Artificial Intelligence: A Snapshot of Its Application in Chronic Inflammatory and Autoimmune Skin Diseases
Abstract
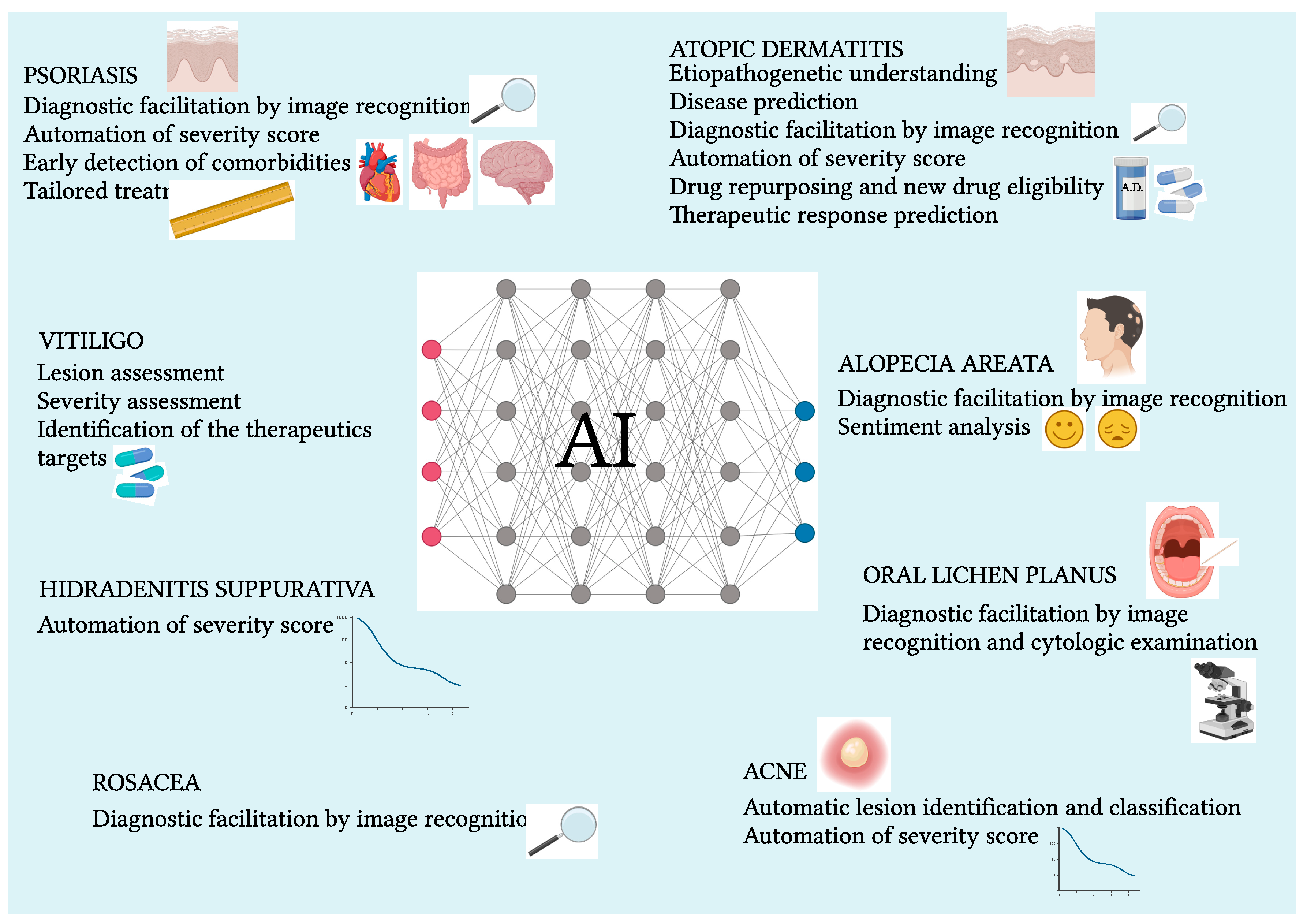
:1. Introduction
2. Results
2.1. Atopic Dermatitis
2.1.1. AI Etiopathogenetic Application
2.1.2. Predictive, Diagnostic, and Classification Performances
2.1.3. A New Concept of AD Severity Scoring
2.1.4. AI in Therapeutic Frontiers in Personalized Medicine
- -
- -
- -
- -
- -
2.2. Psoriasis
2.2.1. Image Analysis
2.2.2. AI-Assisted Severity Scores and Comorbidities
2.2.3. AI-Based Therapies and Efficacy Prediction
- -
- -
- -
- -
2.3. Alopecia Areata
2.4. Vitiligo
- -
- -
- A multi-target strategy for vitiligo assessment is an object of study [92]
2.5. Hidradenitis Suppurativa
- -
- AIHS4 can evaluate the severity of HS like expert clinicians, indicating its potential integration into CAD systems [94].
2.6. Acne
- -
- -
- The application of DL techniques, particularly CNNs, has revolutionized the acne severity assessment. Models like AcneNet, utilizing deep residual neural networks, have achieved a remarkable overall accuracy of over 94% [107].
2.7. Rosacea
- -
2.8. Lichen
- -
- -
- Except for immunobullous diseases, clinical applications of AI are being widely investigated for all other major chronic dermatoses.
- -
- Application areas involve the improved understanding of etiopathogenesis, prediction of disease onset, automation of diagnosis and differential diagnostic by image recognition, identification of new biomarkers for diagnostic and prognostic purposes, characterization of pheno-endotypes and subtypes of disease, early identification of comorbidities, automation of disease severity staging, drug repositioning, identification of new drug candidates, and prediction of therapeutic response.
- -
- The dermatoses of major interest so far are atopic dermatitis, psoriasis, and acne.
- -
- Future and challenging AI application goals towards a “precision intelligence” concern the ever-increasing mastery of epigenetic datasets for the unequaled clinical-therapeutic cognitive evolution of discussed dermatoses.
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Borgia, F.; Custurone, P.; Li Pomi, F.; Vaccaro, M.; Alessandrello, C.; Gangemi, S. IL-33 and IL-37: A Possible Axis in Skin and Allergic Diseases. Int. J. Mol. Sci. 2022, 24, 372. [Google Scholar] [CrossRef]
- Zhang, J.; Zhong, F.; He, K.; Ji, M.; Li, S.; Li, C. Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review. Diagnostics 2023, 13, 3506. [Google Scholar] [CrossRef] [PubMed]
- Du, A.X.; Emam, S.; Gniadecki, R. Review of Machine Learning in Predicting Dermatological Outcomes. Front. Med. 2020, 7, 266. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Tonacci, A.; Sciaccotta, R.; Genovese, S.; Musolino, C.; Pioggia, G.; Gangemi, S. Machine Learning and Deep Learning Applications in Multiple Myeloma Diagnosis, Prognosis, and Treatment Selection. Cancers 2022, 14, 606. [Google Scholar] [CrossRef] [PubMed]
- Tartarisco, G.; Tonacci, A.; Minciullo, P.L.; Billeci, L.; Pioggia, G.; Incorvaia, C.; Gangemi, S. The Soft Computing-Based Approach to Investigate Allergic Diseases: A Systematic Review. Clin. Mol. Allergy 2017, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Khoury, P.; Srinivasan, R.; Kakumanu, S.; Ochoa, S.; Keswani, A.; Sparks, R.; Rider, N.L. A Framework for Augmented Intelligence in Allergy and Immunology Practice and Research—A Work Group Report of the AAAAI Health Informatics, Technology, and Education Committee. J. Allergy Clin. Immunol. Pr. 2022, 10, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- MacMath, D.; Chen, M.; Khoury, P. Artificial Intelligence: Exploring the Future of Innovation in Allergy Immunology. Curr. Allergy Asthma Rep. 2023, 23, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Duverdier, A.; Custovic, A.; Tanaka, R.J. Data-driven Research on Eczema: Systematic Characterization of the Field and Recommendations for the Future. Clin. Transl. Allergy 2022, 12, e12170. [Google Scholar] [CrossRef]
- Takahashi, K.; Hayasawa, H.; Tomita, M. A Predictive Model for Affect of Atopic Dermatitis in Infancy by Neural Network and Multiple Logistic Regression. Arerugi 1999, 48, 1222–1229. [Google Scholar]
- Ko, K.I.; Merlet, J.J.; DerGarabedian, B.P.; Zhen, H.; Suzuki-Horiuchi, Y.; Hedberg, M.L.; Hu, E.; Nguyen, A.T.; Prouty, S.; Alawi, F.; et al. NF-ΚB Perturbation Reveals Unique Immunomodulatory Functions in Prx1 + Fibroblasts That Promote Development of Atopic Dermatitis. Sci. Transl. Med. 2022, 14, eabj0324. [Google Scholar] [CrossRef]
- Aggarwal, S.L.P. Data Augmentation in Dermatology Image Recognition Using Machine Learning. Ski. Res. Technol. 2019, 25, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Ding, L.; Sivaprasad, U.; Geh, E.; Biagini Myers, J.; Bernstein, J.A.; Khurana Hershey, G.K.; Mersha, T.B. Multiple Transcriptome Data Analysis Reveals Biologically Relevant Atopic Dermatitis Signature Genes and Pathways. PLoS ONE 2015, 10, e0144316. [Google Scholar] [CrossRef]
- Martínez, B.A.; Shrotri, S.; Kingsmore, K.M.; Bachali, P.; Grammer, A.C.; Lipsky, P.E. Machine Learning Reveals Distinct Gene Signature Profiles in Lesional and Nonlesional Regions of Inflammatory Skin Diseases. Sci. Adv. 2022, 8, eabn4776. [Google Scholar] [CrossRef]
- Borgia, F.; Li Pomi, F.; Vaccaro, M.; Alessandrello, C.; Papa, V.; Gangemi, S. Oxidative Stress and Phototherapy in Atopic Dermatitis: Mechanisms, Role, and Future Perspectives. Biomolecules 2022, 12, 1904. [Google Scholar] [CrossRef]
- Yamamoto-Hanada, K.; Kawakami, E.; Saito-Abe, M.; Sato, M.; Mitsubuchi, H.; Oda, M.; Katoh, T.; Sanefuji, M.; Ohga, S.; Kuwajima, M.; et al. Exploratory Analysis of Plasma Cytokine/Chemokine Levels in 6-Year-Old Children from a Birth Cohort Study. Cytokine 2020, 130, 155051. [Google Scholar] [CrossRef]
- Smith, A.M.; Walsh, J.R.; Long, J.; Davis, C.B.; Henstock, P.; Hodge, M.R.; Maciejewski, M.; Mu, X.J.; Ra, S.; Zhao, S.; et al. Standard Machine Learning Approaches Outperform Deep Representation Learning on Phenotype Prediction from Transcriptomics Data. BMC Bioinform. 2020, 21, 119. [Google Scholar] [CrossRef]
- Berna, R.; Mitra, N.; Hoffstad, O.; Wan, J.; Margolis, D.J. Identifying Phenotypes of Atopic Dermatitis in a Longitudinal United States Cohort Using Unbiased Statistical Clustering. J. Investig. Dermatol. 2020, 140, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Li, J.; Kong, N.; Kim, J.-H.; Kim, B.-S.; Lee, M.-J.; Park, Y.M.; Lee, S.-Y.; Hong, S.-J.; Sul, J.H. Accurate Diagnosis of Atopic Dermatitis by Combining Transcriptome and Microbiota Data with Supervised Machine Learning. Sci. Rep. 2022, 12, 290. [Google Scholar] [CrossRef]
- Dev, K.; Ho, C.J.H.; Bi, R.; Yew, Y.W.; S, D.U.; Attia, A.B.E.; Moothanchery, M.; Guan, S.T.T.; Olivo, M. Machine Learning Assisted Handheld Confocal Raman Micro-Spectroscopy for Identification of Clinically Relevant Atopic Eczema Biomarkers. Sensors 2022, 22, 4674. [Google Scholar] [CrossRef] [PubMed]
- Hogarty, D.T.; Su, J.C.; Phan, K.; Attia, M.; Hossny, M.; Nahavandi, S.; Lenane, P.; Moloney, F.J.; Yazdabadi, A. Artificial Intelligence in Dermatology—Where We Are and the Way to the Future: A Review. Am. J. Clin. Dermatol. 2020, 21, 41–47. [Google Scholar] [CrossRef]
- De, A.; Sarda, A.; Gupta, S.; Das, S. Use of Artificial Intelligence in Dermatology. Indian. J. Dermatol. 2020, 65, 352. [Google Scholar] [CrossRef]
- Hsiao, Y.-P.; Chiu, C.-W.; Lu, C.-W.; Nguyen, H.T.; Tseng, Y.S.; Hsieh, S.-C.; Wang, H.-C. Identification of Skin Lesions by Using Single-Step Multiframe Detector. J. Clin. Med. 2021, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.; Chen, B.; Santiago, R.; Young, J.; Eder, L.; Chan, A.-W.; Pope, E.; Tu, K.; Jaakkimainen, L.; Drucker, A.M. Accuracy of Algorithms to Identify People with Atopic Dermatitis in Ontario Routinely Collected Health Databases. J. Investig. Dermatol. 2021, 141, 1840–1843. [Google Scholar] [CrossRef]
- Guimarães, P.; Batista, A.; Zieger, M.; Kaatz, M.; Koenig, K. Artificial Intelligence in Multiphoton Tomography: Atopic Dermatitis Diagnosis. Sci. Rep. 2020, 10, 7968. [Google Scholar] [CrossRef]
- Wu, H.; Yin, H.; Chen, H.; Sun, M.; Liu, X.; Yu, Y.; Tang, Y.; Long, H.; Zhang, B.; Zhang, J.; et al. A Deep Learning, Image Based Approach for Automated Diagnosis for Inflammatory Skin Diseases. Ann. Transl. Med. 2020, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Czajkowska, J.; Badura, P.; Korzekwa, S.; Płatkowska-Szczerek, A.; Słowińska, M. Deep Learning-Based High-Frequency Ultrasound Skin Image Classification with Multicriteria Model Evaluation. Sensors 2021, 21, 5846. [Google Scholar] [CrossRef]
- Czajkowska, J.; Badura, P.; Korzekwa, S.; Płatkowska-Szczerek, A. Deep Learning Approach to Skin Layers Segmentation in Inflammatory Dermatoses. Ultrasonics 2021, 114, 106412. [Google Scholar] [CrossRef]
- Czajkowska, J.; Badura, P.; Korzekwa, S.; Płatkowska-Szczerek, A. Automated Segmentation of Epidermis in High-Frequency Ultrasound of Pathological Skin Using a Cascade of DeepLab V3+ Networks and Fuzzy Connectedness. Comput. Med. Imaging Graph. 2022, 95, 102023. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wen, H.-J.; Guo, Y.-L.L.; Wei, T.-Y.; Wang, W.-C.; Tsai, S.-F.; Tseng, V.S.; Wang, S.-L.J. Prenatal Exposure to Air Pollutants and Childhood Atopic Dermatitis and Allergic Rhinitis Adopting Machine Learning Approaches: 14-Year Follow-up Birth Cohort Study. Sci. Total Environ. 2021, 777, 145982. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, D.; Xu, Z.; Heidari, A.A.; Chen, H.; Jiang, X.; Liu, Z.; Wang, M.; Zhou, Q.; Xu, S. BSRWPSO-FKNN: A Boosted PSO with Fuzzy K-Nearest Neighbor Classifier for Predicting Atopic Dermatitis Disease. Front. Neuroinform 2023, 16, 1063048. [Google Scholar] [CrossRef]
- Yanagisawa, Y.; Shido, K.; Kojima, K.; Yamasaki, K. Convolutional Neural Network-Based Skin Image Segmentation Model to Improve Classification of Skin Diseases in Conventional and Non-Standardized Picture Images. J. Dermatol. Sci. 2023, 109, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Chen, G.; Zhang, Z.; He, M.; Li, H.; Yan, F. Construction and Verification of Atopic Dermatitis Diagnostic Model Based on Pyroptosis Related Biological Markers Using Machine Learning Methods. BMC Med. Genom. 2023, 16, 138. [Google Scholar] [CrossRef] [PubMed]
- Proper, S.P.; Azouz, N.P.; Mersha, T.B. Achieving Precision Medicine in Allergic Disease: Progress and Challenges. Front. Immunol. 2021, 12, 720746. [Google Scholar] [CrossRef] [PubMed]
- Maintz, L.; Welchowski, T.; Herrmann, N.; Brauer, J.; Kläschen, A.S.; Fimmers, R.; Schmid, M.; Bieber, T.; Schmid-Grendelmeier, P.; Traidl-Hoffmann, C.; et al. Machine Learning–Based Deep Phenotyping of Atopic Dermatitis. JAMA Dermatol. 2021, 157, 1414. [Google Scholar] [CrossRef] [PubMed]
- Hurault, G.; Domínguez-Hüttinger, E.; Langan, S.M.; Williams, H.C.; Tanaka, R.J. Personalized Prediction of Daily Eczema Severity Scores Using a Mechanistic Machine Learning Model. Clin. Exp. Allergy 2020, 50, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, D.A.; Feizpour, A.; Evans, C.L. Quantifying Inflammatory Response and Drug-Aided Resolution in an Atopic Dermatitis Model with Deep Learning. J. Investig. Dermatol. 2023, 143, 1430–1438.e4. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.J.H.; Yew, Y.W.; Dinish, U.S.; Kuan, A.H.Y.; Wong, M.K.W.; Bi, R.; Dev, K.; Li, X.; Singh, G.; Moothanchery, M.; et al. Handheld Confocal Raman Spectroscopy (CRS) for Objective Assessment of Skin Barrier Function and Stratification of Severity in Atopic Dermatitis (AD) Patients. J. Dermatol. Sci. 2020, 98, 20–25. [Google Scholar] [CrossRef]
- Bang, C.H.; Yoon, J.W.; Ryu, J.Y.; Chun, J.H.; Han, J.H.; Lee, Y.B.; Lee, J.Y.; Park, Y.M.; Lee, S.J.; Lee, J.H. Automated Severity Scoring of Atopic Dermatitis Patients by a Deep Neural Network. Sci. Rep. 2021, 11, 6049. [Google Scholar] [CrossRef] [PubMed]
- Medela, A.; Mac Carthy, T.; Aguilar Robles, S.A.; Chiesa-Estomba, C.M.; Grimalt, R. Automatic SCOring of Atopic Dermatitis Using Deep Learning: A Pilot Study. JID Innov. 2022, 2, 100107. [Google Scholar] [CrossRef]
- Park, S.; Saw, S.N.; Li, X.; Paknezhad, M.; Coppola, D.; Dinish, U.S.; Ebrahim Attia, A.B.; Yew, Y.W.; Guan Thng, S.T.; Lee, H.K.; et al. Model Learning Analysis of 3D Optoacoustic Mesoscopy Images for the Classification of Atopic Dermatitis. Biomed. Opt. Express 2021, 12, 3671. [Google Scholar] [CrossRef]
- Patella, V.; Florio, G.; Palmieri, M.; Bousquet, J.; Tonacci, A.; Giuliano, A.; Gangemi, S. Atopic Dermatitis Severity during Exposure to Air Pollutants and Weather Changes with an Artificial Neural Network (ANN) Analysis. Pediatr. Allergy Immunol. 2020, 31, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.I.; Lee, D.; Han, B.; Lee, J.S.; Hong, J.Y.; Chung, J.H.; Lee, D.H.; Na, J.-I. Practical Training Approaches for Discordant Atopic Dermatitis Severity Datasets: Merging Methods With Soft-Label and Train-Set Pruning. IEEE J. Biomed. Health Inf. 2023, 27, 166–175. [Google Scholar] [CrossRef]
- Patrick, M.T.; Raja, K.; Miller, K.; Sotzen, J.; Gudjonsson, J.E.; Elder, J.T.; Tsoi, L.C. Drug Repurposing Prediction for Immune-Mediated Cutaneous Diseases Using a Word-Embedding–Based Machine Learning Approach. J. Investig. Dermatol. 2019, 139, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Spinazze, P.; Bottle, A.; Car, J. Digital Health Sensing for Personalized Dermatology. Sensors 2019, 19, 3426. [Google Scholar] [CrossRef]
- Moreau, A.; Anderer, P.; Ross, M.; Cerny, A.; Almazan, T.H.; Peterson, B. Detection of Nocturnal Scratching Movements in Patients with Atopic Dermatitis Using Accelerometers and Recurrent Neural Networks. IEEE J. Biomed. Health Inf. 2018, 22, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Matsushita, S.; Shimizu, N.; Masuko, S.; Yamamoto, M.; Murata, T. Automated Detection of Mouse Scratching Behaviour Using Convolutional Recurrent Neural Network. Sci. Rep. 2021, 11, 658. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qin, D.; Jin, L.; Liang, G. Caffeoyl Malic Acid Is a Potential Dual Inhibitor Targeting TNFα/IL-4 Evaluated by a Combination Strategy of Network Analysis-Deep Learning-Molecular Simulation. Comput. Biol. Med. 2022, 145, 105410. [Google Scholar] [CrossRef] [PubMed]
- Clayton, K.; Vallejo, A.; Sirvent, S.; Davies, J.; Porter, G.; Reading, I.C.; Lim, F.; Ardern-Jones, M.R.; Polak, M.E. Machine Learning Applied to Atopic Dermatitis Transcriptome Reveals Distinct Therapy-dependent Modification of the Keratinocyte Immunophenotype*. Br. J. Dermatol. 2021, 184, 913–922. [Google Scholar] [CrossRef]
- Miyano, T.; Tanaka, R.J. Identification of Keratinocyte Subpopulations in Transcriptome to Evaluate Drug Effects in Atopic Dermatitis. Br. J. Dermatol. 2021, 184, 798–799. [Google Scholar] [CrossRef]
- Wu, J.J.; Hong, C.; Merola, J.F.; Gruben, D.; Güler, E.; Feeney, C.; Bhambri, A.; Myers, D.E.; DiBonaventura, M. Predictors of Nonresponse to Dupilumab in Patients with Atopic Dermatitis. Ann. Allergy Asthma Immunol. 2022, 129, 354–359.e5. [Google Scholar] [CrossRef]
- McMullen, E.P.; Syed, S.A.; Espiritu, K.D.; Grewal, R.S.; Elder, G.A.; Morita, P.P.; Drucker, A.M. The Therapeutic Applications of Machine Learning in Atopic Dermatitis: A Scoping Review. J. Cutan. Med. Surg. 2023, 27, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, C.; Uki, S.; Kaitoh, K.; Iwata, M.; Yamanishi, Y. De Novo Drug Design Based on Patient Gene Expression Profiles via Deep Learning. Mol. Inf. 2023, 42, 2300064. [Google Scholar] [CrossRef] [PubMed]
- Zvulunov, A.; Lenevich, S.; Migacheva, N. A Mobile Health App for Facilitating Disease Management in Children with Atopic Dermatitis: Feasibility and Impact Study. JMIR Dermatol. 2023, 6, e49278. [Google Scholar] [CrossRef] [PubMed]
- Lakdawala, N.; Channa, L.; Gronbeck, C.; Lakdawala, N.; Weston, G.; Sloan, B.; Feng, H. Assessing the Accuracy and Comprehensiveness of ChatGPT in Offering Clinical Guidance for Atopic Dermatitis and Acne Vulgaris. JMIR Dermatol. 2023, 6, e50409. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, K.; DeSena, G.; Laageide, L.; El-Feghaly, J.; Lio, P. From Eczema to Anxiety: How Artificial Intelligence Shapes Parental Perspectives. Pediatr. Dermatol. 2023, 40, 964–965. [Google Scholar] [CrossRef] [PubMed]
- Papa, V.; Li Pomi, F.; Borgia, F.; Genovese, S.; Pioggia, G.; Gangemi, S. “Mens Sana in Cute Sana”—A State of the Art of Mutual Etiopathogenetic Influence and Relevant Pathophysiological Pathways between Skin and Mental Disorders: An Integrated Approach to Contemporary Psychopathological Scenarios. Cells 2023, 12, 1828. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, V.K.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. Computer-Aided Diagnosis of Psoriasis Skin Images with HOS, Texture and Color Features: A First Comparative Study of Its Kind. Comput. Methods Programs Biomed. 2016, 126, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, V.K.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. Reliability Analysis of Psoriasis Decision Support System in Principal Component Analysis Framework. Data Knowl. Eng. 2016, 106, 1–17. [Google Scholar] [CrossRef]
- Shrivastava, V.K.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. Reliable and Accurate Psoriasis Disease Classification in Dermatology Images Using Comprehensive Feature Space in Machine Learning Paradigm. Expert. Syst. Appl. 2015, 42, 6184–6195. [Google Scholar] [CrossRef]
- Zhao, S.; Xie, B.; Li, Y.; Zhao, X.; Kuang, Y.; Su, J.; He, X.; Wu, X.; Fan, W.; Huang, K.; et al. Smart Identification of Psoriasis by Images Using Convolutional Neural Networks: A Case Study in China. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 518–524. [Google Scholar] [CrossRef]
- Verma, A.K.; Pal, S.; Kumar, S. Classification of Skin Disease Using Ensemble Data Mining Techniques. Asian Pac. J. Cancer Prev. 2019, 20, 1887–1894. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, J.; Hwang, M.; Kim, M.; Jin Jo, S.; Je, M.; Jang, J.E.; Lee, D.H.; Hwang, J.Y. Smartphone-Based Multispectral Imaging and Machine-Learning Based Analysis for Discrimination between Seborrheic Dermatitis and Psoriasis on the Scalp. Biomed. Opt. Express 2019, 10, 879. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Syed, M.N.; Bernardis, E.; Gelfand, J.M. Machine Learning Applications in the Evaluation and Management of Psoriasis: A Systematic Review. J. Psoriasis Psoriatic Arthritis 2020, 5, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, Y.; Zhou, R.; Liu, W. Three-Dimensional Skin CT Based on Intelligent Algorithm in the Analysis of Skin Lesion Sites Features in Children with Psoriasis. Comput. Math. Methods Med. 2022, 2022, 8195243. [Google Scholar] [CrossRef] [PubMed]
- George, Y.; Aldeen, M.; Garnavi, R. Psoriasis Image Representation Using Patch-Based Dictionary Learning for Erythema Severity Scoring. Comput. Med. Imaging Graph. 2018, 66, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Raina, A.; Hennessy, R.; Rains, M.; Allred, J.; Hirshburg, J.M.; Diven, D.G.; Markey, M.K. Objective Measurement of Erythema in Psoriasis Using Digital Color Photography with Color Calibration. Ski. Res. Technol. 2016, 22, 375–380. [Google Scholar] [CrossRef] [PubMed]
- George, Y.; Aldeen, M.; Garnavi, R. Automatic Scale Severity Assessment Method in Psoriasis Skin Images Using Local Descriptors. IEEE J. Biomed. Health Inf. 2020, 24, 577–585. [Google Scholar] [CrossRef]
- Ahmad Fadzil, M.H.; Prakasa, E.; Asirvadam, V.S.; Nugroho, H.; Affandi, A.M.; Hussein, S.H. 3D Surface Roughness Measurement for Scaliness Scoring of Psoriasis Lesions. Comput. Biol. Med. 2013, 43, 1987–2000. [Google Scholar] [CrossRef] [PubMed]
- Meienberger, N.; Anzengruber, F.; Amruthalingam, L.; Christen, R.; Koller, T.; Maul, J.T.; Pouly, M.; Djamei, V.; Navarini, A.A. Observer-independent Assessment of Psoriasis-affected Area Using Machine Learning. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1362–1368. [Google Scholar] [CrossRef]
- Fink, C.; Fuchs, T.; Enk, A.; Haenssle, H.A. Design of an Algorithm for Automated, Computer-Guided PASI Measurements by Digital Image Analysis. J. Med. Syst. 2018, 42, 248. [Google Scholar] [CrossRef]
- Karampinis, E.; Papadopoulou, M.-M.; Chaidaki, K.; Georgopoulou, K.-E.; Magaliou, S.; Roussaki Schulze, A.V.; Bogdanos, D.P.; Zafiriou, E. Plaque Psoriasis Exacerbation and COVID-19 Vaccination: Assessing the Characteristics of the Flare and the Exposome Parameters. Vaccines 2024, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.T.; Stuart, P.E.; Raja, K.; Gudjonsson, J.E.; Tejasvi, T.; Yang, J.; Chandran, V.; Das, S.; Callis-Duffin, K.; Ellinghaus, E.; et al. Genetic Signature to Provide Robust Risk Assessment of Psoriatic Arthritis Development in Psoriasis Patients. Nat. Commun. 2018, 9, 4178. [Google Scholar] [CrossRef] [PubMed]
- Love, T.J.; Cai, T.; Karlson, E.W. Validation of Psoriatic Arthritis Diagnoses in Electronic Medical Records Using Natural Language Processing. Semin. Arthritis Rheum. 2011, 40, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Queiro, R.; Seoane-Mato, D.; Laiz, A.; Agirregoikoa, E.G.; Montilla, C.; Park, H.-S.; Pinto-Tasende, J.A.; Bethencourt Baute, J.J.; Ibáñez, B.J.; Toniolo, E.; et al. Minimal Disease Activity (MDA) in Patients with Recent-Onset Psoriatic Arthritis: Predictive Model Based on Machine Learning. Arthritis Res. Ther. 2022, 24, 153. [Google Scholar] [CrossRef] [PubMed]
- Munger, E.; Choi, H.; Dey, A.K.; Elnabawi, Y.A.; Groenendyk, J.W.; Rodante, J.; Keel, A.; Aksentijevich, M.; Reddy, A.S.; Khalil, N.; et al. Application of Machine Learning to Determine Top Predictors of Noncalcified Coronary Burden in Psoriasis: An Observational Cohort Study. J. Am. Acad. Dermatol. 2020, 83, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Florek, A.G.; Wang, C.J.; Armstrong, A.W. Treatment Preferences and Treatment Satisfaction among Psoriasis Patients: A Systematic Review. Arch. Dermatol. Res. 2018, 310, 271–319. [Google Scholar] [CrossRef] [PubMed]
- Tomalin, L.E.; Kim, J.; Correa da Rosa, J.; Lee, J.; Fitz, L.J.; Berstein, G.; Valdez, H.; Wolk, R.; Krueger, J.G.; Suárez-Fariñas, M. Early Quantification of Systemic Inflammatory Proteins Predicts Long-Term Treatment Response to Tofacitinib and Etanercept. J. Investig. Dermatol. 2020, 140, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Damiani, G.; Conic, R.; Pigatto, P.; Carrera, C.; Franchi, C.; Cattaneo, A.; Malagoli, P.; Uppala, R.; Linder, D.; Bragazzi, N.; et al. Predicting Secukinumab Fast-Responder Profile in Psoriatic Patients: Advanced Application of Artificial-Neural-Networks (ANNs). J. Drugs Dermatol. 2020, 19, 1241–1246. [Google Scholar] [CrossRef]
- Gottlieb, A.B.; Mease, P.J.; Kirkham, B.; Nash, P.; Balsa, A.C.; Combe, B.; Rech, J.; Zhu, X.; James, D.; Martin, R.; et al. Secukinumab Efficacy in Psoriatic Arthritis. JCR J. Clin. Rheumatol. 2021, 27, 239–247. [Google Scholar] [CrossRef]
- Pournara, E.; Kormaksson, M.; Nash, P.; Ritchlin, C.T.; Kirkham, B.W.; Ligozio, G.; Pricop, L.; Ogdie, A.; Coates, L.C.; Schett, G.; et al. Clinically Relevant Patient Clusters Identified by Machine Learning from the Clinical Development Programme of Secukinumab in Psoriatic Arthritis. RMD Open 2021, 7, e001845. [Google Scholar] [CrossRef]
- Zhu, C.-Y.; Wang, Y.-K.; Chen, H.-P.; Gao, K.-L.; Shu, C.; Wang, J.-C.; Yan, L.-F.; Yang, Y.-G.; Xie, F.-Y.; Liu, J. A Deep Learning Based Framework for Diagnosing Multiple Skin Diseases in a Clinical Environment. Front. Med. 2021, 8, 626369. [Google Scholar] [CrossRef] [PubMed]
- Motolese, A.; Ceccarelli, M.; Macca, L.; Li Pomi, F.; Ingrasciotta, Y.; Nunnari, G.; Guarneri, C. Novel Therapeutic Approaches to Psoriasis and Risk of Infectious Disease. Biomedicines 2022, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, S.; Okada, Y. GREP: Genome for REPositioning Drugs. Bioinformatics 2019, 35, 3821–3823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Nie, Y. Prediction of the Risk of Alopecia Areata Progressing to Alopecia Totalis and Alopecia Universalis: Biomarker Development with Bioinformatics Analysis and Machine Learning. Dermatology 2022, 238, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, C.S.; Khan, S.J.; Chaudhry, B.; Aijaz, S.F.; Hassan, U. Classification Framework for Healthy Hairs and Alopecia Areata: A Machine Learning (ML) Approach. Comput. Math. Methods Med. 2021, 2021, 1102083. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J.W.; Choe, S.J.; Yang, S.; Koh, S.B.; Ahn, Y.S.; Lee, W.-S. Clinically Applicable Deep Learning Framework for Measurement of the Extent of Hair Loss in Patients with Alopecia Areata. JAMA Dermatol. 2020, 156, 1018. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Dai, Z.; Christiano, A.M. Regulatory Network Analysis Defines Unique Drug Mechanisms of Action and Facilitates Patient-Drug Matching in Alopecia Areata Clinical Trials. Comput. Struct. Biotechnol. J. 2021, 19, 4751–4758. [Google Scholar] [CrossRef] [PubMed]
- Böhm, M.; Schunter, J.A.; Fritz, K.; Salavastru, C.; Dargatz, S.; Augustin, M.; Tanew, A. S1 Guideline: Diagnosis and Therapy of Vitiligo. JDDG J. Der Dtsch. Dermatol. Ges. 2022, 20, 365–378. [Google Scholar] [CrossRef]
- Hillmer, D.; Merhi, R.; Boniface, K.; Taieb, A.; Barnetche, T.; Seneschal, J.; Hagedorn, M. Evaluation of Facial Vitiligo Severity with a Mixed Clinical and Artificial Intelligence Approach. J. Investig. Dermatol. 2024, 144, 351–357.e4. [Google Scholar] [CrossRef]
- Guo, L.; Yang, Y.; Ding, H.; Zheng, H.; Yang, H.; Xie, J.; Li, Y.; Lin, T.; Ge, Y. A Deep Learning-Based Hybrid Artificial Intelligence Model for the Detection and Severity Assessment of Vitiligo Lesions. Ann. Transl. Med. 2022, 10, 590. [Google Scholar] [CrossRef]
- Li Pomi, F.; Papa, V.; Borgia, F.; Vaccaro, M.; Allegra, A.; Cicero, N.; Gangemi, S. Rosmarinus Officinalis and Skin: Antioxidant Activity and Possible Therapeutical Role in Cutaneous Diseases. Antioxidants 2023, 12, 680. [Google Scholar] [CrossRef]
- Wang, J.; Luo, L.; Ding, Q.; Wu, Z.; Peng, Y.; Li, J.; Wang, X.; Li, W.; Liu, G.; Zhang, B.; et al. Development of a Multi-Target Strategy for the Treatment of Vitiligo via Machine Learning and Network Analysis Methods. Front. Pharmacol. 2021, 12, 754175. [Google Scholar] [CrossRef] [PubMed]
- Li Pomi, F.; Macca, L.; Motolese, A.; Ingrasciotta, Y.; Berretta, M.; Guarneri, C. Neoplastic Implications in Patients Suffering from Hidradenitis Suppurativa under Systemic Treatments. Biomedicines 2021, 9, 1594. [Google Scholar] [CrossRef]
- Hernández Montilla, I.; Medela, A.; Mac Carthy, T.; Aguilar, A.; Gómez Tejerina, P.; Vilas Sueiro, A.; González Pérez, A.M.; Vergara de la Campa, L.; Luna Bastante, L.; García Castro, R.; et al. Automatic International Hidradenitis Suppurativa Severity Score System (AIHS4): A Novel Tool to Assess the Severity of Hidradenitis Suppurativa Using Artificial Intelligence. Ski. Res. Technol. 2023, 29, e13357. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, D. Advancing Dermatological Care: A Comprehensive Narrative Review of Tele-Dermatology and MHealth for Bridging Gaps and Expanding Opportunities beyond the COVID-19 Pandemic. Healthcare 2023, 11, 1911. [Google Scholar] [CrossRef] [PubMed]
- Martora, F.; Fabbrocini, G.; Megna, M.; Scalvenzi, M.; Battista, T.; Villani, A.; Potestio, L. Teledermatology for Common Inflammatory Skin Conditions: The Medicine of the Future? Life 2023, 13, 1037. [Google Scholar] [CrossRef]
- Borgia, F.; Li Pomi, F.; Alessandrello, C.; Gangemi, S. Comment on Marasca et al. Teledermatology and Inflammatory Skin Conditions during COVID-19 Era: New Perspectives and Applications. J. Clin. Med. 2022, 11, 1511. J. Clin. Med. 2022, 11, 4063. [Google Scholar] [CrossRef]
- Jusuf, N.K.; Putra, I.B.; Puteri Rangkuti, A.D. Assessing Acne Severity: Teledermatology Versus Face to Face Consultations during the COVID-19 Pandemic. J. Clin. Aesthet. Dermatol. 2023, 16, 30–34. [Google Scholar]
- Li, A.; Fang, R.; Sun, Q. Artificial Intelligence for Grading in Acne Vulgaris: Current Situation and Prospect. J. Cosmet. Dermatol. 2022, 21, 865–866. [Google Scholar] [CrossRef]
- Li, J.; Du, D.; Zhang, J.; Liu, W.; Wang, J.; Wei, X.; Xue, L.; Li, X.; Diao, P.; Zhang, L.; et al. Development and Validation of an Artificial Intelligence-Powered Acne Grading System Incorporating Lesion Identification. Front. Med. 2023, 10, 1255704. [Google Scholar] [CrossRef]
- Min, S.; Kong, H.; Yoon, C.; Kim, H.C.; Suh, D.H. Development and Evaluation of an Automatic Acne Lesion Detection Program Using Digital Image Processing. Ski. Res. Technol. 2013, 19, e423–e432. [Google Scholar] [CrossRef] [PubMed]
- Melina, A.; Dinh, N.N.; Tafuri, B.; Schipani, G.; Nisticò, S.; Cosentino, C.; Amato, F.; Thiboutot, D.; Cherubini, A. Artificial Intelligence for the Objective Evaluation of Acne Investigator Global Assessment. J. Drugs Dermatol. 2018, 17, 1006–1009. [Google Scholar] [PubMed]
- Lim, Z.V.; Akram, F.; Ngo, C.P.; Winarto, A.A.; Lee, W.Q.; Liang, K.; Oon, H.H.; Thng, S.T.G.; Lee, H.K. Automated Grading of Acne Vulgaris by Deep Learning with Convolutional Neural Networks. Ski. Res. Technol. 2020, 26, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Alamdari, N.; Tavakolian, K.; Alhashim, M.; Fazel-Rezai, R. Detection and Classification of Acne Lesions in Acne Patients: A Mobile Application. In Proceedings of the 2016 IEEE International Conference on Electro Information Technology (EIT), Grand Forks, ND, USA, 19–21 May 2016; pp. 0739–0743. [Google Scholar]
- Maroni, G.; Ermidoro, M.; Previdi, F.; Bigini, G. Automated Detection, Extraction and Counting of Acne Lesions for Automatic Evaluation and Tracking of Acne Severity. In Proceedings of the 2017 IEEE Symposium Series on Computational Intelligence (SSCI), Honolulu, HI, USA, 27 November–1 December 2017; pp. 1–6. [Google Scholar]
- Shen, X.; Zhang, J.; Yan, C.; Zhou, H. An Automatic Diagnosis Method of Facial Acne Vulgaris Based on Convolutional Neural Network. Sci. Rep. 2018, 8, 5839. [Google Scholar] [CrossRef] [PubMed]
- Junayed, M.S.; Jeny, A.A.; Atik, S.T.; Neehal, N.; Karim, A.; Azam, S.; Shanmugam, B. AcneNet—A Deep CNN Based Classification Approach for Acne Classes. In Proceedings of the 2019 12th International Conference on Information & Communication Technology and System (ICTS), Surabaya, Indonesia, 18 July 2019; pp. 203–208. [Google Scholar]
- Seité, S.; Khammari, A.; Benzaquen, M.; Moyal, D.; Dréno, B. Development and Accuracy of an Artificial Intelligence Algorithm for Acne Grading from Smartphone Photographs. Exp. Dermatol. 2019, 28, 1252–1257. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, L.; Wu, Q.; Zhang, M.; Zeng, R.; Ding, H.; Zheng, H.; Xie, J.; Li, Y.; Ge, Y.; et al. Construction and Evaluation of a Deep Learning Model for Assessing Acne Vulgaris Using Clinical Images. Dermatol. Ther. 2021, 11, 1239–1248. [Google Scholar] [CrossRef]
- Liu, S.; Fan, Y.; Duan, M.; Wang, Y.; Su, G.; Ren, Y.; Huang, L.; Zhou, F. AcneGrader: An Ensemble Pruning of the Deep Learning Base Models to Grade Acne. Ski. Res. Technol. 2022, 28, 677–688. [Google Scholar] [CrossRef]
- Binol, H.; Plotner, A.; Sopkovich, J.; Kaffenberger, B.; Niazi, M.K.K.; Gurcan, M.N. Ros-NET: A Deep Convolutional Neural Network for Automatic Identification of Rosacea Lesions. Ski. Res. Technol. 2020, 26, 413–421. [Google Scholar] [CrossRef]
- Zhao, Z.; Wu, C.-M.; Zhang, S.; He, F.; Liu, F.; Wang, B.; Huang, Y.; Shi, W.; Jian, D.; Xie, H.; et al. A Novel Convolutional Neural Network for the Diagnosis and Classification of Rosacea: Usability Study. JMIR Med. Inf. 2021, 9, e23415. [Google Scholar] [CrossRef]
- Bobrova, M.; Taranik, M.; Kopanitsa, G. Using Neural Networks for Diagnosing in Dermatology. Stud. Health Technol. Inf. 2019, 261, 211–216. [Google Scholar]
- Tiwari, A.; Gupta, N.; Singla, D.; Ranjan Swain, J.; Gupta, R.; Mehta, D.; Kumar, S. Artificial Intelligence’s Use in the Diagnosis of Mouth Ulcers: A Systematic Review. Cureus 2023, 15, e45187. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the Accuracy of Human Readers versus Machine-Learning Algorithms for Pigmented Skin Lesion Classification: An Open, Web-Based, International, Diagnostic Study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-H.; Jeon, E.-H.; Lee, J.-Y.; Kim, Y.-S.; Yoon, H.-J.; Hong, S.-P.; Lee, J.-H. The Potential of Interleukin 12 Receptor Beta 2 (IL12RB2) and Tumor Necrosis Factor Receptor Superfamily Member 8 (TNFRSF8) Gene as Diagnostic Biomarkers of Oral Lichen Planus (OLP). Acta Odontol. Scand. 2015, 73, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Kistenev, Y.V.; Borisov, A.V.; Titarenko, M.A.; Baydik, O.D.; Shapovalov, A.V. Diagnosis of Oral Lichen Planus from Analysis of Saliva Samples Using Terahertz Time-Domain Spectroscopy and Chemometrics. J. Biomed. Opt. 2018, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Farah, C.S.; Shearston, K.; Kujan, O. A Machine-learning Algorithm for the Reliable Identification of Oral Lichen Planus. J. Oral. Pathol. Med. 2021, 50, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Achararit, P.; Manaspon, C.; Jongwannasiri, C.; Phattarataratip, E.; Osathanon, T.; Sappayatosok, K. Artificial Intelligence-Based Diagnosis of Oral Lichen Planus Using Deep Convolutional Neural Networks. Eur. J. Dent. 2023, 17, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Keser, G.; Bayrakdar, İ.Ş.; Pekiner, F.N.; Çelik, Ö.; Orhan, K. A Deep Learning Algorithm for Classification of Oral Lichen Planus Lesions from Photographic Images: A Retrospective Study. J. Stomatol. Oral. Maxillofac. Surg. 2023, 124, 101264. [Google Scholar] [CrossRef]
- P, P.J.; Prasad, S.S.; Manohar, N. Genital and Extragenital Lichen Sclerosus et Atrophicus: A Case Series Written Using ChatGPT. Cureus 2023, 15, e38987. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li Pomi, F.; Papa, V.; Borgia, F.; Vaccaro, M.; Pioggia, G.; Gangemi, S. Artificial Intelligence: A Snapshot of Its Application in Chronic Inflammatory and Autoimmune Skin Diseases. Life 2024, 14, 516. https://doi.org/10.3390/life14040516
Li Pomi F, Papa V, Borgia F, Vaccaro M, Pioggia G, Gangemi S. Artificial Intelligence: A Snapshot of Its Application in Chronic Inflammatory and Autoimmune Skin Diseases. Life. 2024; 14(4):516. https://doi.org/10.3390/life14040516
Chicago/Turabian StyleLi Pomi, Federica, Vincenzo Papa, Francesco Borgia, Mario Vaccaro, Giovanni Pioggia, and Sebastiano Gangemi. 2024. "Artificial Intelligence: A Snapshot of Its Application in Chronic Inflammatory and Autoimmune Skin Diseases" Life 14, no. 4: 516. https://doi.org/10.3390/life14040516
APA StyleLi Pomi, F., Papa, V., Borgia, F., Vaccaro, M., Pioggia, G., & Gangemi, S. (2024). Artificial Intelligence: A Snapshot of Its Application in Chronic Inflammatory and Autoimmune Skin Diseases. Life, 14(4), 516. https://doi.org/10.3390/life14040516











