Parasitism-Induced Changes in Microbial Eukaryotes of Peruvian Alpaca Gastrointestinal Tract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal and Sample Collection
2.2. Parasitological Study of Feces
2.3. DNA Extraction and Sequencing
2.4. Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
3.1. Alpha Diversity of the Gut Microbiota in the Alpaca Population
3.2. Beta Diversity of the Gut Microbiota in the Alpaca Population
3.3. Correlation between Biochemical Parameters and the Alpha Diversity of Protozoan Composition in the Microbiota
3.4. Composition Taxonomic of the Gut Microbiota
3.5. Detection of Modifications in the Taxonomic Composition Linked to the Influence of Parasites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosadio, A.R.; Maturrano, H.L.; Pérez, J.D.; Luna, E.L. El complejo entérico neonatal en alpacas andinas. Rev. Investig. Vet. Perú 2012, 23, 261–271. [Google Scholar] [CrossRef]
- Cordero, F.A.; Contreras, P.J.; Mayhua, M.P.; Jurado, E.M.; Castrejón, V.M. Correlaciones Fenotípicas Entre Características Productivas En Alpacas Huacaya. Rev. Investig. Vet. Perú 2011, 22, 15–21. [Google Scholar] [CrossRef]
- Quispe, E.C.; Rodríguez, T.C.; Iñiguez, L.R.; Mueller, J.P. Producción de Fibra de Alpaca, Llama, Vicuña y Guanaco En Sudamérica. Anim. Genet. Resour. Inf. 2009, 45, 1–14. [Google Scholar] [CrossRef]
- Bustinza Choque, A.V.; Machaca Machaca, V.; Cano Fuentes, V.; Quispe Coaquira, J. Evolución y Desarrollo de Las Razas de Alpaca: Suri y Huacaya. Rev. Investig. Vet. Perú 2021, 32, e19876. [Google Scholar] [CrossRef]
- Whitehead, C.E. Neonatal Diseases in Llamas and Alpacas. Vet. Clin. North Am. Food Anim. Pract. 2009, 25, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Manchego, A.; Rocha, C.B.; Fornells, L.A.; Silva, R.C.; Mendes, G.S.; Dias, H.G.; Sandoval, N.; Pezo, D.; Santos, N. Outbreak of Diarrhea among Preweaning Alpacas (Vicugna pacos) in the Southern Peruvian Highland. J. Infect. Dev. Ctries. 2016, 10, 269–274. [Google Scholar] [CrossRef]
- Heller, M.C.; Chigerwe, M. Diagnosis and Treatment of Infectious Enteritis in Neonatal and Juvenile Ruminants. Vet. Clin. North Am. Food Anim. Pract. 2018, 34, 101–117. [Google Scholar] [CrossRef]
- Cebra, C.K.; Mattson, D.E.; Baker, R.J.; Sonn, R.J.; Dearing, P.L. Potential Pathogens in Feces from Unweaned Llamas and Alpacas with Diarrhea. J. Am. Vet. Med. Assoc. 2003, 223, 1806–1808. [Google Scholar] [CrossRef]
- Fowler, M.E. Medicine and Surgery of South American Camelids: Llama, Alpaca, Vicuña, Guanaco; Iowa State University Press: Ames, IA, USA, 1989. [Google Scholar]
- Cheney, J.M.; Allen, G.T. Parasitism in Llamas. Vet. Clin. North Am. Food Anim. Pract. 1989, 5, 217–225. [Google Scholar] [CrossRef]
- Whitehead, C.E.; Anderson, D.E. Neonatal Diarrhea in Llamas and Alpacas. Small Rumin. Res. 2006, 61, 207–215. [Google Scholar] [CrossRef]
- Guerrero, C.A. Coccidia (Protozoa: Eimeriidae) of the Alpaca Lama pacos. J. Protozool. 1967, 14, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, C.A.; Hernández, J.; Bazalar, H.; Alva, J. Eimeria macusaniensis n. sp. (Protozoa: Eimeriidae) of the Alpaca Lama Pacos. J. Protozool. 1971, 18, 162–163. [Google Scholar] [CrossRef] [PubMed]
- Santin, M. Cryptosporidium and Giardia in Ruminants. Vet. Clin. North Am. Food Anim. Pract. 2020, 36, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Kiorpes, A.L.; Kirkpatrick, C.E.; Bowman, D.D. Isolation of Giardia from a Llama and from Sheep. Can. J. Vet. Res. 1987, 51, 277–280. [Google Scholar] [PubMed]
- Gomez-Puerta, L.A.; Lopez-Urbina, M.T.; Alarcon, V.; Cama, V.; Gonzalez, A.E.; Xiao, L. Occurrence of Giardia duodenalis Assemblages in Alpacas in the Andean Region. Parasitol. Int. 2014, 63, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Couso, H.; Ortega-Mora, L.M.; Aguado-Martínez, A.; Rosadio-Alcántara, R.; Maturrano-Hernández, L.; Luna-Espinoza, L.; Zanabria-Huisa, V.; Pedraza-Díaz, S. Presence and Molecular Characterisation of Giardia and Cryptosporidium in Alpacas (Vicugna pacos) from Peru. Vet. Parasitol. 2012, 187, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Koehler, A.V.; Rashid, M.H.; Zhang, Y.; Vaughan, J.L.; Gasser, R.B.; Jabbar, A. First Cross-Sectional, Molecular Epidemiological Survey of Cryptosporidium, Giardia and Enterocytozoon in Alpaca (Vicugna pacos) in Australia. Parasit. Vectors 2018, 11, 498. [Google Scholar] [CrossRef]
- Trout, J.M.; Santín, M.; Fayer, R. Detection of Assemblage A, Giardia duodenalis and Eimeria spp. in Alpacas on Two Maryland Farms. Vet. Parasitol. 2008, 153, 203–208. [Google Scholar] [CrossRef]
- Partida-Rodríguez, O.; Serrano-Vázquez, A.; Nieves-Ramírez, M.E.; Moran, P.; Rojas, L.; Portillo, T.; González, E.; Hernández, E.; Finlay, B.B.; Ximenez, C. Human Intestinal Microbiota: Interaction between Parasites and the Host Immune Response. Arch. Med. Res. 2017, 48, 690–700. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, Z.; Cao, L.; Hu, S.; Zhang, Z.; Qin, B.; Guo, Z.; Nie, K. Upregulation of Chicken TLR4, TLR15 and MyD88 in Heterophils and Monocyte-Derived Macrophages Stimulated with Eimeria tenella In Vitro. Exp. Parasitol. 2013, 133, 427–433. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, K.; Zou, M.; Sun, Y.; Peng, X. Gga-miR-451 Negatively Regulates Mycoplasma gallisepticum (HS Strain)-Induced Inflammatory Cytokine Production via Targeting YWHAZ. Int. J. Mol. Sci. 2018, 19, 1191. [Google Scholar] [CrossRef] [PubMed]
- Cotton, J.A.; Amat, C.B.; Buret, A.G. Disruptions of Host Immunity and Inflammation by Giardia duodenalis: Potential Consequences for Co-Infections in the Gastro-Intestinal Tract. Pathogens 2015, 4, 764–792. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Isaacson, R.E. The Pig Gut Microbial Diversity: Understanding the Pig Gut Microbial Ecology through the next Generation High Throughput Sequencing. Vet. Microbiol. 2015, 177, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Hadziavdic, K.; Lekang, K.; Lanzen, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA Gene for Designing Universal Eukaryote Specific Primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M.; Knudsen, C.; Beaumont, M.; Rodriguez, J.; Neyrinck, A.M.; Bindels, L.B. Contribution of the Gut Microbiota to the Regulation of Host Metabolism and Energy Balance: A Focus on the Gut–Liver Axis. Proc. Nutr. Soc. 2019, 78, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Bowman, D.D. Georgi. Parasitología Para Veterinarios; Elsevier Health Sciences: Amsterdam, The Netherlands, 2022; ISBN 978-84-13-82357-7. [Google Scholar]
- Barriga, O.O. Las Enfermedades Parasitarias de los Animales Domésticos en la América Latina; Editorial Germinal: Santiago de Chile, Chile, 2002; ISBN 978-956-291-398-0. [Google Scholar]
- Cebra, C.K.; Stang, B.V. Comparison of Methods to Detect Gastrointestinal Parasites in Llamas and Alpacas. J. Am. Vet. Med. Assoc. 2008, 232, 733–741. [Google Scholar] [CrossRef]
- Vega, L.; Jaimes, J.; Morales, D.; Martínez, D.; Cruz-Saavedra, L.; Muñoz, M.; Ramírez, J.D. Microbial Communities’ Characterization in Urban Recreational Surface Waters Using Next Generation Sequencing. Microb. Ecol. 2021, 81, 847–863. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Xu, S.; Zhan, L.; Tang, W.; Wang, Q.; Dai, Z.; Zhou, L.; Feng, T.; Chen, M.; Wu, T.; Hu, E.; et al. MicrobiotaProcess: A comprehensive R package for deep mining microbiome. Innovation 2023, 4, 100388. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational multivariate analysis of variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 1–15. ISBN 978-1-118-44511-2. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R. Vegan: Community Ecology Package. R Package Version 2.6-4. 2022. Available online: http://cran.r-project.org/package=vegan (accessed on 9 November 2023).
- Richard, M.L.; Sokol, H. The gut mycobiota: Insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef]
- Cholewińska, P.; Czyż, K.; Nowakowski, P.; Wyrostek, A. The microbiome of the digestive system of ruminants—A review. Anim. Health Res. Rev. 2020, 21, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.A.; Petri, S.E.; Schneider, B.N.; Reichman, D.J.; Jiang, N.; Begum, S.; Watanabe, K.; Jansen, C.S.; Elliott, K.P.; Burgess, S.L.; et al. Role of the Gut Microbiota of Children in Diarrhea Due to the Protozoan Parasite Entamoeba histolytica. J. Infect. Dis. 2016, 213, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Herrera, G.; Vega, L.; Patarroyo, M.A.; Ramírez, J.D.; Muñoz, M. Gut microbiota composition in health-care facility-and community-onset diarrheic patients with Clostridioides difficile Infection. Sci. Rep. 2021, 11, 10849. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.; Travers, P.; Walport, M.; Shlomchik, M.J. Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001; ISBN 978-0-8153-3642-6. [Google Scholar]
- Alzate, J.F.; Toro-Londoño, M.; Cabarcas, F.; Garcia-Montoya, G.; Galvan-Diaz, A. Contrasting microbiota profiles observed in children carrying either Blastocystis spp. or the commensal amoebas Entamoeba coli or Endolimax nana. Sci. Rep. 2020, 10, 15354. [Google Scholar] [CrossRef]
- Pernthaler, J. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 2005, 3, 537–546. [Google Scholar] [CrossRef]
- Johnke, J.; Cohen, Y.; de Leeuw, M.; Kushmaro, A.; Jurkevitch, E.; Chatzinotas, A. Multiple micro-predators controlling bacterial communities in the environment. Curr. Opin. Biotechnol. 2014, 27, 185–190. [Google Scholar] [CrossRef]
- Zhu, D.; Lu, L.; Zhang, Z.; Qi, D.; Zhang, M.; O’Connor, P.; Wei, F.; Zhu, Y.-G. Insights into the roles of fungi and protist in the giant panda gut microbiome and antibiotic resistome. Environ. Int. 2021, 155, 106703. [Google Scholar] [CrossRef]
- Mann, A.E.; Mazel, F.; Lemay, M.A.; Morien, E.; Billy, V.; Kowalewski, M.; Di Fiore, A.; Link, A.; Goldberg, T.L.; Tecot, S.; et al. Biodiversity of protists and nematodes in the wild nonhuman primate gut. ISME J. 2020, 14, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Vega, L.; Herrera, G.; Muñoz, M.; Patarroyo, M.A.; Maloney, J.G.; Santín, M.; Ramírez, J.D. Gut microbiota profiles in diarrheic patients with co-occurrence of Clostridioides difficile and Blastocystis. PLoS ONE 2021, 16, e0248185. [Google Scholar] [CrossRef] [PubMed]
- Gil, F.F.; Ventura, L.L.A.; Fonseca, J.F.; Saniago, H.C.; Busatti, H.; Santos, J.F.G.; Gomes, M.A. Hematological profile in natural progression of giardiasis: Kinetics of experimental infection in gerbils. J. Infect. Dev. Ctries. 2018, 12, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Rakhshandehroo, E.; Nazifi, S.; Razavi, S.M.; Ghane, M.; Alavi, A.M. Caprine coccidiosis: The effects of induced infection on certain blood parameters. Vet. Fak. Sveučilišta U Zagrebu 2013, 83, 623–631. [Google Scholar]
- Ruiz, A.; Matos, L.; Muñoz, M.C.; Hermosilla, C.; Molina, J.M.; Andrada, M.; Rodríguez, F.; Pérez, D.; López, A.; Guedes, A.; et al. Isolation of an Eimeria ninakohlyakimovae field strain (Canary Islands) and analysis of its infection characteristics in goat kids. Res. Vet. Sci. 2013, 94, 277–284. [Google Scholar] [CrossRef]
- Cebra, C. Disorders of the Digestive System. In Llama Alpaca Care; Elsevier: Toronto, ON, Canada, 2014; pp. 477–536. [Google Scholar] [CrossRef]
- Palacios, E.C.; Tabacchi, N.L.; Chavera, C.A.; López, U.T.; Santillán, A.G.; Sandoval, C.N.; Pezo, C.D.; Perales, C.R. Eimeriosis En Crías de Alpacas: Estudio Anátomo Histopatológico. Rev. Investig. Vet. Perú 2004, 15, 174–178. [Google Scholar]
- Azad, E.; Fehr, K.B.; Derakhshani, H.; Forster, R.; Acharya, S.; Khafipour, E.; McGeough, E.; McAllister, T.A. Interrelationships of fiber-associated anaerobic fungi and bacterial communities in the rumen of bloated cattle grazing alfalfa. Microorganisms 2020, 8, 1543. [Google Scholar] [CrossRef]
- Ramírez, A.L.; Herrera, G.; Muñoz, M.; Vega, L.; Cruz-Saavedra, L.; García-Corredor, D.; Pulido-Medellín, M.; Bulla-Castañeda, D.M.; Giraldo, J.C.; Bernal, M.C.; et al. Describing the intestinal microbiota of Holstein Fasciola-positive and -negative cattle from a hyperendemic area of fascioliasis in central Colombia. PLoS Negl. Trop. Dis. 2021, 15, e0009658. [Google Scholar] [CrossRef]
- Wang, H.; Li, P.; Liu, X.; Zhang, C.; Lu, Q.; Xi, D.; Yang, R.; Wang, S.; Bai, W.; Yang, Z.; et al. The Composition of Fungal Communities in the Rumen of Gayals (Bos frontalis), Yaks (Bos grunniens), and Yunnan and Tibetan Yellow Cattle (Bos taurs). Pol. J. Microbiol. 2019, 68, 505–514. [Google Scholar] [CrossRef]
- Chen, X.; An, M.; Zhang, W.; Li, K.; Kulyar, M.F.-E.-A.; Duan, K.; Zhou, H.; Wu, Y.; Wan, X.; Li, J.; et al. Integrated Bacteria-Fungi Diversity Analysis Reveals the Gut Microbial Changes in Buffalo with Mastitis. Front. Vet. Sci. 2022, 9, 918541. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.-B.; Meng, J.-X.; Ma, H.; Liu, R.; Qin, Y.; Qin, Y.-F.; Geng, H.-L.; Ni, H.-B.; Zhang, X.-X. Description of Gut Mycobiota Composition and Diversity of Caprinae Animals. Microbiol. Spectr. 2023, 11, e0242422. [Google Scholar] [CrossRef] [PubMed]
- Fliegerova, K.O.; Podmirseg, S.M.; Vinzelj, J.; Grilli, D.J.; Kvasnová, S.; Schierová, D.; Sechovcová, H.; Mrázek, J.; Siddi, G.; Arenas, G.N.; et al. The effect of a high-grain diet on the rumen microbiome of goats with a special focus on anaerobic fungi. Microorganisms 2021, 9, 157. [Google Scholar] [CrossRef]
- Sánchez-Herencia, D.; Mamani-Mango, G.; Coila-Añasco, P. Eimeria control in baby alpacas using Toltrazuril as a prophylactic measure in humid Puna. J. Selva Andina Anim. Sci. 2021, 8, 82–89. [Google Scholar] [CrossRef]
- Dubey, J.P. A review of Coccidiosis in South American camelids. Parasitol. Res. 2018, 117, 1999–2013. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.D.; Mongi, F.; Sánchez, A.; Ramírez, J.D. Blastocystis and urticaria: Examination of subtypes and morphotypes in an unusual clinical manifestation. Acta Trop. 2015, 148, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, C.; Elmhadi, M.; Zhang, H.; Han, Y.; Shen, B.; He, B.L.; Liu, X.Y.; Wang, H.R. Thiamine ameliorates metabolic disorders induced by a long-term high-concentrate diet and promotes rumen epithelial development in goats. J. Dairy Sci. 2021, 104, 11522–11536. [Google Scholar] [CrossRef]
- Zhao, G.H.; Hu, X.F.; Liu, T.L.; Hu, R.S.; Yu, Z.Q.; Yang, W.B.; Wu, Y.L.; Yu, S.K.; Song, J.K. Molecular Characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol. Res. 2017, 116, 2327–2333. [Google Scholar] [CrossRef]
- Deng, L.; Yao, J.; Chen, S.; He, T.; Chai, Y.; Zhou, Z.; Shi, X.; Liu, H.; Zhong, Z.; Fu, H.; et al. First identification and molecular subtyping of Blastocystis sp. in zoo animals in southwestern China. Parasit. Vectors 2021, 14, 11. [Google Scholar] [CrossRef]
- Gao, W.-W.; Ma, Y.-T.; Ma, Y.-Y.; Li, R.-L.; Li, J.; Zheng, F.-G.; Zheng, W.-B.; Liu, Q.; Zhu, X.-Q. First report of Eimeria and Entamoeba infection in alpacas (Vicugna pacos) in Shanxi Province, northern China. Parasitol. Res. 2021, 120, 2031–2035. [Google Scholar] [CrossRef]
- Qi, T.; Zheng, W.; Guo, L.; Sun, Y.; Li, J.; Kang, M. First description of Blastocystis sp. and Entamoeba sp. infecting zoo animals in the Qinghai-Tibetan plateau area, China. Front. Cell. Infect. Microbiol. 2023, 13, 1212617. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.; Zhang, W.; Kulyar, M.F.-A.; Ullah, K.; Han, Z.; Qin, J.; Bi, C.; Wang, Y.; Li, K. Comparative analysis of gut microbiota in healthy and diarrheic yaks. Microb. Cell Factories 2022, 21, 111. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Liu, Y.; Nie, C.; Chen, C.; Niu, J.; Zhang, W. Comparison of bacterial and fungal community structure and potential function analysis of Yak feces before and after weaning. BioMed Res. Int. 2022, 2022, 6297231. [Google Scholar] [CrossRef] [PubMed]
- Mapperson, R.R.; Kotiw, M.; Davis, R.A.; Dearnaley, J.D.W. The diversity and antimicrobial activity of Preussia sp. endophytes isolated from australian dry rainforests. Curr. Microbiol. 2014, 68, 30–37. [Google Scholar] [CrossRef]
- Paudel, B.; Bhattarai, K.; Bhattarai, H.D. Antimicrobial and antioxidant activities of two polyketides from lichen-endophytic fungus Preussia sp. Z. Naturforschung C J. Biosci. 2018, 73, 161–163. [Google Scholar] [CrossRef]
- Macauley-Patrick, S.; Fazenda, M.L.; McNeil, B.; Harvey, L.M. Heterologous protein production using the Pichia pastoris expression system. Yeast Chichester Engl. 2005, 22, 249–270. [Google Scholar] [CrossRef]

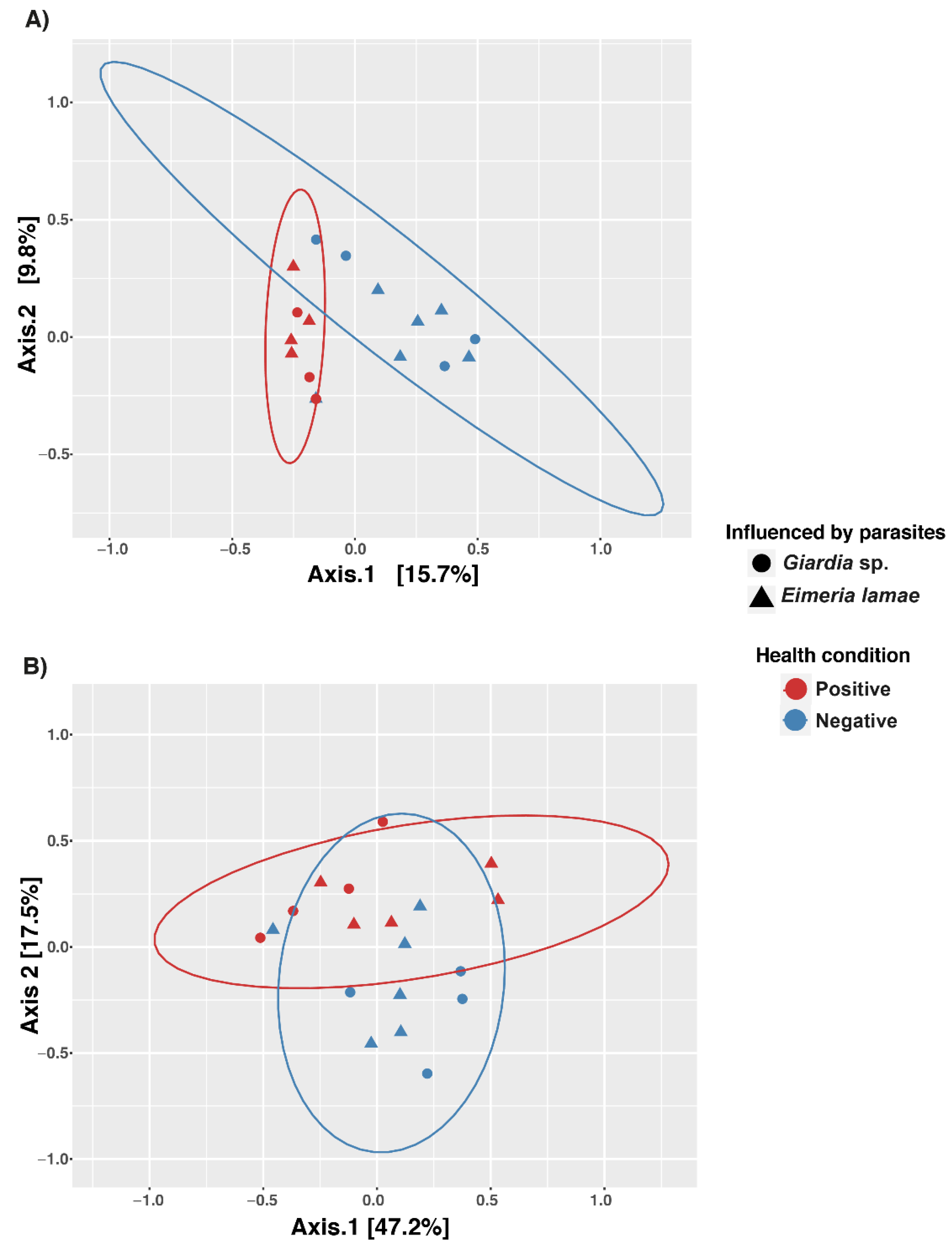
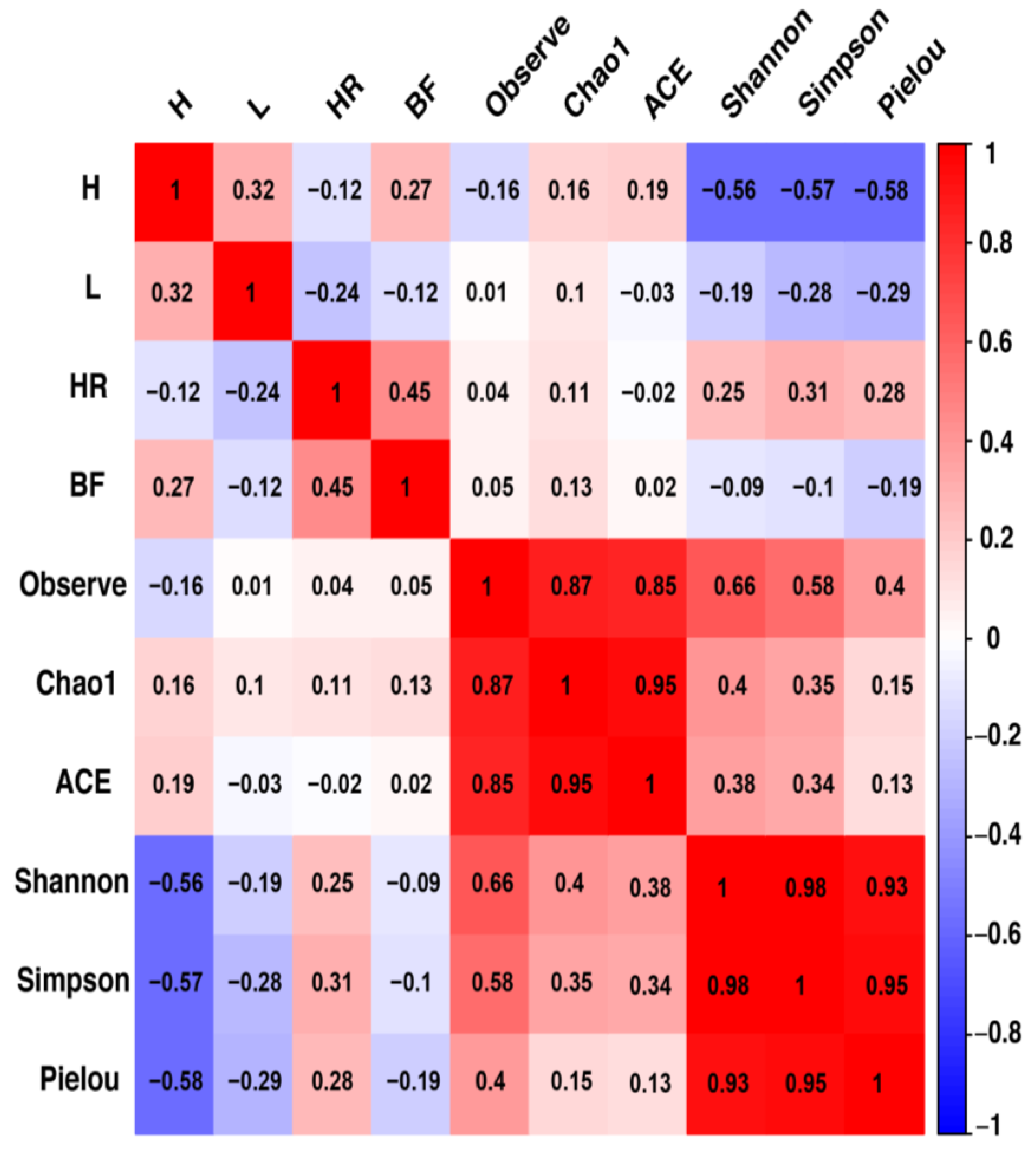
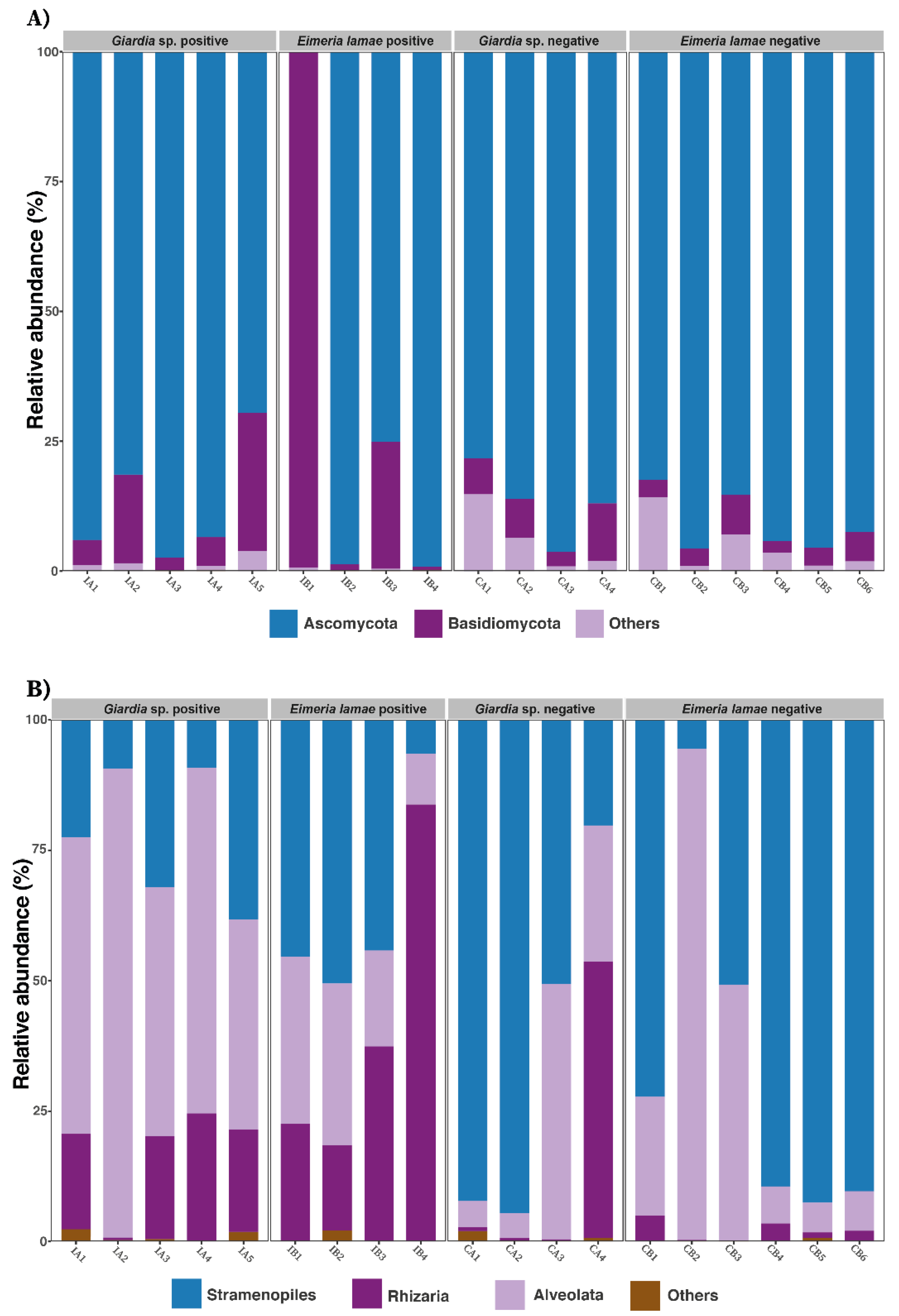

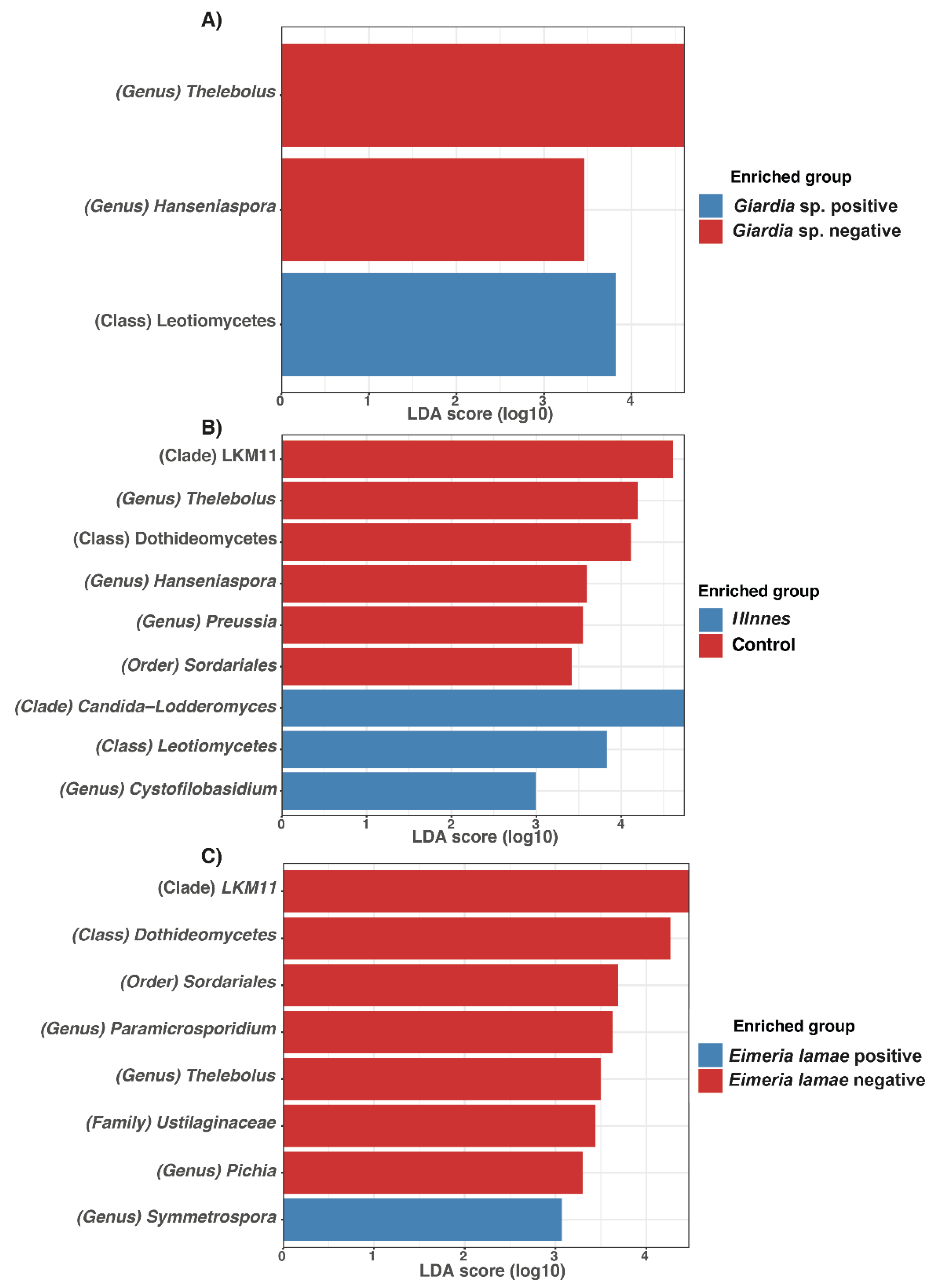
| Items | Df | SumOfSqs | R2 | F | Pr (>F) |
|---|---|---|---|---|---|
| Protists | |||||
| Influenced by parasites | 1 | 0.3052 | 0.04171 | 0.8475 | 0.7089 |
| Health status | 1 | 1.1938 | 0.16313 | 3.3146 | 0.0003 *** |
| Influenced by parasites: Health status | 1 | 0.4165 | 0.05691 | 1.1564 | 0.2005 |
| Fungi | |||||
| Influenced by parasites | 1 | 0.17297 | 0.07116 | 1.537 | 0.11 |
| Health status | 1 | 0.27745 | 0.11414 | 2.4654 | 0.0092 ** |
| Influenced by parasites: Health status | 1 | 0.17985 | 0.07399 | 1.5982 | 0.0906 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez, D.; Zapata, C.; Romero, Y.; Flores-Huarco, N.H.; Oros, O.; Alvarado, W.; Quilcate, C.; Guevara-Alvarado, H.M.; Estrada, R.; Coila, P. Parasitism-Induced Changes in Microbial Eukaryotes of Peruvian Alpaca Gastrointestinal Tract. Life 2024, 14, 187. https://doi.org/10.3390/life14020187
Sanchez D, Zapata C, Romero Y, Flores-Huarco NH, Oros O, Alvarado W, Quilcate C, Guevara-Alvarado HM, Estrada R, Coila P. Parasitism-Induced Changes in Microbial Eukaryotes of Peruvian Alpaca Gastrointestinal Tract. Life. 2024; 14(2):187. https://doi.org/10.3390/life14020187
Chicago/Turabian StyleSanchez, Diana, Celso Zapata, Yolanda Romero, Nils H. Flores-Huarco, Oscar Oros, Wigoberto Alvarado, Carlos Quilcate, Hada M. Guevara-Alvarado, Richard Estrada, and Pedro Coila. 2024. "Parasitism-Induced Changes in Microbial Eukaryotes of Peruvian Alpaca Gastrointestinal Tract" Life 14, no. 2: 187. https://doi.org/10.3390/life14020187
APA StyleSanchez, D., Zapata, C., Romero, Y., Flores-Huarco, N. H., Oros, O., Alvarado, W., Quilcate, C., Guevara-Alvarado, H. M., Estrada, R., & Coila, P. (2024). Parasitism-Induced Changes in Microbial Eukaryotes of Peruvian Alpaca Gastrointestinal Tract. Life, 14(2), 187. https://doi.org/10.3390/life14020187






