; Tabrizi, S.J.; Smeeth, L. Prevalence of adult Huntington’s disease in the UK based on diagnoses recorded in general practice records. J. Neurol. Neurosurg. Psychiatry 2013, 84, 1156–1160. [Google Scholar] [CrossRef]
- Squitieri, F.; Griguoli, A.; Capelli, G.; Porcellini, A.; D’Alessio, B. Epidemiology of Huntington disease: First post-HTT gene analysis of prevalence in Italy. Clin. Genet. 2016, 89, 367–370. [Google Scholar] [CrossRef]
- Hoogeveen, A.T.; Willemsen, R.; Meyer, N.; de Roolj, K.E.; Roos, R.A.C.; van Ommen, G.-J.B.; Galjaard, H. Characterization and localization of the Huntington disease gene product. Hum. Mol. Genet. 1993, 2, 2069–2073. [Google Scholar] [CrossRef]
- Faquih, T.O.; Aziz, N.A.; Gardiner, S.L.; Li-Gao, R.; de Mutsert, R.; Milaneschi, Y.; Trompet, S.; Jukema, J.W.; Rosendaal, F.R.; van Hylckama Vlieg, A.; et al. Normal range CAG repeat size variations in the HTT gene are associated with an adverse lipoprotein profile partially mediated by body mass index. Hum. Mol. Genet. 2023, 32, 1741–1752. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.L.; Saft, C.; Nopoulos, P.C. Association of CAG Repeat Length in the Huntington Gene with Cognitive Performance in Young Adults. Neurology 2021, 96, e2407–e2413. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Harding, R.J.; Loppnau, P.; Ackloo, S.; Lemak, A.; Hutchinson, A.; Hunt, B.; Holehouse, A.S.; Ho, J.C.; Fan, L.; Toledo-Sherman, L.; et al. Design and characterization of mutant and wildtype huntingtin proteins produced from a toolkit of scalable eukaryotic expression systems. J. Biol. Chem. 2019, 294, 6986–7001. [Google Scholar] [CrossRef]
- Matlahov, I.; van der Wel, P.C. Conformational studies of pathogenic expanded polyglutamine protein deposits from Huntington’s disease. Exp. Biol. Med. 2019, 244, 1584–1595. [Google Scholar] [CrossRef]
- Li, S.-H.; Gutekunst, C.-A.; Hersch, S.M.; Li, X.-J. Interaction of Huntingtin-Associated Protein with Dynactin P150Glued. J. Neurosci. 1998, 18, 1261–1269. [Google Scholar] [CrossRef]
- Gauthier, L.R.; Charrin, B.C.; Borrell-Pagès, M.; Dompierre, J.P.; Rangone, H.; Cordelières, F.P.; De Mey, J.; MacDonald, M.E.; Leßmann, V.; Humbert, S.; et al. Huntingtin Controls Neurotrophic Support and Survival of Neurons by Enhancing BDNF Vesicular Transport along Microtubules. Cell 2004, 118, 127–138. [Google Scholar] [CrossRef]
- Martí, E. RNA toxicity induced by expanded CAG repeats in Huntington’s disease. Brain Pathol. 2016, 26, 779–786. [Google Scholar] [CrossRef]
- McColgan, P.; Thobhani, A.; Boak, L.; Schobel, S.A.; Nicotra, A.; Palermo, G.; Trundell, D.; Zhou, J.; Schlegel, V.; Ducray, P.S.; et al. Tominersen in Adults with Manifest Huntington’s Disease. N. Engl. J. Med. 2023, 389, 2203–2205. [Google Scholar] [CrossRef]
- Qassem, S.; Breier, D.; Naidu, G.S.; Hazan-Halevy, I.; Peer, D. Unlocking the therapeutic potential of locked nucleic acids through lipid nanoparticle delivery. Mol. Ther. Nucleic Acids 2024, 35, 102224. [Google Scholar] [CrossRef]
- Kim, J.; Hu, C.; El Achkar, C.M.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zheng, X.; Lin, Y.; Li, C.; Liu, Z.; Li, J.; Tu, Z.; Zhao, Y.; Huang, C.; Chen, Y.; et al. Cas9-mediated replacement of expanded CAG repeats in a pig model of Huntington’s disease. Nat. Biomed. Eng. 2023, 7, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Lalan, M.; Barve, K. Intranasal delivery: An attractive route for the administration of nucleic acid based therapeutics for CNS disorders. Front. Pharmacol. 2022, 13, 974666. [Google Scholar] [CrossRef] [PubMed]
- Borgonetti, V.; Galeotti, N. Intranasal delivery of an antisense oligonucleotide to the RNA-binding protein HuR relieves nerve injury-induced neuropathic pain. Pain 2021, 162, 1500–1510. [Google Scholar] [CrossRef]
- Ferguson, M.W.; Kennedy, C.J.; Palpagama, T.H.; Waldvogel, H.J.; Faull, R.L.M.; Kwakowsky, A. Current and Possible Future Therapeutic Options for Huntington’s Disease. J. Cent. Nerv. Syst. Dis. 2022, 14, 645–658. [Google Scholar] [CrossRef]
- Rayner, S.; Brignac, S.; Bumeister, R.; Belosludtsev, Y.; Ward, T.; Grant, O.; O’Brien, K.; Evans, G.A.; Garner, H.R. MerMade: An Oligodeoxyribonucleotide Synthesizer for High Throughput Oligonucleotide Production in Dual 96-Well Plates. Genome Res. 1998, 8, 741–747. [Google Scholar] [CrossRef]
- White, H.A. Manual Oligonucleotide Synthesis Using the Phosphoramidite Method. In New Nucleic Acid Techniques; Humana Press: New Jersey, NJ, USA, 1988; pp. 193–214. [Google Scholar] [CrossRef]
- Nakamori, M.; Gourdon, G.; Thornton, C.A. Stabilization of Expanded (CTG)•(CAG) Repeats by Antisense Oligonucleotides. Mol. Ther. 2011, 19, 2222–2227. [Google Scholar] [CrossRef]
- Nakamori, M.; Pearson, C.E.; Thornton, C.A. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)•(CAG) repeats. Hum. Mol. Genet. 2011, 20, 580–588. [Google Scholar] [CrossRef]
- Dragileva, E.; Hendricks, A.; Teed, A.; Gillis, T.; Lopez, E.T.; Friedberg, E.C.; Kucherlapati, R.; Edelmann, W.; Lunetta, K.L.; MacDonald, M.E.; et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009, 33, 37–47. [Google Scholar] [CrossRef]
- Wheeler, T.M.; Sobczak, K.; Lueck, J.D.; Osborne, R.J.; Lin, X.; Dirksen, R.T.; Thornton, C.A. Reversal of RNA Dominance by Displacement of Protein Sequestered on Triplet Repeat RNA. Science 2009, 325, 336–339. [Google Scholar] [CrossRef]
- Mulders, S.A.M.; van den Broek, W.J.A.A.; Wheeler, T.M.; Croes, H.J.E.; van Kuik-Romeijn, P.; de Kimpe, S.J.; Furling, D.; Platenburg, G.J.; Gourdon, G.; Thornton, C.A.; et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. USA 2009, 106, 13915–13920. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Träger, U.; Wild, E.J.; Grueninger, S.; Farmer, R.; Landles, C.; Scahill, R.I.; Lahiri, N.; Haider, S.; Macdonald, D.; et al. Mutant huntingtin fragmentation in immune cells tracks Huntington’s disease progression. J. Clin. Investig. 2012, 122, 3731–3736. [Google Scholar] [CrossRef] [PubMed]
- Goldrosen, M.H.; Gannon, P.J.; Lutz, M.; Holyoke, E.D. Isolation of human peripheral blood lymphocytes: Modification of a double discontinuous density gradient of Ficoll-Hypaque. J. Immunol. Methods 1977, 14, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Conroy, F.; Miller, R.; Alterman, J.F.; Hassler, M.R.; Echeverria, D.; Godinho, B.M.D.C.; Knox, E.G.; Sapp, E.; Sousa, J.; Yamada, K.; et al. Chemical engineering of therapeutic siRNAs for allele-specific gene silencing in Huntington’s disease models. Nat. Commun. 2022, 13, 5802. [Google Scholar] [CrossRef]
- Datson, N.A.; González-Barriga, A.; Kourkouta, E.; Weij, R.; van de Giessen, J.; Mulders, S.; Kontkanen, O.; Heikkinen, T.; Lehtimäki, K.; van Deutekom, J.C.T. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS ONE 2017, 12, e0171127. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Kelley, R.L.; Meller, V.H.; Gordadze, P.R.; Roman, G.; Davis, R.L.; Kuroda, M.I. Epigenetic Spreading of the Drosophila Dosage Compensation Complex from roX RNA Genes into Flanking Chromatin. Cell 1999, 98, 513–522. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.-J.; Lee, J.T. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef]
- Svoboda, P. Key Mechanistic Principles and Considerations Concerning RNA Interference. Front. Plant Sci. 2020, 11, 1237. [Google Scholar] [CrossRef]
- Ocampo-Ortega, S.A.; Cabrera-Becerra, S.E.; Sierra-Sanchez, V.M.; García-Rubio, V.G.; Blancas-Napoles, C.M.; Romero-Nava, R.; Huang, F.; Hong, E.; Aguilera-Méndez, A.; Villafaña, S. Attenuation of Pulmonary Damage Associated with COPD in a Cadmium-Exposed Model Due to the Administration of a siRNA Targeting PAD4. Sci. Pharm. 2024, 92, 12. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Olson, E.N. MEF2: A central regulator of diverse developmental programs. Development 2007, 134, 4131–4140. [Google Scholar] [CrossRef] [PubMed]
- Kilanowska, A.; Studzińska, S. In vivo and in vitro studies of antisense oligonucleotides. RSC Adv. 2020, 10, 34501–34516. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Nagpal, G.; Dhanda, S.K.; Raghava, G.P.S. Prediction of Immunomodulatory potential of an RNA sequence for designing non-toxic siRNAs and RNA-based vaccine adjuvants. Sci. Rep. 2016, 6, 20678. [Google Scholar] [CrossRef]
- Lopez-Fraga, M.; Jimenez, A.I.; Valcarel, T.M. siRNA and Their Use in Methods and Compositions for the Treatment and/or Prevention of Eye Conditions. U.S. Patent No. 9,018,183, 28 April 2015. [Google Scholar]
- Luo, K.Q.; Chang, D.C. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 2004, 318, 303–310. [Google Scholar] [CrossRef]
- Tsakiri, M.; Zivko, C.; Demetzos, C.; Mahairaki, V. Lipid-based nanoparticles and RNA as innovative neuro-therapeutics. Front. Pharmacol. 2022, 13, 900610. [Google Scholar] [CrossRef]
- Springer, A.D.; Dowdy, S.F. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid. Ther. 2018, 28, 109–118. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, W.; Li, M.; Zheng, A. Exosome-Based Carrier for RNA Delivery: Progress and Challenges. Pharmaceutics 2023, 15, 598. [Google Scholar] [CrossRef]



| Attribute | Details |
|---|---|
| Gender | Female |
| Age | 36 years |
| Children | None |
| Caregiver Dependence | Yes |
| CAG Repetitions | 54 (mutant allele)/19 (wild-type allele) |
| Year of Diagnosis | 2013 |
| Family History | Paternal grandmother, paternal aunt, father |
| Clinical Phenotype | Choreic phenotype |
| UHDRS Motor Score | 16/28 (Moderate to Severe) |
| UHDRS Cognition Score | 2/12 (Mild) |
| UHDRS Behavior Score | 6–8/12 (Moderate) |
| UHDRS Functionality Score | 7–8/8 (Severe) |
| UHDRS Total Score | 31–34/60 (Moderate to Severe) |
| Target RNA | Energy (kcal/mol) | Hybridization Energy (kcal/mol) | Unfolding Energy—Target (kcal/mol) | Unfolding Energy—Query (kcal/mol) | Position—Target | Position—Query | Position Seed—Target | Position Seed—Query |
|---|---|---|---|---|---|---|---|---|
| Huntingtin transcript > 30 CAG repeats | −154.92 | −232.98 | 37.28 | 40.78 | 46—140 | 1—95 | 46—52 | 89—95 |
| Huntingtin transcript variant 2 | −116.53 | −189.58 | 32.27 | 40.78 | 46—138 | 3—95 | 46—52 | 89—95 |
| Huntingtin transcript variant 1 | −107.63 | −181.6 | 33.19 | 40.78 | 46—141 | 3—95 | 46—52 | 89—95 |
| Oligo Name | Sequence | # of Bases | Molecular Weight (MW) | Melting Temperature (Tm, °C) | µg/OD |
|---|---|---|---|---|---|
| HTT expression F | GCTGCACCGACCAAAGAAAG | 20 | 6129 | 57 | 30.55 |
| HTT expression R | GTTCCATAGCGATGCCCAGA | 20 | 6102 | 57.2 | 31.45 |
| HPRT1 HOMO F | GACCAGTCAACAGGGGACAT | 20 | 6160.1 | 56.9 | 30.26 |
| HPRT1 HOMO R | GCTTGCGACCTTGACCATCT | 20 | 6044 | 57.8 | 34.22 |
| Number of Nucleotides | Sequence | Off-Targets | Query Cover |
|---|---|---|---|
| 95 nts | CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGAAGG | No significant similarity | NA |
| 80 nts | CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGAAGG | No significant similarity | NA |
| 65 nts | CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGAAGG | No significant similarity | NA |
| 50 nts | CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGAAGG | No significant similarity | NA |
| 35 nts | CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGAAGG | No significant similarity | NA |
| 20 nts | CTGCTGCTGCTGCTGGAAGG | Myocyte enhancer factor 2A (MEF2A) | 95% |
| Range of Positions | Sequence | Immunomodulatory Score |
|---|---|---|
| 1–64 | CUGCUGCUGCUGCUGCUGCUGCUGCUGCUG | 5.8 |
| 65–66 | UGCCUGCUGCUGCUGCUGCUGGAAG | 5.4 |
| 67 | GCUGCUGCUGCUGCUGCUGCUGGAAGG | 5.2 |
| 68 | CUGCUGCUGCUGCUGCUGCUGGAA | 4.7 |
| 69 | UGCCUGCUGCUGCUGCUGCUGGAA | 4.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocampo-Ortega, S.A.; Sierra-Sanchez, V.M.; Blancas-Napoles, C.M.; González-Carteño, A.; Mera-Jiménez, E.; Macías-Pérez, M.E.; Hernandez-Guerra, A.; Romero-Nava, R.; Huang, F.; Hong, E.; et al. Evaluation of an Antisense Oligonucleotide Targeting CAG Repeats: A Patient-Customized Therapy Study for Huntington’s Disease. Life 2024, 14, 1607. https://doi.org/10.3390/life14121607
Ocampo-Ortega SA, Sierra-Sanchez VM, Blancas-Napoles CM, González-Carteño A, Mera-Jiménez E, Macías-Pérez ME, Hernandez-Guerra A, Romero-Nava R, Huang F, Hong E, et al. Evaluation of an Antisense Oligonucleotide Targeting CAG Repeats: A Patient-Customized Therapy Study for Huntington’s Disease. Life. 2024; 14(12):1607. https://doi.org/10.3390/life14121607
Chicago/Turabian StyleOcampo-Ortega, Sergio Adrian, Vivany Maydel Sierra-Sanchez, Citlali Margarita Blancas-Napoles, Asdrúbal González-Carteño, Elvia Mera-Jiménez, Martha Edith Macías-Pérez, Adriana Hernandez-Guerra, Rodrigo Romero-Nava, Fengyang Huang, Enrique Hong, and et al. 2024. "Evaluation of an Antisense Oligonucleotide Targeting CAG Repeats: A Patient-Customized Therapy Study for Huntington’s Disease" Life 14, no. 12: 1607. https://doi.org/10.3390/life14121607
APA StyleOcampo-Ortega, S. A., Sierra-Sanchez, V. M., Blancas-Napoles, C. M., González-Carteño, A., Mera-Jiménez, E., Macías-Pérez, M. E., Hernandez-Guerra, A., Romero-Nava, R., Huang, F., Hong, E., & Villafaña, S. (2024). Evaluation of an Antisense Oligonucleotide Targeting CAG Repeats: A Patient-Customized Therapy Study for Huntington’s Disease. Life, 14(12), 1607. https://doi.org/10.3390/life14121607








