Genetic Analysis of Adaptive Traits in Spring Wheat in Northeast China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Trials
2.2. Phenotyping and Statistical Analysis
2.3. Population Structure and Linkage Disequilibrium
2.4. Association Mapping and the Identification of Candidate Genes
2.5. Establishment and Verification of KASP Markers
2.6. Identification of the Candidate Genes of Adaptive Related Traits
3. Results
3.1. Phenotypic Evaluation
3.2. Population Structure and LD Decay Analysis
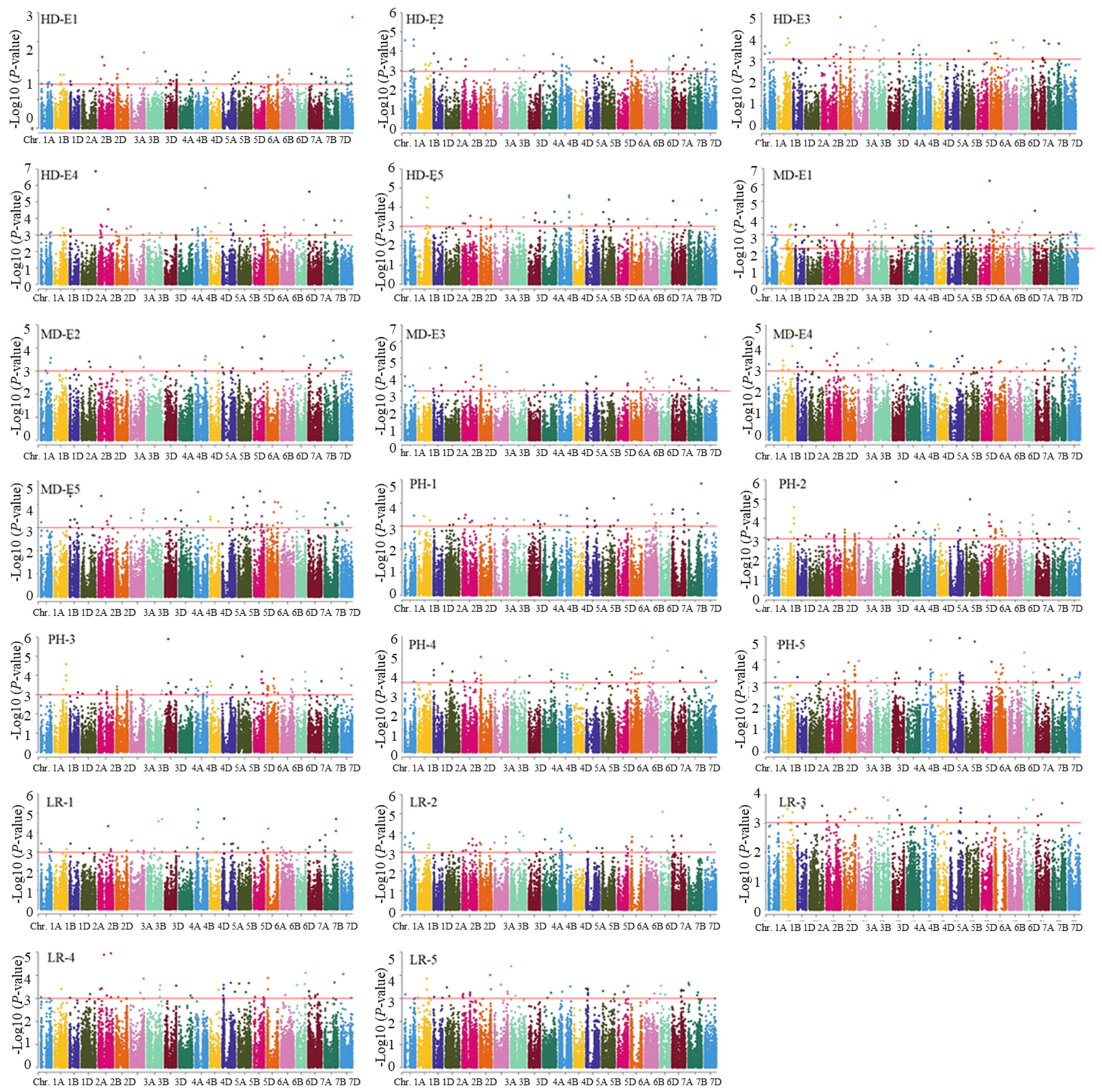
3.3. Genome-Wide Association Studies
3.4. Candidate Genes
3.5. KASP Validation
4. Discussion
4.1. Twenty-Eight Novel Loci for Adiptive Traits in Common Wheat Were Identfied
4.2. Heading Date
4.3. Maturing Date
4.4. Plant Height
4.5. Lodging Resistance
4.6. Candidate Gene Analysis
4.7. Potential Implications on Wheat Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hyles, J.; Bloomfield, M.T.; Hunt, J.R.; Trethowan, R.M.; Trevaskis, B. Phenology and related traits for wheat adaptation. Heredity 2020, 125, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, Y.; Xin, W.; Wei, Y.; Zhang, J.; Guo, L. Wheat breeding in northern China: Achievements and technical advances. Crop J. 2019, 7, 718–729. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; Liu, J.; Zhang, Y.; Cao, S.; He, Z.; Rasheed, A.; Jin, H.; Zhang, C.; Yan, J.; et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2020, 19, 168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hou, J.; Liu, H.; Li, T.; Wang, K.; Hao, C.; Liu, H.; Zhang, X. TaBT1, affecting starch synthesis and thousand kernel weight, underwent strong selection during wheat improvement. J. Exp. Bot. 2019, 70, 1497–1511. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wen, W.; He, Z.; Liu, J.; Jin, H.; Cao, S.; Geng, H.; Yan, J.; Zhang, P.; Wan, Y.; et al. Genome-wide linkage mapping of yield related traits in three Chinese bread wheat populations using high-density SNP markers. Theor. Appl. Genet. 2018, 131, 1903–1924. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, Z.; Wu, L.; Bai, B.; Wen, W.; Xie, C.; Xia, X. Genome-wide linkage mapping of QTL for black point reaction in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2179–2190. [Google Scholar] [CrossRef]
- Chen, Z.; Cheng, X.; Chai, L.; Wang, Z.; Du, D.; Wang, Z.; Bian, R.; Zhao, A.; Xin, M.; Guo, W.; et al. Pleiotropic QTL influencing spikelet number and heading date in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 1825–1838. [Google Scholar] [CrossRef]
- Cui, F.; Li, J.; Ding, A.; Zhao, C.; Wang, L.; Wang, X.; Li, S.; Bao, Y.; Li, X.; Feng, D.; et al. Conditional QTL mapping for plant height with respect to the length of the spike and internode in two mapping populations of wheat. Theor. Appl. Genet. 2011, 122, 1517–1536. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, H.; Qiao, L.; Miao, L.; Yan, D.; Liu, P.; Zhao, G.; Jia, J.; Gao, L. Wheat MADS-box gene TaSEP3-D1 negatively regulates heading date. Crop J. 2021, 9, 1115–1123. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–19. [Google Scholar] [CrossRef]
- Quan, X.; Dong, L.J.; Zhang, N.; Xie, C.; Li, H.; Xia, X.; He, W.; Qin, Y. Genome-wide association study uncover the genetic architecture of salt tolerance-related traits in common wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 563. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, J.; Liu, W.; Yan, W.; Sun, Y.; Che, J.; Tian, C.; Zhang, H.; Yu, L. The genetic architecture of grain yield in spring wheat based on genome-wide association study. Front. Genet. 2021, 12, 728472. [Google Scholar] [CrossRef]
- Wang, S.C.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Le Gouis, J.; Bordes, J.; Ravel, C.; Heumez, E.; Faure, S.; Praud, S.; Galic, N.; Remoué, C.; Balfourier, F.; Allard, V.; et al. Genome-wide association analysis to identify chromosomal regions determining components of earliness in wheat. Theor. Appl. Genet. 2012, 124, 597–611. [Google Scholar] [CrossRef]
- Luján Basile, S.M.; Ramírez, I.A.; Crescente, J.M.; Conde, M.B.; Demichelis, M.; Abbate, P.; Rogers, W.J.; Pontaroli, A.C.; Helguera, M.; Vanzetti, L.S. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019, 19, 553. [Google Scholar] [CrossRef]
- Fu, D.; Szűcs, P.; Yan, L.; Helguera, M.; Skinner, J.S.; Von-Zitzewitz, J. Large deletions within the first intron in Vrn-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genom. 2005, 273, 54–65. [Google Scholar] [CrossRef]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 2012, 7, 33234. [Google Scholar] [CrossRef]
- Xiang, R.; Semagn, K.; Iqbal, M.; Chen, H.; Yang, R.C.; Spaner, D. Phenotypic performance and associated QTL of ‘Peace’בCDC Stanley’mapping population under conventional and organic management systems. Crop Sci. 2021, 61, 3469–3483. [Google Scholar] [CrossRef]
- Semagn, K.; Iqbal, M.; Chen, H.; Perez-Lara, E.; Bemister, D.H.; Xiang, R.; Zou, J.; Asif, M.; Kamran, A.; N’Diaye, A.; et al. Physical mapping of QTL associated with agronomic and end-use quality traits in spring wheat under conventional and organic management systems. Theor. Appl. Genet. 2021, 134, 3699–3719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Islam, M.S.; Zhao, Y.; Anwar, M.; Alhabbar, Z.; She, M.; Yang, R.; Juhasz, A.; Tang, G.; Chen, J.; et al. Non-escaping frost tolerant QTL linked genetic loci at reproductive stage in six wheat DH populations. Crop J. 2022, 10, 147–165. [Google Scholar] [CrossRef]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high- density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Ma, D.; Yin, G.; Rasheed, A.; Dong, Y.; Xiao, Y.; Xia, X.; Wu, X.; He, Z. Genetic progress in grain yield and physiological traits in Chinese wheat cultivars of Southern Yellow and Huai Valley since 1950. Crop Sci. 2017, 57, 760–773. [Google Scholar] [CrossRef]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring. Front. Plant Sci. 2015, 6, 1099. [Google Scholar] [CrossRef]
- Sun, C.W.; Zhang, F.Y.; Yan, X.F.; Zhang, X.F.; Dong, Z.D.; Cui, D.Q.; Chen, F. Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol. J. 2017, 15, 953–969. [Google Scholar] [CrossRef]
- Ellis, M.H.; Rebetzke, G.J.; Azanza, F.; Richards, R.A.; Spielmeyer, W. Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor. Appl. Genet. 2005, 111, 423–430. [Google Scholar] [CrossRef]
- Piñera-Chavez, F.J.; Berry, P.M.; Foulkes, M.J.; Sukumaran, S.; Reynolds, M.P. Identifying quantitative trait loci for lodging-associated traits in the wheat doubled-haploid population Avalon × Cadenza. Crop Sci. 2021, 61, 2371–2386. [Google Scholar] [CrossRef]
- Cui, F.; Zhao, C.; Ding, A.; Li, J.; Wang, L.; Li, X.; Wang, H. Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor. Appl. Genet. 2014, 127, 659–675. [Google Scholar] [CrossRef]
- Dreccer, M.F.; Macdonald, B.; Farnsworth, C.A.; Paccapelo, M.V.; Awasi, M.A.; Condon, A.G.; Forrest, K.; Lee Long, I.; McIntyre, C.L. Multi-donor× elite-based populations reveal QTL for low-lodging wheat. Theor. Appl. Genet. 2022, 135, 1685–1703. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Wang, X.; Wang, X.; Zhou, F.; Xu, X.; Wu, B.; Yao, J.; Lv, D.; Yang, M.; Song, X.; et al. Application of 50K chip-based genetic map to QTL mapping of stem-related traits in wheat. Crop Pasture Sci. 2021, 72, 105–112. [Google Scholar] [CrossRef]
- Blackburn, A.; Sidhu, G.; Schillinger, W.F.; Skinner, D.; Gill, K. QTL mapping using GBS and SSR genotyping reveals genomic regions controlling wheat coleoptile length and seedling emergence. Euphytica 2021, 217, 45. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Xu, W.; Hu, T.; Ma, J.; Zhang, Y.; Hou, J.; Hao, C.; Zhang, X.; Li, T. TaGW2L, a GW2-like RING finger E3 ligase, positively regulates heading date in common wheat (Triticum aestivum L.). Crop J. 2022, 10, 972–979. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Widrig, V.; Herren, G.; Wicker, T.; Zbinden, H.; Gronnier, J.; Spörri, L.; Praz, C.R.; Heuberger, M.; Kolodziej, M.C.; et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 2021, 7, 327–341. [Google Scholar] [CrossRef]
- Singh, K.; Saripalli, G.; Gautam, T.; Prasad, P.; Jain, N.; Balyan, H.S.; Gupta, P.K. BS-Seq reveals major role of differential CHH methylation during leaf rust resistance in wheat (Triticum aestivum L.). Mol. Genet. Genomics. 2022, 279, 731–749. [Google Scholar] [CrossRef]
- Agarwal, P.; Kapoor, S.; Tyagi, A.K. Transcription factors regulating the progression of monocot and dicotseed development. Bioessays 2011, 33, 189–202. [Google Scholar] [CrossRef]
| Source of Variation | df | F-Value | |||
|---|---|---|---|---|---|
| HD | MD | PH | LR | ||
| Genotypes | 250 | 158.9 ** | 189.6 ** | 136.9 ** | 19.8 ** |
| Environments | 3 | 426.6 ** | 362.6 ** | 523.6 ** | 78.6 ** |
| Replicates (nested in environments) | 2 | 15.2 ** | 12.6 ** | 25.3 ** | 4.3 ** |
| Genotypes × Environments | 749 | 6.3 ** | 8.9 ** | 6.9 ** | 2.1 ** |
| Error | 1425 | ||||
| Trait | Chromosome | Representive SNP | Start (Mb) | End (Mb) | R2 | p-Value | Environment | Favorable Allele | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Minimum | Maximum | |||||||
| LR | 1A | AX-111014967 | 125.2 | 134.0 | 4.5 | 4.8 | 1.9 × 10−5 | 3.9 × 10−5 | E1, E2, BLUP | A |
| LR | 1A | AX-110910455 | 275.1 | 282.2 | 6.8 | 7.2 | 8.3 × 10−7 | 5.8 × 10−6 | E1, E2, BLUP | C |
| LR | 1D | AX-111722231 | 416.6 | 421.9 | 6.5 | 7.1 | 4.0 × 10−7 | 1.6 × 10−5 | E1, E2, BLUP | C |
| LR | 3A | AX-109947801 | 619.5 | 627.7 | 4.5 | 4.6 | 2.9 × 10−5 | 5.4 × 10−5 | E1, E2, BLUP | C |
| LR | 3D | AX-95160344 | 512.3 | 517.8 | 5.2 | 5.3 | 3.4 × 10−6 | 5.6 × 10−6 | E1, E2, BLUP | G |
| LR | 5A | AX-109934392 | 535.5 | 540.0 | 4.3 | 4.6 | 4.5 × 10−5 | 3.9 × 10−5 | E1, E2, E4, BLUP | G |
| LR | 5B | AX-109731422 | 420.1 | 424.2 | 6.0 | 6.3 | 3.0 × 10−6 | 8.8 × 10−6 | E1, E2, E4, BLUP | G |
| LR | 5D | AX-109351618 | 9.5 | 11.9 | 4.9 | 11.2 | 5.1 × 10−9 | 1.9 × 10−5 | E1, E2, E4, BLUP | G |
| LR | 6A | AX-111533379 | 681.1 | 684.5 | 4.4 | 4.9 | 6.1 × 10−5 | 7.3 × 10−5 | E1, E2, E4, BLUP | G |
| LR | 6D | AX-109643661 | 7.4 | 10.7 | 4.4 | 4.8 | 4.6 × 10−5 | 5.1 × 10−5 | E1, E2, E4, BLUP | G |
| MD | 1D | AX-110001111 | 16.1 | 19.3 | 5.7% | 7.1% | 1.7 × 10−4 | 8.3 × 10−4 | E1, E2, E4, BLUP | G |
| MD | 2A | AX-110505632 | 379.9 | 410.9 | 5.6% | 7.8% | 9.3 × 10−5 | 9.7 × 10−4 | E1, E3 | G |
| MD | 2D | AX-108981520 | 643.7 | 650.1 | 5.4% | 10.8% | 6.7 × 10−6 | 9.8 × 10−4 | E2, BLUP | A |
| MD | 4D | AX-111354892 | 14.7 | 15.4 | 4.4% | 7.1% | 2.0 × 10−5 | 7.1 × 10−4 | E1, E3, E2, E4, BLUP | C |
| MD | 5B | AX-108905558 | 532.8 | 533.0 | 5.8% | 7.6% | 6.9 × 10−5 | 8.8 × 10−4 | E1, E3, E2, BLUP | C |
| MD | 6A | AX-110672099 | 594.8 | 599.8 | 6.2% | 10.0% | 4.2 × 10−6 | 5.1 × 10−4 | E1, E3, E2, E4, BLUP | C |
| MD | 6D | AX-95088462 | 470.3 | 471.0 | 6.2% | 8.5% | 3.0 × 10−5 | 4.2 × 10−4 | E1, E3, E2, E4, BLUP | G |
| MD | 7D | AX-110495278 | 104.6 | 104.9 | 5.9% | 7.8% | 3.0 × 10−4 | 9.0 × 10−4 | E1 | G |
| PH | 1A | AX-111018634 | 513.0 | 513.0 | 5.8% | 8.9% | 2.3 × 10−5 | 8.3 × 10−4 | E1, E3, E2, E4, BLUP | G |
| PH | 1D | AX-110483115 | 48.1 | 58.1 | 5.6% | 9.8% | 8.6 × 10−6 | 9.6 × 10−4 | E2, E4, BLUP | G |
| PH | 2B | AX-110481288 | 439.2 | 439.5 | 6.1% | 8.8% | 2.4 × 10−5 | 5.5 × 10−4 | E1, E3, E1, E4, BLUP | C |
| PH | 2D | AX-111031463 | 19.2 | 22.3 | 5.8% | 7.3% | 1.4 × 10−4 | 7.8 × 10−4 | E1, E3, E2, E4, BLUP | C |
| PH | 2D | AX-111096297 | 33.0 | 35.7 | 5.9% | 10.7% | 5.9 × 10−6 | 6.7 × 10−4 | E1, E3, E1, E2, E4, BLUP | C |
| PH | 3A | AX-111570251 | 69.3 | 70.7 | 6.0% | 8.3% | 4.8 × 10−5 | 6.1 × 10−4 | E1, E3, E4, BLUP | C |
| PH | 3B | AX-109500452 | 506.9 | 506.9 | 5.7% | 8.5% | 4.9 × 10−5 | 9.8 × 10−4 | E1, E3, E1, E2, E4, BLUP | A |
| PH | 4D | AX-109908071 | 26.0 | 28.8 | 6.0% | 6.8% | 3.4 × 10−4 | 6.8 × 10−4 | E1, E3, E1, E2, E4, BLUP | A |
| PH | 5B | AX-110384536 | 571.5 | 593.5 | 6.1% | 8.0% | 5.7 × 10−5 | 5.7 × 10−4 | E1, E3, E1, E2, E4, BLUP | T |
| PH | 5D | AX-110036274 | 562.0 | 562.7 | 5.8% | 7.4% | 1.2 × 10−4 | 7.5 × 10−4 | E1, E3, E1, E2, E4, BLUP | G |
| PH | 6A | AX-89610547 | 564.4 | 573.5 | 5.7% | 7.5% | 1.2 × 10−4 | 9.5 × 10−4 | E1, E3, E2, BLUP | G |
| PH | 7A | AX-109036056 | 720.0 | 722.3 | 5.8% | 7.1% | 2.2 × 10−4 | 8.9 × 10−4 | E1, E3, E1, BLUP | G |
| PH | 7B | AX-94434165 | 0.1 | 0.1 | 5.8% | 7.4% | 1.3 × 10−4 | 7.5 × 10−4 | E1, E3, E1, E4, BLUP | G |
| PH | 7D | AX-110052150 | 71.6 | 76.9 | 5.7% | 9.6% | 9.6 × 10−6 | 9.3 × 10−4 | E1, E3, E2, E4, BLUP | G |
| PH | 7D | AX-94842881 | 376.1 | 379.8 | 6.0% | 7.1% | 1.8 × 10−4 | 7.1 × 10−4 | E1, E3, E1, E2, E4, BLUP | G |
| HD | 1A | AX-111187227 | 297.7 | 297.7 | 6.0% | 7.6% | 1.1 × 10−4 | 6.6 × 10−4 | E1, E3, E2, BLUP | G |
| HD | 1B | AX-111469159 | 676.2 | 676.7 | 5.8% | 8.8% | 2.6 × 10−5 | 9.4 × 10−4 | E1, E3, E2, BLUP | C |
| HD | 2A | AX-111155549 | 209.2 | 415.3 | 6.1% | 6.6% | 6.7 × 10−4 | 6.9 × 10−4 | E1, E3, E2, BLUP | C |
| HD | 2D | AX-110203406 | 15.7 | 56.6 | 6.0% | 18.9% | 6.3 × 10−10 | 6.7 × 10−4 | E1, E3, E1, E2, E4, BLUP | C |
| HD | 3A | AX-111055674 | 596.2 | 601.5 | 5.7% | 7.7% | 1.4 × 10−4 | 9.6 × 10−4 | E1, E3, E1, E2, E4, BLUP | C |
| HD | 4A | AX-108880805 | 659.2 | 660.1 | 5.7% | 7.8% | 1.5 × 10−4 | 9.4 × 10−4 | E1, E2, BLUP | C |
| HD | 5A | AX-108774016 | 553.0 | 553.4 | 5.8% | 9.5% | 1.0 × 10−5 | 8.2 × 10−4 | E1, E2, E4, BLUP | G |
| HD | 5B | AX-111138644 | 521.0 | 521.7 | 5.7% | 6.8% | 3.1 × 10−4 | 9.4 × 10−4 | E1, E2, E4, BLUP | G |
| HD | 6A | AX-95073334 | 425.8 | 431.2 | 6.0% | 9.4% | 1.1 × 10−5 | 7.3 × 10−4 | E2, E4, BLUP | G |
| HD | 6D | AX-110918412 | 464.9 | 465.3 | 5.7% | 10.7% | 4.4 × 10−6 | 9.0 × 10−4 | E1, E3, E2, BLUP | C |
| Candidate Gene | Chromosome | Start (bp) | End (bp) | Annotation |
|---|---|---|---|---|
| TraesCS1A01G164400 | 1A | 296212311 | 296215426 | E3 ubiquitin-protein ligase |
| TraesCS1A01G343000 | 1A | 531717134 | 531722196 | ATP-binding cassette transporter |
| TraesCS1B01G392000 | 1B | 624970835 | 624972070 | B3 transcription factor family |
| TraesCS2A01G248400 | 2A | 371044868 | 371046934 | E3 ubiquitin-protein ligase family |
| TraesCS2D01G591000 | 2D | 646593035 | 646594771 | Serine/threonine-protein kinase |
| TraesCS6A01G356200 | 6A | 587551821 | 587553888 | ATP-binding cassette transporter |
| TraesCS7A01G520000 | 7A | 704387759 | 704388700 | Calcium-dependent lipid-binding family |
| Marker | SNP | Genotype | Number | Phenotype | p-Value |
|---|---|---|---|---|---|
| Kasp_2D_PH | AX-111096297 | CC | 36 | 78.5 cm (PH) | 0.049 * |
| GG | 115 | 74.0 cm (PH) | |||
| Kasp_6D_HD | AX-110918412 | AA | 105 | 72.0 d (HD) | 0.023 * |
| CC | 45 | 69.9 d (HD) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Li, Y.; Liu, W.; Sun, Y.; Tang, J.; Che, J.; Yang, S.; Wang, X.; Zhang, R. Genetic Analysis of Adaptive Traits in Spring Wheat in Northeast China. Life 2024, 14, 168. https://doi.org/10.3390/life14020168
Zhang H, Li Y, Liu W, Sun Y, Tang J, Che J, Yang S, Wang X, Zhang R. Genetic Analysis of Adaptive Traits in Spring Wheat in Northeast China. Life. 2024; 14(2):168. https://doi.org/10.3390/life14020168
Chicago/Turabian StyleZhang, Hongji, Yuyao Li, Wenlin Liu, Yan Sun, Jingquan Tang, Jingyu Che, Shuping Yang, Xiangyu Wang, and Rui Zhang. 2024. "Genetic Analysis of Adaptive Traits in Spring Wheat in Northeast China" Life 14, no. 2: 168. https://doi.org/10.3390/life14020168
APA StyleZhang, H., Li, Y., Liu, W., Sun, Y., Tang, J., Che, J., Yang, S., Wang, X., & Zhang, R. (2024). Genetic Analysis of Adaptive Traits in Spring Wheat in Northeast China. Life, 14(2), 168. https://doi.org/10.3390/life14020168





