Capillary-Electrophoresis-Based Species Barcoding of Big Cats: CR-mtDNA-Length Polymorphism
Abstract
:1. Introduction
2. Materials and Methods
2.1. PCR Composition and PCR Conditions for Fragment Analysis
2.2. PCR Composition and PCR Conditions for Agarose Gel Electrophoresis and Sanger Sequencing
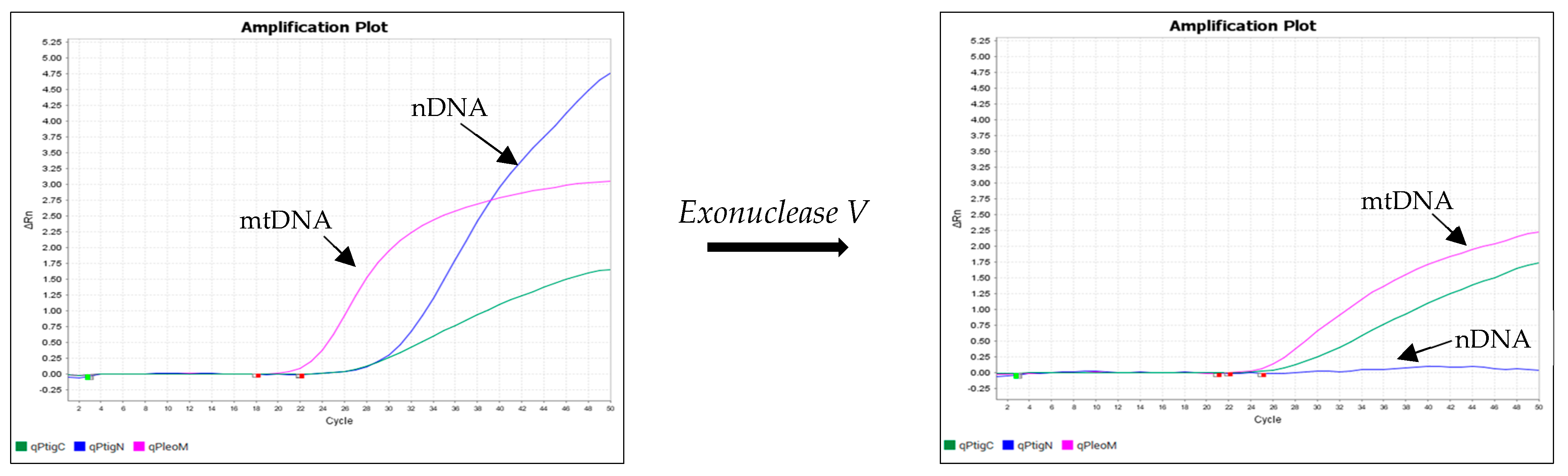
2.3. The Effect of Exonuclease V Treatment
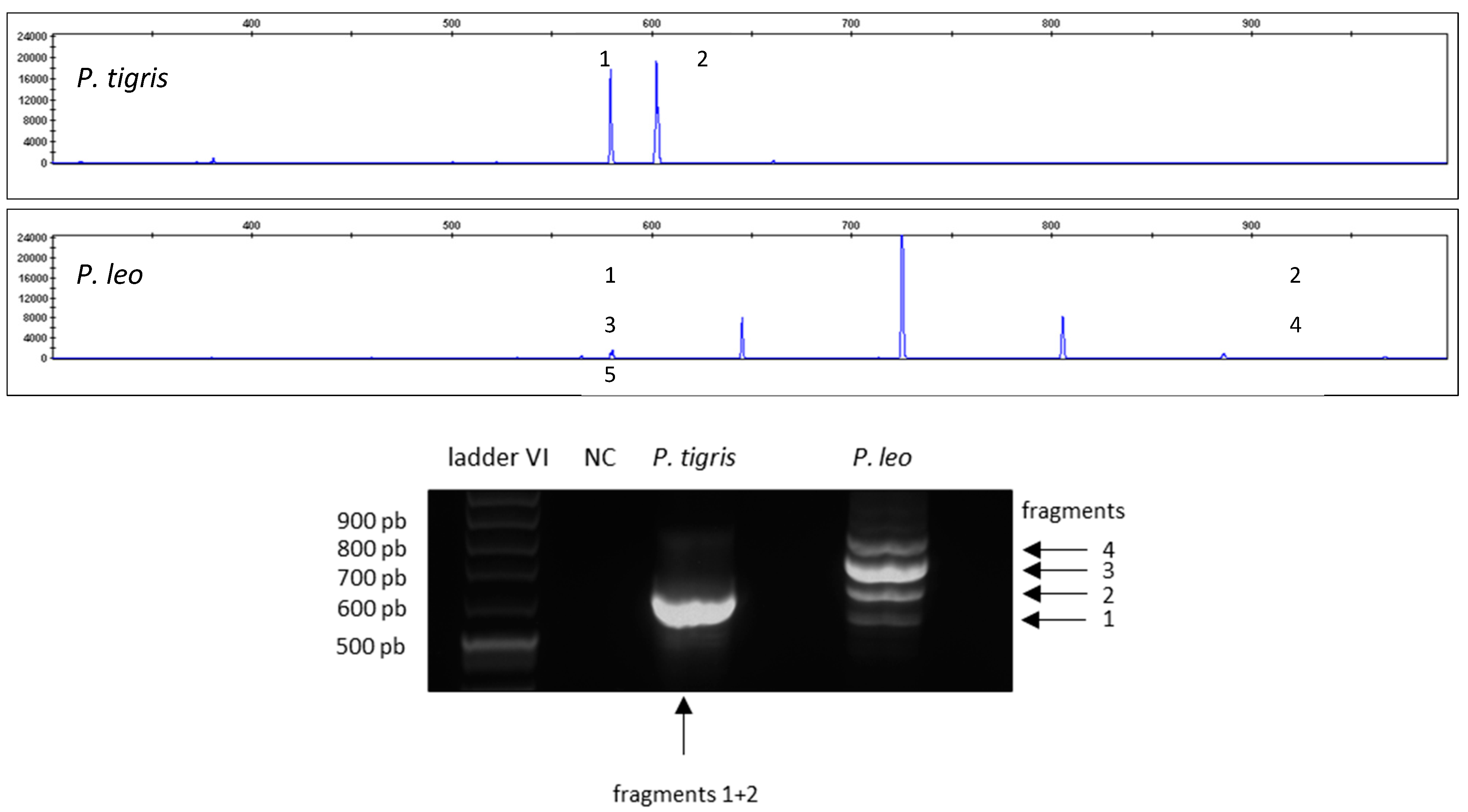
3. Results
Results of the Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheung, H.; Doughty, H.; Hinsley, A.; Hsu, E.; Lee, T.M.; Milner-Gulland, E.; Possingham, H.P.; Biggs, D. Understanding Traditional Chinese Medicine to strengthen conservation outcomes. People Nat. 2021, 3, 115–128. [Google Scholar] [CrossRef]
- Secretariat, C.; de l’Environnement, M.I. Convention on International Trade in Endangered Species of Wild Fauna and Flora; Citeseer: Princeton, NJ, USA, 2011. [Google Scholar]
- Pečnikar, Ž.F.; Buzan, E.V. 20 years since the introduction of DNA barcoding: From theory to application. J. Appl. Genet. 2014, 55, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Pompanon, F.; Taberlet, P. DNA barcoding for ecologists. Trends Ecol. Evol. 2009, 24, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, Z.; Li, C.; Ping, X.; Cui, S.; Tang, S.; Chu, H.; Liu, B. Identification of ungulates used in a traditional Chinese medicine with DNA barcoding technology. Ecol. Evol. 2015, 5, 1818–1825. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Gan, S.; Zhang, J.; Fan, Y.; Li, B.; Wan, L.; Nie, J.; Wang, X.; Chen, J. Application of DNA Barcoding for the Identification of Snake Gallbladders as a Traditional Chinese Medicine. Rev. Bras. Farmacogn. 2022, 32, 663–668. [Google Scholar] [CrossRef]
- Zhang, H.; Yao, H.; Cui, L.; Du, H.; Lin, Z.; Gao, X.; Lang, X.; Song, J.; Luo, K.; Shi, L.; et al. Application of COI-based DNA barcoding for identifying animal medical materials in the Chinese pharmacopoeia. World Sci. Technol. Mod. Tradit. Chin. Med. 2013, 12, 371–380. [Google Scholar]
- Yang, F.; Ding, F.; Chen, H.; He, M.; Zhu, S.; Ma, X.; Jiang, L.; Li, H. DNA barcoding for the identification and authentication of animal species in traditional medicine. Evid. Based Complement. Altern. Med. 2018, 2018, 5160254. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Sun, W.; Li, J.; Yao, H.; Shi, Y.; Wang, P.; Huang, B.; Shi, L.; Liu, D.; Hu, Z. Identifying the species of seeds in traditional Chinese medicine using DNA barcoding. Front. Pharmacol. 2018, 9, 701. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, Q.; Qiu, S.; Dai, J.; Gao, X. DNA barcoding: An efficient technology to authenticate plant species of traditional Chinese medicine and recent advances. Chin. Med. 2022, 17, 112. [Google Scholar] [CrossRef]
- Gong, L.; Qiu, X.H.; Huang, J.; Xu, W.; Bai, J.Q.; Zhang, J.; Su, H.; Xu, C.M.; Huang, Z.H. Constructing a DNA barcode reference library for southern herbs in China: A resource for authentication of southern Chinese medicine. PLoS ONE 2018, 13, e0201240. [Google Scholar] [CrossRef]
- Miao, L.; Xi-Wen, L.; Bao-Sheng, L.; Lu, L.; Yue-Ying, R. Species identification of poisonous medicinal plant using DNA barcoding. Chin. J. Nat. Med. 2019, 17, 585–590. [Google Scholar]
- Menotti-Raymond, M.; David, V.A.; Stephens, J.C.; Lyons, L.A.; O’Brien, S.J. Genetic individualization of domestic cats using feline STR loci for forensic applications. J. Forensic Sci. 1997, 42, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, A.P.; Rohleder, U.; Eichmann, C.; Pfeiffer, I.; Parson, W.; Schleenbecker, U. A proposal for standardization in forensic canine DNA typing: Allele nomenclature of six canine-specific STR loci. J. Forensic Sci. 2006, 51, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Potoczniak, M.J.; Chermak, M.; Quarino, L.; Tobe, S.S.; Conte, J. Development of a multiplex, PCR-based genotyping assay for African and Asian elephants for forensic purposes. Int. J. Leg. Med. 2020, 134, 55–62. [Google Scholar] [CrossRef]
- Singh, A.; Priyambada, P.; Jabin, G.; Singh, S.K.; Joshi, B.D.; Venkatraman, C.; Chandra, K.; Sharma, L.K.; Thakur, M. Pangolin Indexing System: Implications in forensic surveillance of large seizures. Int. J. Leg. Med. 2020, 134, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.K. RhODIS®(The Rhinoceros DNA Index System): The Application of Simple Forensic and Genetic Tools Help Conserve African Rhinoceros. In Wildlife Biodiversity Conservation; Springer: Berlin/Heidelberg, Germany, 2021; pp. 463–485. [Google Scholar]
- Vaněk, D.; Ehler, E.; Vaňková, L. Development of DNA quantitation and STR typing systems for Panthera tigris species determination and individual identification in forensic casework. Eur. J. Environ. Sci. 2021, 11, 113–118. [Google Scholar] [CrossRef]
- Wilkinson, M.J.; Szabo, C.; Ford, C.S.; Yarom, Y.; Croxford, A.E.; Camp, A.; Gooding, P. Replacing Sanger with Next Generation Sequencing to improve coverage and quality of reference DNA barcodes for plants. Sci. Rep. 2017, 7, 46040. [Google Scholar] [CrossRef]
- Haider, N.; Nabulsi, I.; Al-Safadi, B. Identification of meat species by PCR-RFLP of the mitochondrial COI gene. Meat Sci. 2012, 90, 490–493. [Google Scholar] [CrossRef]
- Noikotr, K.; Chaveerach, A.; Pinthong, K.; Tanomtong, A.; Sudmoon, R.; Tanee, T. RAPD and barcode analyses of groupers of the genus Epinephelus. Genet. Mol. Res 2013, 12, 5721–5732. [Google Scholar] [CrossRef]
- Hoffman, J.; Clark, M.; Amos, W.; Peck, L. Widespread amplification of amplified fragment length polymorphisms (AFLPs) in marine Antarctic animals. Polar Biol. 2012, 35, 919–929. [Google Scholar] [CrossRef]
- Lahiff, S.; Glennon, M.; Lyng, J.; Smith, T.; Maher, M.; Shilton, N. Species-specific PCR for the identification of ovine, porcine and chicken species in meat and bone meal (MBM). Mol. Cell. Probes 2001, 15, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Shin, S.E.; Ko, K.S.; Park, S.H. Identification of forensically important Calliphoridae and Sarcophagidae species collected in Korea using SNaPshot multiplex system targeting the cytochrome c oxidase subunit i gene. BioMed Res. Int. 2018, 2018, 2953892. [Google Scholar] [CrossRef] [PubMed]
- Denyingyhot, A.; Phraephaisarn, C.; Vesaratchavest, M.; Dahlan, W.; Keeratipibul, S. A new tool for quality control to monitor contamination of six non-halal meats in food industry by multiplex high-resolution melting analysis (HRMA). NFS J. 2021, 25, 31–40. [Google Scholar] [CrossRef]
- Friedenberger, A.; Doyle, C.; Couillard, L.; Kyle, C.J. The bear necessities: A sensitive qPCR assay for bear DNA detection from bile and derived products to complement wildlife forensic enforcement. Forensic Sci. Int. Genet. 2023, 67, 102935. [Google Scholar] [CrossRef] [PubMed]
- Mori, C.; Matsumura, S. Development and validation of simultaneous identification of 26 mammalian and poultry species by a multiplex assay. Int. J. Leg. Med. 2021, 136, 1–12. [Google Scholar] [CrossRef]
- Pereira, F.; Carneiro, J.; Matthiesen, R.; van Asch, B.; Pinto, N.; Gusmao, L.; Amorim, A. Identification of species by multiplex analysis of variable-length sequences. Nucleic Acids Res. 2010, 38, e203. [Google Scholar] [CrossRef] [PubMed]
- Pun, K.M.; Albrecht, C.; Castella, V.; Fumagalli, L. Species identification in mammals from mixed biological samples based on mitochondrial DNA control region length polymorphism. Electrophoresis 2009, 30, 1008–1014. [Google Scholar] [CrossRef]
- Vankova, L.; Vanek, D. DNA-based identification of big cats and traditional Chinese medicine artifacts in the Czech Republic. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 122–124. [Google Scholar] [CrossRef]
- Warchol, G.L. The transnational illegal wildlife trade. In Transnational Environmental Crime; Routledge: London, UK, 2017; pp. 379–396. [Google Scholar]
- Petrossian, G.A.; Pires, S.F.; van Uhm, D.P. An overview of seized illegal wildlife entering the United States. Glob. Crime 2016, 17, 181–201. [Google Scholar] [CrossRef]
- Bagatharia, S.B.; Joshi, M.N.; Pandya, R.V.; Pandit, A.S.; Patel, R.P.; Desai, S.M.; Sharma, A.; Panchal, O.; Jasmani, F.P.; Saxena, A.K. Complete mitogenome of asiatic lion resolves phylogenetic status within Panthera. BMC Genom. 2013, 14, 572. [Google Scholar] [CrossRef]
- Lopez, J.V.; Cevario, S.; O’Brien, S.J. Complete nucleotide sequences of the domestic cat (Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics 1996, 33, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Hewitt, G.M. Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol. Evol. 1996, 11, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Shankaranarayanan, P.; Banerjee, M.; Kacker, R.K.; Aggarwal, R.K.; Singh, L. Genetic variation in Asiatic lions and Indian tigers. Electrophoresis 1997, 18, 1693–1700. [Google Scholar] [CrossRef]
- Webster, L.M.; Prigge, T.-L.; Frankham, G.J. A guide for the validation of DNA based species identification in forensic casework. Forensic Sci. Int. Anim. Environ. 2024, 5, 100080. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef]
- Mori, C.; Matsumura, S. Current issues for mammalian species identification in forensic science: A review. Int. J. Leg. Med. 2021, 135, 3–12. [Google Scholar] [CrossRef]
- Egeland, T.; Salas, A. A statistical framework for the interpretation of mtDNA mixtures: Forensic and medical applications. PLoS ONE 2011, 6, e26723. [Google Scholar] [CrossRef]
- Mandape, S.N.; Smart, U.; King, J.L.; Muenzler, M.; Kapema, K.B.; Budowle, B.; Woerner, A.E. MMDIT: A tool for the deconvolution and interpretation of mitochondrial DNA mixtures. Forensic Sci. Int. Genet. 2021, 55, 102568. [Google Scholar] [CrossRef]
- Holland, M.M.; McQuillan, M.R.; O’Hanlon, K.A. Second generation sequencing allows for mtDNA mixture deconvolution and high resolution detection of heteroplasmy. Croat. Med. J. 2011, 52, 299–313. [Google Scholar] [CrossRef]
- Kim, H.; Erlich, H.A.; Calloway, C.D. Analysis of mixtures using next generation sequencing of mitochondrial DNA hypervariable regions. Croat. Med. J. 2015, 56, 208–217. [Google Scholar] [CrossRef]
- Churchill, J.D.; Stoljarova, M.; King, J.L.; Budowle, B. Massively parallel sequencing-enabled mixture analysis of mitochondrial DNA samples. Int. J. Leg. Med. 2018, 132, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Wisner, M.; Erlich, H.; Shih, S.; Calloway, C. Resolution of mitochondrial DNA mixtures using a probe capture next generation sequencing system and phylogenetic-based software. Forensic Sci. Int. Genet. 2021, 53, 102531. [Google Scholar] [CrossRef]
- Schultz, J.A.; Hebert, P.D. Do pseudogenes pose a problem for metabarcoding marine animal communities? Mol. Ecol. Resour. 2022, 22, 2897–2914. [Google Scholar] [CrossRef]
- Cruaud, P.; Rasplus, J.-Y.; Rodriguez, L.J.; Cruaud, A. High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy. Sci. Rep. 2017, 7, 41948. [Google Scholar] [CrossRef]
- Gross, R.; Nilsson, J.; Schmitz, M. A new species-specific nuclear DNA marker for identification of hybrids between Atlantic salmon and brown trout. J. Fish Biol. 1996, 49, 537–540. [Google Scholar]
- Van de Putte, A.P.; Van Houdt, J.; Maes, G.; Janko, K.; Koubbi, P.; Rock, J.; Volckaert, F. Species identification in the trematomid family using nuclear genetic markers. Polar Biol. 2009, 32, 1731–1741. [Google Scholar] [CrossRef]
- Purugganan, M.; Wessler, S. Transposon signatures: Species-specific molecular markers that utilize a class of multiple-copy nuclear DNA. Mol. Ecol. 1995, 4, 265–270. [Google Scholar] [CrossRef]
- Bosmali, I.; Lagiotis, G.; Stavridou, E.; Haider, N.; Osathanunkul, M.; Pasentsis, K.; Madesis, P. Novel authentication approach for coffee beans and the brewed beverage using a nuclear-based species-specific marker coupled with high resolution melting analysis. LWT 2021, 137, 110336. [Google Scholar] [CrossRef]
- Eberle, J.; Ahrens, D.; Mayer, C.; Niehuis, O.; Misof, B. A plea for standardized nuclear markers in metazoan DNA taxonomy. Trends Ecol. Evol. 2020, 35, 336–345. [Google Scholar] [CrossRef]
- Dietz, L.; Eberle, J.; Mayer, C.; Kukowka, S.; Bohacz, C.; Baur, H.; Espeland, M.; Huber, B.A.; Hutter, C.; Mengual, X. Standardized nuclear markers improve and homogenize species delimitation in Metazoa. Methods Ecol. Evol. 2023, 14, 543–555. [Google Scholar] [CrossRef]
- Pereira, S.L.; Baker, A.J. Low number of mitochondrial pseudogenes in the chicken (Gallus gallus) nuclear genome: Implications for molecular inference of population history and phylogenetics. BMC Evol. Biol. 2004, 4, 17. [Google Scholar] [CrossRef]
- Nacer, D.F.; do Amaral, F.R. Striking pseudogenization in avian phylogenetics: Numts are large and common in falcons. Mol. Phylogenetics Evol. 2017, 115, 1–6. [Google Scholar] [CrossRef]
- Richly, E.; Leister, D. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 2004, 21, 1081–1084. [Google Scholar] [CrossRef]
- Hazkani-Covo, E.; Zeller, R.M.; Martin, W. Molecular poltergeists: Mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010, 6, e1000834. [Google Scholar] [CrossRef]
- Cihlar, J.C.; Strobl, C.; Lagacé, R.; Muenzler, M.; Parson, W.; Budowle, B. Distinguishing mitochondrial DNA and NUMT sequences amplified with the precision ID mtDNA whole genome panel. Mitochondrion 2020, 55, 122–133. [Google Scholar] [CrossRef]
- Soto-Calderón, I.D.; Clark, N.J.; Wildschutte, J.V.H.; DiMattio, K.; Jensen-Seaman, M.I.; Anthony, N.M. Identification of species-specific nuclear insertions of mitochondrial DNA (numts) in gorillas and their potential as population genetic markers. Mol. Phylogenetics Evol. 2014, 81, 61–70. [Google Scholar] [CrossRef]
- Wolff, J.N.; Shearman, D.C.; Brooks, R.C.; Ballard, J.W. Selective enrichment and sequencing of whole mitochondrial genomes in the presence of nuclear encoded mitochondrial pseudogenes (numts). PLoS ONE 2012, 7, e37142. [Google Scholar] [CrossRef]
- Morgan, K.I.; Ewart, K.M.; Nguyen, T.Q.; Sitam, F.T.; Ouitavon, K.; Lightson, A.L.; Kotze, A.; McEwing, R. Avoiding common numts to provide reliable species identification for tiger parts. Forensic Sci. Int. Rep. 2021, 3, 100166. [Google Scholar] [CrossRef]
- Kunz, D.; Tay, W.T.; Elfekih, S.; Gordon, K.H.J.; De Barro, P.J. Take out the rubbish–Removing NUMTs and pseudogenes from the Bemisia tabaci cryptic species mtCOI database. bioRxiv 2019. [Google Scholar] [CrossRef]
- Ožana, S.; Dolný, A.; Pánek, T. Nuclear copies of mitochondrial DNA as a potential problem for phylogenetic and population genetic studies of Odonata. Syst. Entomol. 2022, 47, 591–602. [Google Scholar] [CrossRef]
- Marshall, C.; Parson, W. Interpreting NUMTs in forensic genetics: Seeing the forest for the trees. Forensic Sci. Int. Genet. 2021, 53, 102497. [Google Scholar] [CrossRef] [PubMed]
- Rossel, S.; Uhlenkott, K.; Peters, J.; Vink, A.; Arbizu, P.M. Evaluating species richness using proteomic fingerprinting and DNA barcoding—A case study on meiobenthic copepods from the Clarion Clipperton Fracture Zone. Mar. Biodivers. 2022, 52, 67. [Google Scholar] [CrossRef]
- Raupach, M.J.; Amann, R.; Wheeler, Q.D.; Roos, C. The application of “-omics” technologies for the classification and identification of animals. Org. Divers. Evol. 2016, 16, 1–12. [Google Scholar] [CrossRef]
- Davidson, N.B.; Koch, N.I.; Sarsby, J.; Jones, E.; Hurst, J.L.; Beynon, R.J. Rapid identification of species, sex and maturity by mass spectrometric analysis of animal faeces. BMC Biol. 2019, 17, 66. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, H.; Wei, Q.; Cao, R.; Zhang, H.; He, Y.; Wang, L. Combining DNA barcoding and HPLC fingerprints to trace species of an important traditional Chinese medicine fritillariae bulbus. Molecules 2019, 24, 3269. [Google Scholar] [CrossRef]




| Species | No. of Samples Tested | Sample Type (Number) |
|---|---|---|
| Panthera leo | 30 | hair (15), blood (15) |
| Panthera tigris | 30 | hair (15), tissue (5), blood (5), excrement (5) |
| Panthera onca | 2 | hair (2) |
| Panthera pardus | 10 | hair (7), blood (3) |
| Panthera uncia | 3 | hair (3) |
| Tigon (P. tigris x P. leo) | 2 | hair (2) |
| Leptailurus serval | 4 | hair (4) |
| Lynx lynx | 4 | hair (2), excrement (2) |
| Felis catus | 2 | buccal swab (2) |
| Homo sapiens | 5 | buccal swab (5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vankova, L.; Vanek, D. Capillary-Electrophoresis-Based Species Barcoding of Big Cats: CR-mtDNA-Length Polymorphism. Life 2024, 14, 497. https://doi.org/10.3390/life14040497
Vankova L, Vanek D. Capillary-Electrophoresis-Based Species Barcoding of Big Cats: CR-mtDNA-Length Polymorphism. Life. 2024; 14(4):497. https://doi.org/10.3390/life14040497
Chicago/Turabian StyleVankova, Lenka, and Daniel Vanek. 2024. "Capillary-Electrophoresis-Based Species Barcoding of Big Cats: CR-mtDNA-Length Polymorphism" Life 14, no. 4: 497. https://doi.org/10.3390/life14040497





