Composition of Proteins Associated with Red Clover (Trifolium pratense) and the Microbiota Identified in Honey
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Honey and Determination of Its Botanical Origin
2.2. Protein Isolation and Preparation for LC–MS
2.3. LC–MSE (DIA)-Based Protein Identification
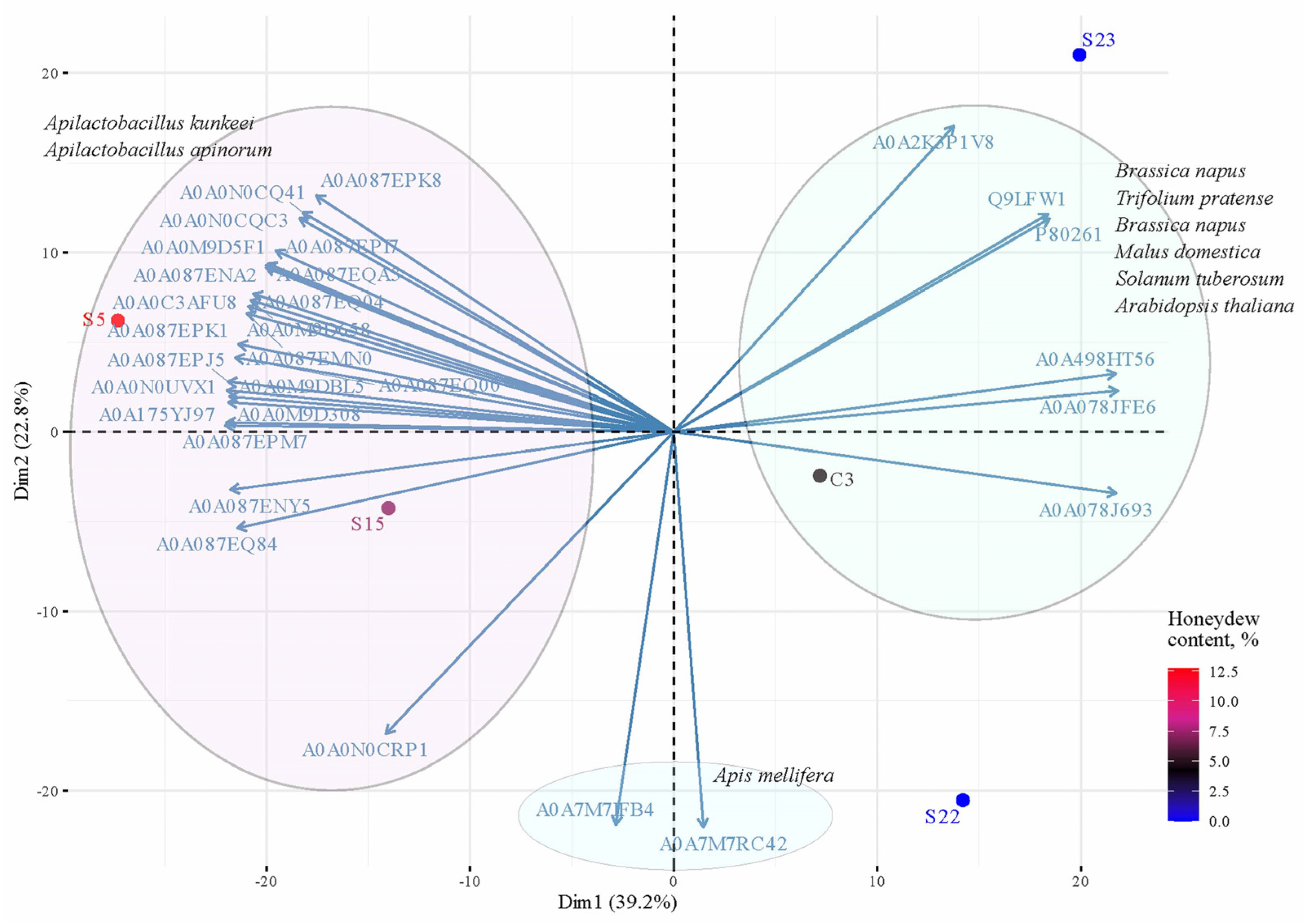
2.4. Statistical Analysis
3. Results
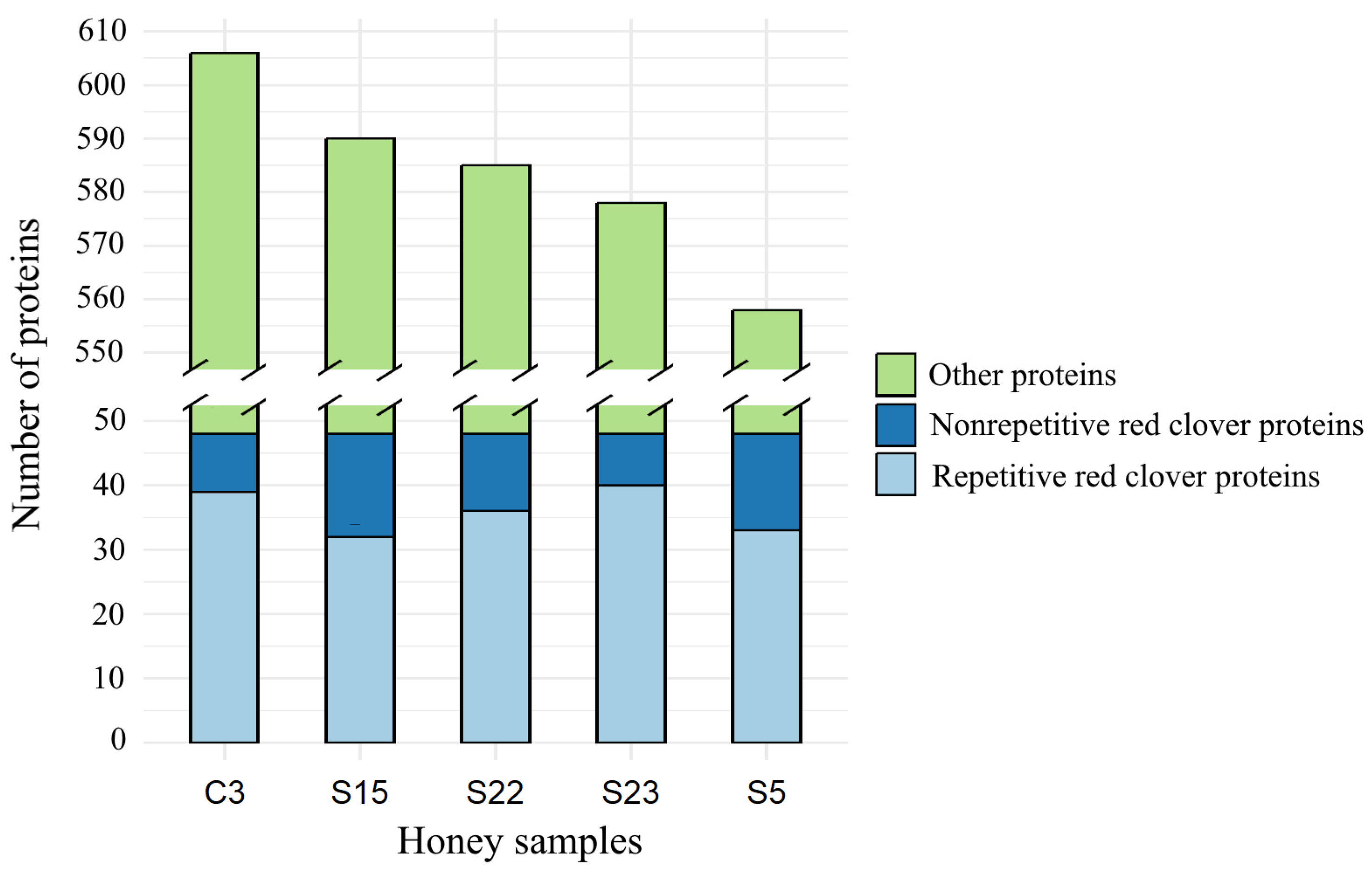
3.1. Comparison of the Protein Number of Monofloral Red Clover Honey with Other Honeys of Different Origins
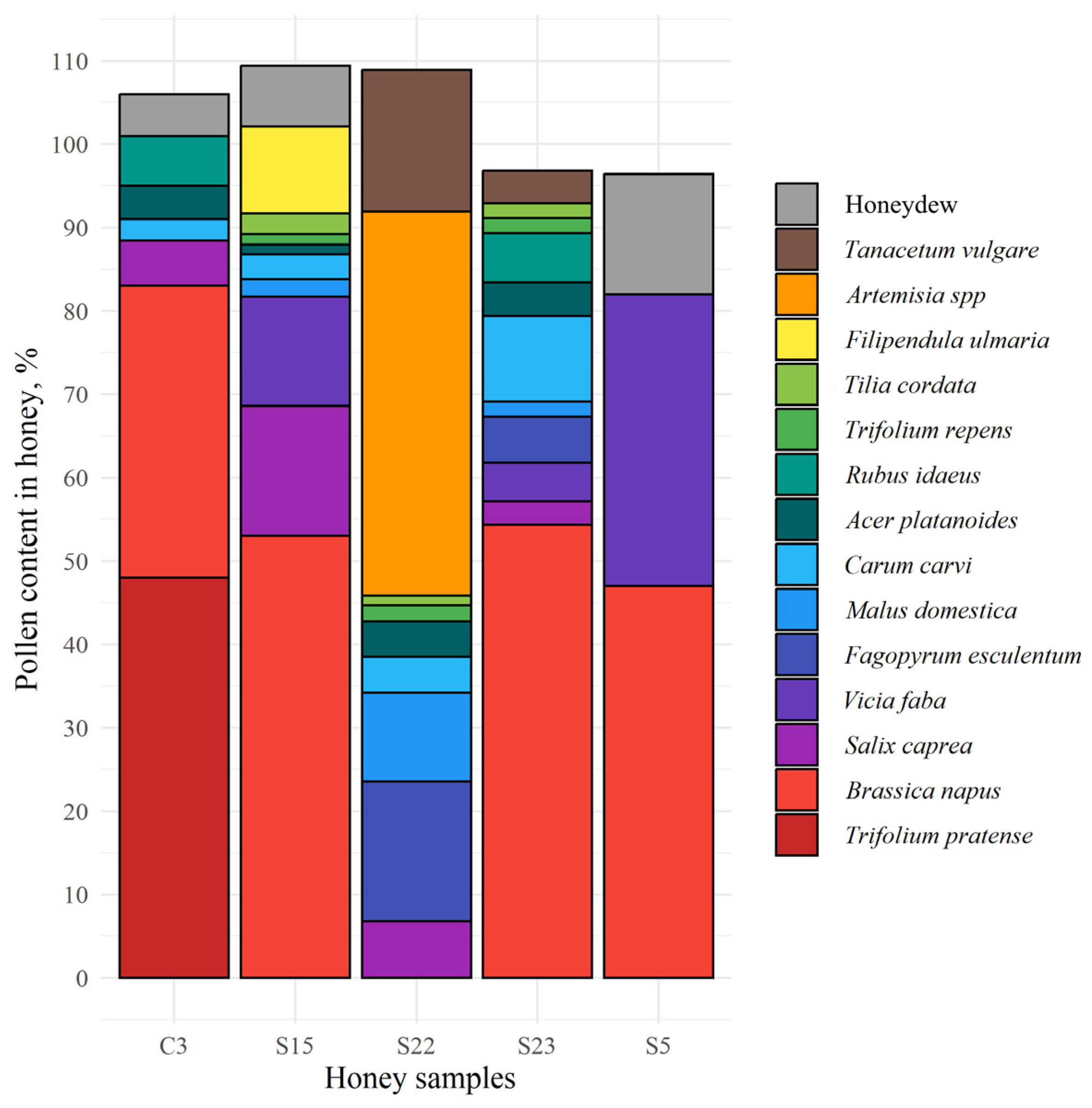
3.2. Pollen Composition of Honey Samples from Different Regions of Lithuania
3.3. Comparison of the Diversity of Plant Proteins Found in Honey Samples
3.4. The Identified Proteins of Aphids and Their Endosymbionts
3.5. Lactic Acid Bacteria in Honey
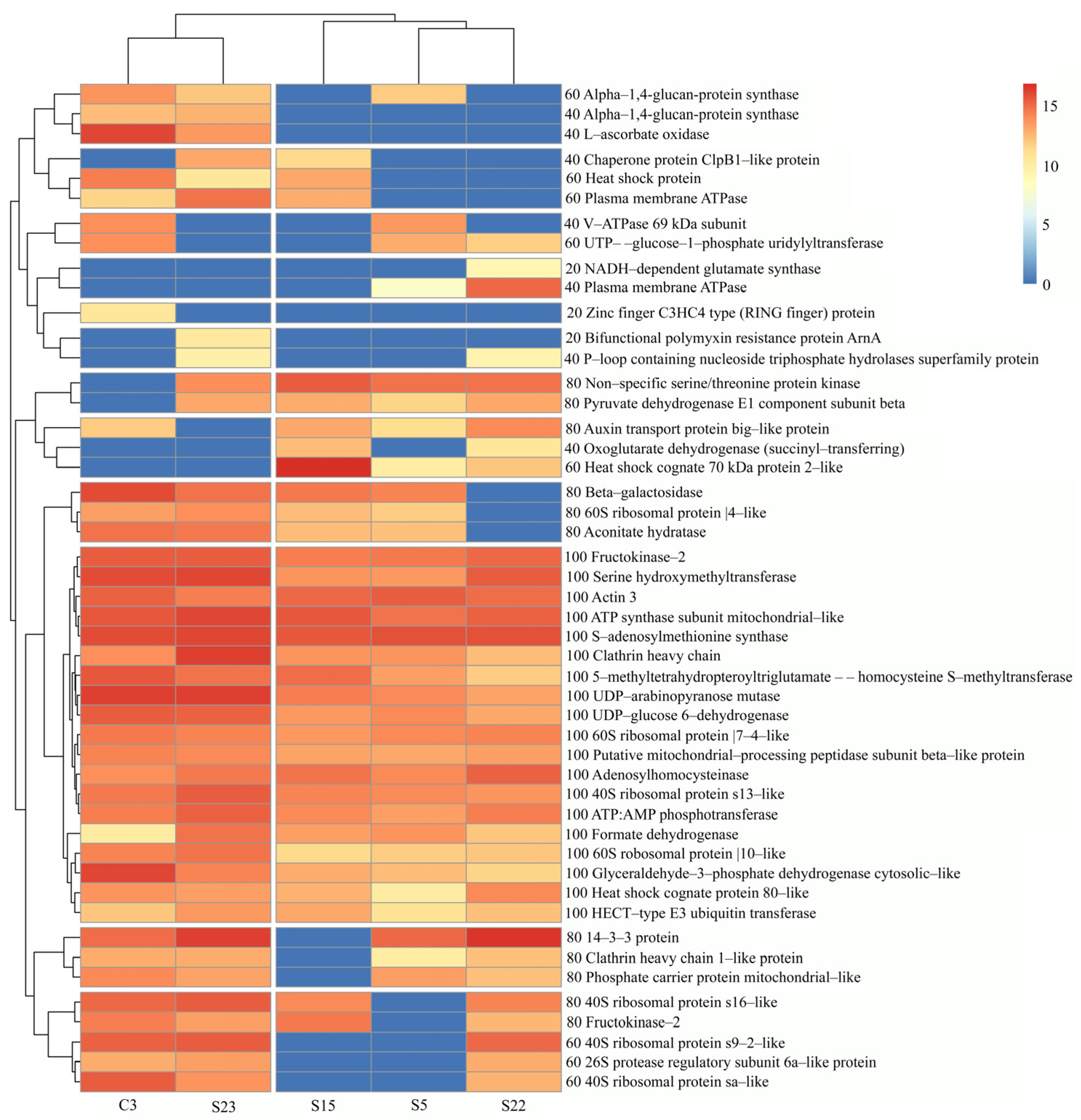
3.6. Gene Ontology (GO) Classification of Red Clover Proteins
3.6.1. Evaluation of Red Clover Proteins According to Biological Processes
3.6.2. Characteristics of Red Clover Proteins Annotated in the Biological Process and Results of Experimental Data
3.6.3. Evaluation of Red Clover Proteins According to Molecular Functions
3.6.4. Evaluation of Red Clover Proteins According to the Cellular Components or Macromolecular Complexes
4. Discussion
5. Conclusions
- Studies of the botanical composition of honey pollen and the identification of honey extracted plant proteins of pollen have provided new data on the composition of proteins. Red clover pollen was found only in monofloral red clover honey, while red-clover-related proteins were found in all honey samples. It can be assumed that bees can reach red clover nectar not only from their own nectarines, but also from the bottom of the flowers.
- Individual honey samples contained repetitive proteins common to all samples studied. Our data show that more repetitive red clover proteins were identified in each honey sample compared to some proteins that were not repeated in honey samples. The common number of proteins found in all honey samples was 39.6% of the total number of proteins found in each honey sample.
- Data from the annotation results show that the most predominant molecular functions of red clover proteins in honey samples were related to ion binding to ATP, while others are metal ion binding; zinc ion binding; copper ion binding; pyridoxal phosphate binding; and thiamine pyrophosphate binding.
- Analysis of the gene ontology of cellular components revealed single cell elements, various membranes, and macromolecular compounds. Some protein complexes that have been identified in the cell composition are histone acetyltransferase complex, proton-transporting V-type ATPase, V1 domain, and clathrin complex.
- Proteins associated with aphids, such as pea aphid, as well as endosymbiont proteins, were identified in honey, among which the largest number of proteins was from Acyrthosiphon pisum.
- In the honey samples tested, the lactic acid bacteria Lactobacillus kunkeei was found in higher concentrations than Apilactobacillus apinorum. Lactiplantibacillus plantarum and Lactiplantibacillus acidophilus were found in small amounts and Lactiplantibacillus acidophilus, Lactiplantibacillus delbrueckii subsp. Bulgaricus was solitary or unidentified in some samples.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoopingarner, R.H.; Waller, G.D. Crop Pollination. In The Hive and the Honeybe; Graham, J.M., Ed.; Dadant and Sons: Hamilton, IL, USA, 1992; pp. 1043–1082. [Google Scholar]
- Carvalho, A.T.; Schlindwein, C. Obligate Association of an Oligolectic Bee and a Seasonal Aquatic Herb in Semi-Arid North-Eastern Brazil. Biol. J. Linn. Soc. 2011, 102, 355–368. [Google Scholar] [CrossRef]
- McGregor, S.E. Clover and Some Relatives. In Insect Pollination of Cultivated Crop Plants; McGregor, S.E., Ed.; USDA-ARS: Washington, DC, USA, 1976; pp. 162–235. [Google Scholar]
- Balžekas, J.A.; Balžekas, J. The Efficiency of the First Hybrid Generation of Different Bee Races Crossed with Caucasian Bees. Zemdirb.—Agric. 1995, 42, 21–29. [Google Scholar]
- Vleugels, T.; Amdahl, H.; Roldán-Ruiz, I.; Cnops, G. Factors Underlying Seed Yield in Red Clover: Review of Current Knowledge and Perspectives. Agronomy 2019, 9, 829. [Google Scholar] [CrossRef]
- Free, J.B. The Behaviour of Bees Visiting Runner Beans (Phaseolus multiflorus). J. Appl. Ecol. 1968, 5, 631–638. [Google Scholar] [CrossRef]
- Vleugels, T.; Ceuppens, B.; Cnops, G.; Lootens, P.; van Parijs, F.R.D.; Smagghe, G.; Roldán-Ruiz, I. Models with Only Two Predictor Variables Can Accurately Predict Seed Yield in Diploid and Tetraploid Red Clover. Euphytica 2016, 209, 507–523. [Google Scholar] [CrossRef]
- Hederström, V.; Rundlöf, M.; Birgersson, G.; Larsson, M.C.; Balkenius, A.; Lankinen, Å. Do Plant Ploidy and Pollinator Tongue Length Interact to Cause Low Seed Yield in Red Clover? Ecosphere 2021, 12, e03416. [Google Scholar] [CrossRef]
- Balžekas, J.A. The Most Productive Inter-Racial Bee Hybrids of the First Generation. Zemdirb.—Agric. 1995, 42, 61–74. [Google Scholar]
- Čeksteryte, V.; Navakauskiene, R.; Treigyte, G.; Jansen, E.; Kurtinaitiene, B.; Dabkevičiene, G.; Balžekas, J. Fatty Acid Profiles of Monofloral Clover Beebread and Pollen and Proteomics of Red Clover (Trifolium pratense) Pollen. Biosci. Biotechnol. Biochem. 2016, 80, 2100–2108. [Google Scholar] [CrossRef][Green Version]
- Treigytė, G.; Zaikova, I.; Matuzevičius, D.; Čeksterytė, V.; Dabkevičienė, G.; Kurtinaitienė, B.; Navakauskienė, R. Comparative Proteomic Analysis of Pollen of Trifolium pratense, T. alexandrinum and T. repens. Zemdirbyste 2014, 101, 453–460. [Google Scholar] [CrossRef][Green Version]
- Smetanska, I.; Alharthi, S.S.; Selim, K.A. Physicochemical, Antioxidant Capacity and Color Analysis of Six Honeys from Different Origin. J. King Saud Univ. Sci. 2021, 33, 101447. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zhu, H. Genetic and Molecular Mechanisms Underlying Symbiotic Specificity in Legume-Rhizobium Interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Sainz, M.; Tabares-da Rosa, S.; Monza, J. The Role of Nitric Oxide in Nitrogen Fixation by Legumes. Front. Plant Sci. 2020, 11, 521. [Google Scholar] [CrossRef]
- Brear, E.M.; Day, D.A.; Smith, P.M.C. Iron: An Essential Micronutrient for the Legume-Rhizobium Symbiosis. Front. Plant Sci. 2013, 4, 359. [Google Scholar] [CrossRef]
- Zhang, D. Plant Seed-Derived Human Transferrin: Expression, Characterisation and Applications. OA Biotechnol. 2013, 2, 17. [Google Scholar] [CrossRef]
- Ragland, M.; Theil, E.C. Ferritin (MRNA, Protein) and Iron Concentrations during Soybean Nodule Development. Plant Mol. Biol. 1993, 21, 555–560. [Google Scholar] [CrossRef]
- Strózycki, P.M.; Legocki, A.B. Leghemoglobins from an Evolutionarily Old Legume, Lupinus luteus. Plant Sci. 1995, 110, 83–93. [Google Scholar] [CrossRef]
- Hoppler, M.; Schönbächler, A.; Meile, L.; Hurrell, R.F.; Walczyk, T. Ferritin-Iron Is Released during Boiling and In Vitro Gastric Digestion. J. Nutr. 2008, 138, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Strozycki, P.M.; Szczurek, A.; Lotocka, B.; Figlerowicz, M.; Legocki, A.B. Ferritins and Nodulation in Lupinus luteus: Iron Management in Indeterminate Type Nodules. J. Exp. Bot. 2007, 58, 3145–3153. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Beatriz, M.; Oliveira, P.P.; Mafra, I. A Comprehensive Review on the Main Honey Authentication Issues: Production and Origin. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1072–1100. [Google Scholar] [CrossRef]
- Siddiqui, A.J.; Musharraf, S.G.; Choudhary, M.I.; Rahman, A.U. Application of Analytical Methods in Authentication and Adulteration of Honey. Food Chem. 2017, 217, 687–698. [Google Scholar] [CrossRef]
- Čeksterytė, V.; Kurtinaitienė, B.; Jaškūnė, K. The Influence of Storage Conditions on Invertase, Glucose Oxidase Activity and Free Acidity of Bee Bread and Bee-Collected Pollen Mixed with Honey and Vegetable Oils. J. Apic. Res. 2020, 59, 862–875. [Google Scholar] [CrossRef]
- Louveaux, J.; Maurizio, A.; Vorwohl, G. Methods of Melissopalynology. Bee World 1978, 59, 139–157. [Google Scholar] [CrossRef]
- Ruoff, K.; Luginbühl, W.; Bogdanov, S.; Bosset, J.O.; Estermann, B.; Ziolko, T.; Amadò, R. Authentication of the Botanical Origin of Honey by Near-Infrared Spectroscopy. J. Agric. Food Chem. 2006, 54, 6867–6872. [Google Scholar] [CrossRef]
- Čeksterytė, V.; Bliznikas, S.; Jaškūnė, K. The Composition of Fatty Acids in Bee Pollen, Royal Jelly, Buckthorn Oil and Their Mixtures with Pollen Preserved for Storage. Foods 2023, 12, 3164. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, S.; Cocuzza, G.E.M. A Survey of the Aphid Fauna in the Italian Regions of Latium and Campania. Redia 2014, 97, 19–47. [Google Scholar]
- Goławska, S. Effect of Various Host-Plants on the Population Growth and Development of the Pea Aphid. J. Plant Prot. Res. 2010, 50, 224–228. [Google Scholar] [CrossRef]
- Shaaban, B.; Seeburger, V.; Schroeder, A.; Lohaus, G. Sugar, Amino Acid and Inorganic Ion Profiling of the Honeydew from Different Hemipteran Species Feeding on Abies alba and Picea abies. PLoS ONE 2020, 15, e0228171. [Google Scholar] [CrossRef]
- Recklies, K.; Peukert, C.; Kölling-Speer, I.; Speer, K. Differentiation of Honeydew Honeys from Blossom Honeys and According to Their Botanical Origin by Electrical Conductivity and Phenolic and Sugar Spectra. J. Agric. Food Chem. 2021, 69, 1329–1347. [Google Scholar] [CrossRef]
- Sabri, A.; Vandermoten, S.; Leroy, P.D.; Haubruge, E.; Hance, T.; Thonart, P.; De Pauw, E.; Francis, F. Proteomic Investigation of Aphid Honeydew Reveals an Unexpected Diversity of Proteins. PLoS ONE 2013, 8, e74656. [Google Scholar] [CrossRef]
- Čeksterytė, V. Augalų Žiedadulkių, Randamų Lietuvos Meduje, Elektroninis Katalogas. [Electronic Catalog of Plant Pollen Found in Lithuanian Honey]; Lithuania Ministry of Agriculture: Vilnius, Lithuania, 2012. [Google Scholar]
- Čeksterytė, V.; Kaupinis, A.; Jaškūnė, K.; Treigytė, G.; Navakauskienė, R. Characteristics of Honey Bee Physiological Proteins Extracted from Faba Bean (Vicia faba L.) Honey. J. Apic. Res. 2023, 62, 1250–1261. [Google Scholar] [CrossRef]
- Wiśniewski, J.; Zougman, A.; Nagaraj, N. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift Time-Specific Collision Energies Enable Deep-Coverage Data-Independent Acquisition Proteomics. Nat. Methods 2013, 11, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Kuharev, J.; Navarro, P.; Distler, U.; Jahn, O.; Tenzer, S. In-Depth Evaluation of Software Tools for Data-Independent Acquisition Based Label-Free Quantification. Proteomics 2015, 15, 3140–3151. [Google Scholar] [CrossRef] [PubMed]
- Borutinskaitė, V.; Treigytė, G.; Matuzevičius, D.; Čeksterytė, V.; Kurtinaitienė, B.; Serackis, A.; Navakauskas, D.; Navakauskienė, R. Proteomic Studies of Honeybee- and Manually-Collected Pollen. Zemdirb.—Agric. 2019, 106, 183–190. [Google Scholar] [CrossRef]
- Čeksterytė, V.; Borutinskaitė, V.; Matuzevičius, D.; Treigytė, G.; Navakauskas, D.; Kurtinaitienė, B.; Navakauskienė, R. Evaluation of Proteome Profiles of Salix spp. Pollen and Relationship Between Glucose Oxi-Dase Activity and Pollen Content in Willow Honey. Balt For. 2019, 25, 83–96. [Google Scholar] [CrossRef]
- Borutinskaitė, V.; Treigytė, G.; Matuzevičius, D.; Zaikova, I.; Čeksterytė, V.; Navakauskas, D.; Kurtinaitienė, B.; Navakauskienė, R. Proteomic Analysis of Pollen and Blossom Honey from Rape Seed Brassica napus L. J. Apic. Sci. 2017, 61, 73–92. [Google Scholar] [CrossRef]
- Myhre, S.; Tveit, H.; Mollestad, T.; Laegreid, A. Additional Gene Ontology Structure for Improved Biological Reasoning. Bioinformatics 2006, 22, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Bargsten, J.W.; Severing, E.I.; Nap, J.P.; Sanchez-Perez, G.F.; van Dijk, A.D. Biological Process Annotation of Proteins across the Plant Kingdom. Curr. Plant Biol. 2014, 1, 73–82. [Google Scholar] [CrossRef]
- Chao, Y.; Yuan, J.; Li, S.; Jia, S.; Han, L.; Xu, L. Analysis of Transcripts and Splice Isoforms in Red Clover (Trifolium pratense L.) by Single-Molecule Long-Read Sequencing. BMC Plant Biol. 2018, 18, 300. [Google Scholar] [CrossRef]
- Braunschmid, V.; Fuerst, S.; Perz, V.; Zitzenbacher, S.; Hoyo, J.; Fernandez-Sanchez, C.; Tzanov, T.; Steinkellner, G.; Gruber, K.; Nyanhongo, G.S.; et al. A Fungal Ascorbate Oxidase with Unexpected Laccase Activity. Int. J. Mol. Sci. 2020, 21, 5754. [Google Scholar] [CrossRef]
- Felton, G.W.; Summers, C.B. Potential Role of Ascorbate Oxidase as a Plant Defense Protein against Insect Herbivory. J. Chem. Ecol. 1993, 19, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Borutinskaitė, V.; Treigytė, G.; Čeksterytė, V.; Kurtinaitienė, B.; Navakauskienė, R. Proteomic Identification and Enzymatic Activity of Buckwheat (Fagopyrum esculentum) Honey Based on Different Assays. J. Food Nutr. Res. 2018, 57, 57–69. [Google Scholar]
- Kretavičius, J.; Kurtinaitienė, B.; Račys, J.; Čeksterytė, V. Inactivation of Glucose Oxidase during Heat-Treatment de-Crystallization of Honey. Zemdirb.—Agric. 2010, 97, 115–122. [Google Scholar]
- Mockaitis, K.; Estelle, M. Integrating Transcriptional Controls for Plant Cell Expansion. Genome Biol. 2004, 5, 245. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, C. Auxin Transport: Why Plants like to Think BIG. Curr. Biol. 2001, 11, R831–R833. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Khan, A.; Hashem, A.; Abd_Allah, E.F.; Al-Harrasi, A. The Molecular Mass and Isoelectric Point of Plant Proteomes. BMC Genom. 2019, 20, 631. [Google Scholar] [CrossRef]
- Grillo, M.A.; Colombatto, S. S-Adenosylmethionine and Its Products. Amino Acids 2008, 34, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Markham, G.D.; Pajares, M.A. Structure-Function Relationships in Methionine Adenosyltransferases. Cell. Mol. Life Sci. 2009, 66, 636–648. [Google Scholar] [CrossRef]
- Bakshi, A.; Shemansky, J.M.; Chang, C.; Binder, B.M. History of Research on the Plant Hormone Ethylene. J. Plant Growth Regul. 2015, 34, 809–827. [Google Scholar] [CrossRef]
- Zeng, Z.; Zeng, X.; Guo, Y.; Wu, Z.; Cai, Z.; Pan, D. Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus acidophilus NCFM to Freeze-Drying. Foods 2022, 11, 1719. [Google Scholar] [CrossRef]
- Coleman, H.D.; Ellis, D.D.; Gilbert, M.; Mansfield, S.D. Up-Regulation of Sucrose Synthase and UDP-Glucose Pyrophosphorylase Impacts Plant Growth and Metabolism. Plant Biotechnol. J. 2006, 4, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Geisler, M.; Ciereszko, I.; Johansson, H. UDP-Glucose Pyrophosphorylase. An Old Protein with New Tricks. Plant Physiol. 2004, 134, 912–918. [Google Scholar] [CrossRef]
- Thoden, J.B.; Holden, H.M. The Molecular Architecture of Glucose-1-Phosphate Uridylyltransferase. Protein Sci. 2007, 16, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Chua, T.K.; Bujnicki, J.M.; Tan, T.-C.; Huynh, F.; Patel, B.K.; Sivaraman, J. The Structure of Sucrose Phosphate Synthase from Halothermothrix orenii Reveals Its Mechanism of Action and Binding Mode. Plant Cell 2008, 20, 1059–1072. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Harada, K.; Miyazawa, S.-I.; Kondo, A.; Fukusaki, E.; Miyake, C. Metabolic Turnover Analysis by a Combination of in Vivo 13C-Labelling from 13CO2 and Metabolic Profiling with CE-MS/MS Reveals Rate-Limiting Steps of the C3 Photosynthetic Pathway in Nicotiana tabacum Leaves. J. Exp. Bot. 2010, 61, 1041–1051. [Google Scholar] [CrossRef]
- Decker, D.; Kleczkowski, L.A. UDP-Sugar Producing Pyrophosphorylases: Distinct and Essential Enzymes with Overlapping Substrate Specificities, Providing de Novo Precursors for Glycosylation Reactions. Front. Plant Sci. 2019, 9, 424694. [Google Scholar] [CrossRef]
- Bar-Peled, M.; O’Neill, M.A. Plant Nucleotide Sugar Formation, Interconversion, and Salvage by Sugar Recycling. Annu. Rev. Plant Biol. 2011, 6, 127–155. [Google Scholar] [CrossRef]
- Orellana, A.; Moraga, C.; Araya, M.; Moreno, A. Overview of Nucleotide Sugar Transporter Gene Family Functions Across Multiple Species. J. Mol. Biol. 2016, 428, 3150–3156. [Google Scholar] [CrossRef]
- Liu, D.; Wang, L.; Liu, C.; Song, X.; He, S.; Zhai, H.; Liu, Q. An Ipomoea Batatas Iron-Sulfur Cluster Scaffold Protein Gene, IbNFU1, Is Involved in Salt Tolerance. PLoS ONE 2014, 9, e93935. [Google Scholar] [CrossRef]
- Olson, J.W.; Agar, J.N.; Johnson, M.K.; Maier, R.J. Characterization of the NifU and NifS Fe−S Cluster Formation Proteins Essential for Viability in Helicobacter pylori. Biochemistry 2000, 39, 16213–16219. [Google Scholar] [CrossRef]
- Yuvaniyama, P.; Agar, J.N.; Cash, V.L.; Johnson, M.K.; Dean, D.R. NifS-Directed Assembly of a Transient [2Fe-2S] Cluster within the NifU Protein. Proc. Natl. Acad. Sci. USA 2000, 97, 599–604. [Google Scholar] [CrossRef]
- Hwang, D.M.; Dempsey, A.; Tan, K.-T.; Liew, C.-C. A Modular Domain of NifU, a Nitrogen Fixation Cluster Protein, Is Highly Conserved in Evolution. J. Mol. Evol. 1996, 43, 536–540. [Google Scholar] [CrossRef]
- Lee, E.H.; Lee, K.; Hwang, K.Y. Structural Characterization and Comparison of the Large Subunits of IPM Isomerase and Homoaconitase from Methanococcus jannaschii. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 922–931. [Google Scholar] [CrossRef]
- Dunn, M.F. Tricarboxylic Acid Cycle and Anaplerotic Enzymes in Rhizobia. FEMS Microbiol. Rev. 1998, 22, 105–123. [Google Scholar] [CrossRef]
- Resendis-Antonio, O.; Hernández, M.; Salazar, E.; Contreras, S.; Batallar, G.M.; Mora, Y.; Encarnación, S. Systems Biology of Bacterial Nitrogen Fixation: High-Throughput Technology and Its Integrative Description with Constraint-Based Modeling. BMC Syst. Biol. 2011, 5, 120. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, B.; Seeburger, V.; Schroeder, A.; Lohaus, G. Suitability of Sugar, Amino Acid, and Inorganic Ion Compositions to Distinguish Fir and Spruce Honey. Eur. Food Res. Technol. 2021, 247, 879–888. [Google Scholar] [CrossRef]
- Capinera, J.L. Green Peach Aphid, Myzus Persicae (Sulzer) (Insecta: Hemiptera: Aphididae); UF/IFAs (University of Florida, Institute of Food and Agricultural Sciences): Panama City, FL, USA, 2001. [Google Scholar]
- Sandhi, R.K.; Reddy, G.V.P. Biology, Ecology, and Management Strategies for Pea Aphid (Hemiptera: Aphididae) in Pulse Crops. J. Integr. Pest Manag. 2020, 11, 18. [Google Scholar] [CrossRef]
- Ashby, J.W.; Fletcher, J.D.; Farrell, J.A.K.; Stufkens, M.R. Observations on Host Preferences and Epidemiology of Aphid Species Associated with Legume Crops. N. Z. J. Agric. Res. 1982, 25, 267–272. [Google Scholar] [CrossRef]
- Paudel, S.; Bechinski, E.J.; Stokes, B.S.; Pappu, H.R.; Eigenbrode, D.S. Deriving Economic Models for Pea Aphid (Hemiptera: Aphididae) as a Direct-Pest and a Virus-Vector on Commercial Lentils. J. Econ. Entomol. 2018, 111, 2225–2232. [Google Scholar] [CrossRef] [PubMed]
- Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops. An Identification and Information Guide, 2nd ed.; John Wiley & Sons: Chichester, UK, 2000. [Google Scholar]
- Osiadacz, B.; Hałaj, R.; Chmura, D. Biodiversity—Economy or Ecology? Long-Term Study of Changes in the Biodiversity of Aphids Living in Steppe-like Grasslands in Central Europe. Eur. J. Entomol. 2017, 114, 140–146. [Google Scholar] [CrossRef]
- Shigenobu, S.; Hattori, H.W.M.; Sakaki, Y.; Ishikawa, H. Genome Sequence of the Endocellular Bacterial Symbiont of Aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Hoy, M.A. Genetic Systems, Genome Evolution, and Genetic Control of Embryonic Development in Insects. In Insect Molecular Genetics: An Introduction to Principles and Applications; Academic Press: San Diego, CA, USA, 2013; p. 546. [Google Scholar]
- Olofsson, T.C.; Butler, È.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vásquez, A. Lactic Acid Bacterial Symbionts in Honeybees—An Unknown Key to Honey’s Antimicrobial and Therapeutic Activities. Int. Wound J. 2016, 13, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Vásquez, A. Detection and Identification of a Novel Lactic Acid Bacterial Flora within the Honey Stomach of the Honeybee Apis mellifera. Curr. Microbiol. 2008, 57, 356–363. [Google Scholar] [CrossRef]
- De Jesus, L.; Aburjaile, F.; Sousa, T.; Felice, A.; Soares, S.; Alcantara, L.; Azevedo, V. Genomic Characterization of Lactobacillus delbrueckii Strains with Probiotics Properties. Front. Bioinform. 2022, 2, 912795. [Google Scholar] [CrossRef] [PubMed]
- Gustaw, K.; Michalak, M.; Polak-Berecka, M.; Waśko, A. Isolation and Characterization of a New Fructophilic Lactobacillus plantarum FPL Strain from Honeydew. Ann. Microbiol. 2018, 68, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Pannella, G.; Lombardi, S.; Ganassi, S.; Testa, B.; Succi, M.; Sorrentino, E.; Petrarca, S.; De Cristofaro, A.; Coppola, R. Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera Ligustica Colonies. Microorganisms 2020, 8, 1578. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Futagawa-Endo, Y.; Dicks, L. Isolation and Characterization of Fructophilic Lactic Acid Bacteria from Fructose-Rich Niches. Syst. Appl. Microbiol. 2009, 32, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Salminen, S. Honeybees and Beehives Are Rich Sources for Fructophilic Lactic Acid Bacteria. Syst. Appl. Microbiol. 2013, 36, 444–448. [Google Scholar] [CrossRef]
- Filannino, P.; Di Cagno, R.; Tlais, A.Z.A.; Cantatore, V.; Gobbetti, M. Fructose-Rich Niches Traced the Evolution of Lactic Acid Bacteria toward Fructophilic Species. Crit. Rev. Microbiol. 2019, 45, 65–81. [Google Scholar] [CrossRef]
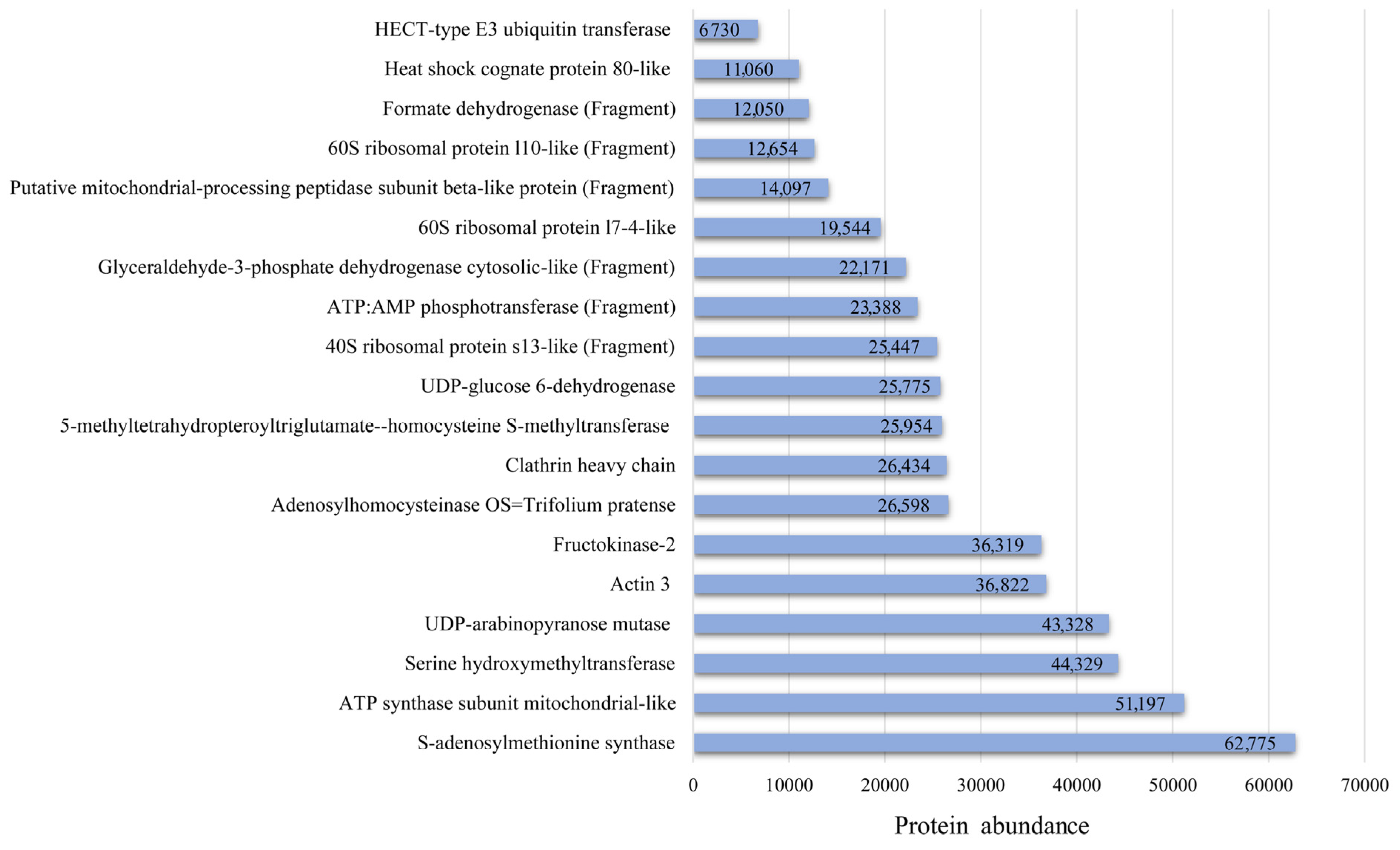
| Accession Number | Entry | Description | Mw (Da) | pI (pH) | NRP 1 | SC 2, (%) |
|---|---|---|---|---|---|---|
| A0A2 K3P3K7 | A0A2K3P3K7_TRIPR | 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase OS = T. pratense OX = 57577 GN = L195_g006435 PE = 3 SV = 1 * | 84,808 | 6.21 | 15 | 22.35 |
| A0A2K3N8B1 | A0A2K3N8B1_TRIPR | Actin 3 OS = Trifolium pratense OX = 57577 GN = actin 3 PE = 3 SV = 1 * | 42,047.0 | 5.08 | 13 | 37.67 |
| A0A2K3PQG8 | A0A2K3PQG8_TRIPR | 14-3-3 protein OS = T. pratense OX = 57577 GN = L195_g014284 PE = 3 SV = 1 | 29,428 | 4.47 | 6 | 28.85 |
| A0A2K3PKL4 | A0A2K3PKL4_TRIPR | UDP-arabinopyranose mutase OS = T. pratense OX = 57577 GN = L195_g012535 PE = 3 SV = 1 * | 40,749.8 | 5.75 | 10 | 38.81 |
| A0A2K3NAQ8 | A0A2K3NAQ8_TRIPR | ATP synthase subunit mitochondrial-like OS = Trifolium pratense OX = 57577 GN = L195_g023402 PE = 3 SV = 1 * | 36,945.3 | 8.68 | 9 | 33.33 |
| A0A2K3P1V8 | A0A2K3P1V8_TRIPR | S-adenosylmethionine synthase OS = T. pratense OX = 57577 GN = L195_g005826 PE = 3 SV = 1 * | 43,170.9 | 5.95 | 9 | 21.03 |
| A0A2K3N8U2 | A0A2K3N8U2_TRIPR | Glyceraldehyde-3-phosphate dehydrogenase cytosolic-like (Fragment) OS = Trifolium pratense OX = 57577 GN = L195_g022684 PE = 3 SV = 1 * | 27,983.8 | 6.12 | 8 | 34.23 |
| A0A2K3MUC7 | A0A2K3MUC7_TRIPR | Adenosylhomocysteinase OS = T. pratense OX = 57577 GN = L195_g017604 PE = 3 SV = 1 * | 53,696.5 | 5.90 | 12 | 27.01 |
| A0A2K3M758 | A0A2K3M758_TRIPR | Alpha-1,4-glucan-protein synthase OS = Trifolium pratense OX = 57577 GN = L195_g042687 PE = 4 SV = 1 | 24,287.1 | 6.33 | 4 | 26.67 |
| A0A2K3M1W9 | AOA2K3M1W9_TRIPR | 40S ribosomal protein s9-2-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g040850 PE = 3 SV = 1 | 16,395.1 | 10.72 | 2 | 7.41 |
| A0A2K3PJ70 | A0A2K3PJ70_TRIPR | Plasma membrane ATPase OS = Trifolium pratense OX = 57577 GN = L195_g012022 PE = 3 SV = 1 | 89,990.8 | 4.85 | 7 | 9.5 |
| A0A2K3NU12 | A0A2K3NU12_TRIPR | Aconitate hydratase OS = T. pratense OX = 57577 GN = L195_g002996 PE = 3 SV = 1 | 107,854.8 | 7.93 | 11 | 14.80 |
| A0A2K3NL25 | A0A2K3NL25_TRIPR | L-ascorbate oxidase OS = T. pratense OX = 57577 GN = L195_g000155 PE = 3 SV = 1 | 62,128.5 | 8.93 | 4 | 8.91 |
| A0A2K3P8U8 | A0A2K3P8U8_TRIPR | Plasma membrane ATPase OS = T. pratense OX = 57577 GN = L195_g008323 PE = 3 SV = 1 | 105,769.6 | 6.27 | 6 | 8.66 |
| A0A2K3L1T3 | A0A2K3L1T3_TRIPR | UDP-glucose 6-dehydrogenase OS = Trifolium pratense OX = 57577 GN = L195_g028356 PE = 3 SV = 1 * | 54,262.1 | 6.34 | 10 | 21.56 |
| A0A2K3NLX6 | A0A2K3NLX6_TRIPR | V-ATPase 69 kDa subunit OS = T. pratense OX = 57577 GN = L195_g000444 PE = 3 SV = 1 | 68,962.7 | 5.08 | 14 | 32.26 |
| A0A2K3LNL7 | A0A2K3LNL7_TRIPR | Heat shock protein OS = T. pratense OX = 57577 GN = L195_g036127 PE = 3 SV = 1 | 59,084.3 | 5.13 | 4 | 7.41 |
| A0A2K3NRS2 | A0A2K3NRS2_TRIPR | Fructokinase-2 OS = T.pratense OX = 57577 GN = L195_g002190 PE = 4 SV = 1 * | 31,966.5 | 5.66 | 6 | 21.69 |
| A0A2K3PGB1 | A0A2K3PGB1_TRIPR | Beta-galactosidase OS = T. pratense OX = 57577 GN = L195_g011002 PE = 3 SV = 1 | 87,315.5 | 8.55 | 5 | 6.33 |
| A0A2K3NMF1 | A0A2K3NMF1_TRIPR | Serine hydroxymethyltransferase OS = T. pratense OX = 57577 GN = L195_g000637 PE = 3 SV = 1 * | 52,275.3 | 6.65 | 6 | 20.81 |
| A0A2K3PJL4 | A0A2K3PJL4_TRIPR | Fructokinase-2 OS = T. pratense OX = 57577 GN = L195_g006176 PE = 4 SV = 1 | 38,560.9 | 4.69 | 3 | 14.08 |
| A0A2K3LKQ2 | A0A2K3LKQ2_TRIPR | 40S ribosomal protein s13-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g035087 PE = 3 SV = 1 * | 13,628.1 | 10.63 | 4 | 31.09 |
| A0A2K3MF29 | A0A2K3MF29_TRIPR | 40S ribosomal protein s16-like OS = T. pratense OX = 57577 GN = L195_g041963 PE = 3 SV = 1 | 16,297.1 | 10.68 | 3 | 17.86 |
| A0A2K3NCQ3 | A0A2K3NCQ3_TRIPR | Heat shock cognate 70 kDa protein 2-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g024109 PE = 4 SV = 1 | 97,918.3 | 5.60 | 2 | 3.46 |
| A0A2K3NWT6 | A0A2K3NWT6_TRIPR | UTP--glucose-1-phosphate uridylyltransferase (Fragment) OS = T. pratense OX = 57577 GN = L195_g004002 PE = 3 SV = 1 | 41,977.1 | 5.67 | 8 | 22.98 |
| A0A2K3PGD6 | A0A2K3PGD6_TRIPR | Alpha-1,4-glucan-protein synthase OS = Trifolium pratense OX = 57577 GN = L195_g011040 PE = 3 SV = 1 | 38,857.7 | 5.62 | 2 | 7.49 |
| A0A2K3JM62 | A0A2K3JM62_TRIPR | 40S ribosomal protein sa-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g048757 PE = 3 SV = 1 | 24,945.21 | 5.07 | 2 | 12.50 |
| A0A2K3NK64 | A0A2K3NK64_TRIPR | Putative mitochondrial-processing peptidase subunit beta-like protein (Fragment) OS = T. pratense OX = 57577 GN = L195_g026750 PE = 4 SV = 1 * | 29,192.50 | 7.31 | 3 | 10.34 |
| A0A2K3PKP0 | A0A2K3PKP0_TRIPR | 60S ribosomal protein l7-4-like OS = T. pratense OX = 57577 GN = L195_g012568 PE = 3 SV = 1 * | 28,587.59 | 10.40 | 6 | 20.90 |
| A0A2K3MW20 | A0A2K3MW20_TRIPR | 26S protease regulatory subunit 6a-like protein OS = T. pratense OX = 57577 GN = L195_g018159 PE = 3 SV = 1 | 42,632.90 | 5.26 | 2 | 8.44 |
| A0A2K3LNN3 | A0A2K3LNN3_TRIPR | 60S ribosomal protein l10-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g036135 PE = 3 SV = 1 * | 19,939.28 | 10.91 | 4 | 21.05 |
| A0A2K3PN04 | A0A2K3PN04_TRIPR | Heat shock cognate protein 80-like OS = T. pratense OX = 57577 GN = L195_g013385 PE = 3 SV = 1 * | 80,416.30 | 4.74 | 4 | 6.44 |
| A0A2K3NMQ3 | A0A2K3NMQ3_TRIPR | Clathrin heavy chain OS = T. pratense OX = 57577 GN = L195_g000733 PE = 3 SV = 1 * | 194,249.43 | 5.12 | 5 | 3.93 |
| A0A2K3NL88 | A0A2K3NL88_TRIPR | Zinc finger C3HC4 type (RING finger) protein (Fragment) OS = T. pratense OX = 57577 GN = L195_g000168 PE = 4 SV = 1 | 498,961.65 | 5.45 | 4 | 1.59 |
| A0A2K3MUW7 | A0A2K3MUW7_TRIPR | ATP:AMP phosphotransferase (Fragment) OS = T. pratense OX = 57577 GN = L195_g017796 PE = 3 SV = 1 * | 23,286.85 | 7.17 | 2 | 9.52 |
| A0A2K3PPZ5 | A0A2K3PPZ5_TRIPR | Formate dehydrogenase (Fragment) OS = T. pratense OX = 57577 GN = L195_g012685 PE = 3 SV = 1 * | 39,037.45 | 6.12 | 2 | 4.52 |
| A0A2K3NV46 | A0A2K3NV46_TRIPR | Phosphate carrier protein mitochondrial-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g003373 PE = 3 SV = 1 | 28,095.87 | 9.88 | 2 | 7.66 |
| A0A2K3P5Y0 | A0A2K3P5Y0_TRIPR | Oxoglutarate dehydrogenase (succinyl-transferring) OS = T. pratense OX = 57577 GN = L195_g007276 PE = 3 SV = 1 | 117,275.66 | 6.61 | 2 | 1.96 |
| A0A2K3NKS8 | A0A2K3NKS8_TRIPR | Non-specific serine/threonine protein kinase OS = T. pratense OX = 57577 GN = L195_g000037 PE = 4 SV = 1 | 451,716.00 | 6.54 | 2 | 1.67 |
| A0A2K3MA51 | A0A2K3MA51_TRIPR | Chaperone protein ClpB1-like protein (Fragment) OS = T. pratense OX = 57577 GN = L195_g043754 PE = 4 SV = 1 | 49,117.09 | 5.40 | 2 | 6.76 |
| A0A2K3PPN3 | A0A2K3PPN3_TRIPR | Bifunctional polymyxin resistance protein ArnA OS = T. pratense OX = 57577 GN = L195_g013962 PE = 3 SV = 1 | 43,325.58 | 6.73 | 2 | 6.82 |
| A0A2K3NQ11 | A0A2K3NQ11_TRIPR | Pyruvate dehydrogenase E1 component subunit beta OS = T. pratense OX = 57577 GN = L195_g001564 PE = 4 SV = 1 | 39,140.65 | 5.61 | 4 | 13.61 |
| A0A2K3NL22 | A0A2K3NL22_TRIPR | Auxin transport protein big-like protein OS = T. pratense OX = 57577 GN = L195_g000147 PE = 3 SV = 1 | 573,903.34 | 5.78 | 6 | 1.30 |
| A0A2K3NLG1 | A0A2K3NLG1_TRIPR | HECT-type E3 ubiquitin transferase OS = T.pratense OX = 57577 GN = L195_g000287 PE = 4 SV = 1 * | 391,925.50 | 4.91 | 2 | 0.59 |
| A0A2K3LUX0 | A0A2K3LUX0_TRIPR | 60S ribosomal protein l4-like (Fragment) OS = T. pratense OX = 57577 GN = L195_g038328 PE = 3 SV = 1 | 40,718.52 | 11,09 | 3 | 8.40 |
| A0A2K3P6Y1 | A0A2K3P6Y1_TRIPR | Clathrin heavy chain 1-like protein (Fragment) OS = T. pratense OX = 57577 GN = L195_g007628 PE = 4 SV = 1 | 140,432.72 | 4.94 | 5 | 6.23 |
| A0A2K3N3T6 | A0A2K3N3T6_TRIPR | P-loop containing nucleoside triphosphate hydrolases superfamily protein (Fragment) OS = T.pratense OX = 57577 GN = L195_g020931 PE = 3 SV = 1 | 69,425.16 | 5.17 | 4 | 9.97 |
| A0A2K3PCD9 | A0A2K3PCD9_TRIPR | NADH-dependent glutamate synthase (Fragment) OS = T. pratense OX = 57577 GN = L195_g009558 PE = 3 SV = 1 | 143,094.74 | 5.94 | 3 | 5.34 |
| Plant, Microbiota, and Bee-Specific Proteins, Determined in Honey | Honey Samples | Total Number | Number of Proteins Expressed in % | ||||
|---|---|---|---|---|---|---|---|
| C3 | S5 | S15 | S22 | S23 | |||
| Number of Proteins in Honey Samples | |||||||
| Proteins associated with the pollen of nectariferousN and anemophilousAN plants | |||||||
| Brassica napusN | 82 | 59 | 69 | 67 | 77 | 354 | 12.1 |
| Trifolium pratenseN | 39 | 33 | 32 | 36 | 40 | 180 | 6.2 |
| Malus domesticaN | 37 | 34 | 29 | 47 | 34 | 181 | 6.2 |
| Salix viminalisN | 31 | 23 | 28 | 28 | 37 | 147 | 5.0 |
| Prunus aviumN | 24 | 22 | 22 | 29 | 26 | 123 | 4.2 |
| Vicia fabaN | 3 | 3 | 3 | 3 | 3 | 15 | 0.5 |
| Cirsium eriophorumN | 1 | 0 | 0 | 0 | 0 | 1 | 0.3 |
| Vicia ramulifloraN | 1 | 1 | 1 | 2 | 0 | 5 | 0.2 |
| Artemisia annuaAN | 45 | 33 | 47 | 50 | 42 | 217 | 7.4 |
| Daucus carota subsp. sativusAN | 36 | 35 | 35 | 37 | 33 | 176 | 6.0 |
| Solanum tuberosumAN | 34 | 28 | 35 | 32 | 35 | 164 | 5.6 |
| Arabidopsis thalianaAN | 35 | 26 | 30 | 31 | 30 | 152 | 5.2 |
| Artemisia keiskeanaAN | 1 | 1 | 1 | 1 | 1 | 5 | 0.2 |
| Proteins associated with Apis mellifera | |||||||
| Apis mellifera | 60 | 59 | 61 | 60 | 60 | 300 | 10.3 |
| Proteins associated with aphidsA, endosiombionts of aphisE, lactic acid bacteriaL | |||||||
| Acyrthosiphon pisumA | 22 | 19 | 25 | 17 | 19 | 102 | 3.5 |
| Aphis craccivoraA | 11 | 12 | 16 | 14 | 10 | 63 | 2.2 |
| Aphis glycinesA | 9 | 11 | 8 | 6 | 4 | 38 | 1.3 |
| Buchnera aphidicola (Aphis fabae)E | 0 | 1 | 1 | 0 | 0 | 2 | 0.07 |
| Buchnera aphidicola (Aphis glycines)E | 0 | 1 | 0 | 0 | 0 | 1 | 0.3 |
| Buchnera aphidicola (Aphis gossypii)E | 0 | 1 | 0 | 0 | 0 | 1 | 0.3 |
| Serratia symbioticaE | 3 | 3 | 3 | 2 | 3 | 14 | 0.5 |
| Arsenophonus endosymbiont of aphis craccivorE | 3 | 3 | 3 | 2 | 2 | 13 | 0.5 |
| Apilactobacillus kunkeeiL | 92 | 105 | 98 | 89 | 87 | 471 | 16.1 |
| Apilactobacillus apinorumL | 27 | 36 | 34 | 23 | 26 | 146 | 5.0 |
| Lactiplantibacillus amylovorusL | 0 | 1 | 0 | 0 | 1 | 2 | 0.7 |
| Lactiplantibacillus plantarumL | 4 | 2 | 3 | 3 | 2 | 14 | 0.5 |
| Lactiplantibacillus acidophilusL | 3 | 2 | 2 | 2 | 3 | 12 | 0.4 |
| Lactiplantibacillus delbrueckii subsp. bulgaricusL | 1 | 2 | 1 | 0 | 1 | 5 | 0.2 |
| Proteins associated with bacteriaB and virusesV, animal-relatedAR | |||||||
| Escherichia coli (strain K12)B | 1 | 1 | 1 | 1 | 1 | 5 | 0.2 |
| Fagopyrum esculentum endornavirus 1V | 0 | 0 | 1 | 1 | 0 | 2 | 0.7 |
| Grand total | 606 | 558 | 590 | 585 | 578 | 2917 | 100.0 |
| Uniprot Accession Number | Protein Name | Species | C3 | S5 | S15 | S22 | S23 |
|---|---|---|---|---|---|---|---|
| A0A078J693 | BnaC03g73810D protein | Brassica napus | 64.0 | 25.4 | 12.0 | 31.9 | 47.6 |
| A0A078JFE6 | Fructose-bisphosphate aldolase | Brassica napus | 36.8 | 3.4 | 2.0 | 17.5 | 47.5 |
| A0A498HT56 | Uncharacterized protein | Malus domestica | 54.1 | 10.0 | 6.7 | 25.3 | 63.1 |
| A0A2K3P1V8 | S-adenosylmethionine synthase | Trifolium pratense | 42.4 | 23.1 | 8.5 | 17.4 | 37.1 |
| Q9LFW1 | UDP-arabinopyranose mutase 2 | Arabidopsis thaliana | 10.0 | 2.5 | 1.1 | 3.2 | 16.6 |
| P80261 | NADH dehydrogenase [ubiquinone] iron–sulfur protein 3 | Solanum tuberosum | 5.9 | 1.8 | 0.7 | 2.0 | 7.5 |
| A0A175YJ97 | AAI domain-containing protein | Daucus carota subsp. sativus | 4.3 | 15.5 | 3.6 | 1.5 | 1.9 |
| A0A087ENY5 | Phosphoglycerate kinase | Apilactobacillus kunkeei | 50.6 | 57.7 | 16.6 | 22.0 | 25.2 |
| A0A087EPJ5 | 50S ribosomal protein L4 | Apilactobacillus kunkeei | 20.7 | 26.0 | 7.3 | 7.4 | 11.6 |
| A0A087EPM7 | 30S ribosomal protein S9 | Apilactobacillus kunkeei | 18.5 | 24.8 | 7.5 | 6.3 | 8.8 |
| A0A087EQ00 | Glutamine synthetase | Apilactobacillus kunkeei | 16.2 | 28.4 | 8.0 | 4.6 | 8.7 |
| A0A087EQA3 | Threonine--tRNA ligase | Apilactobacillus kunkeei | 14.7 | 20.9 | 5.1 | 4.0 | 9.7 |
| A0A0M9DBL5 | 50S ribosomal protein L5 | Apilactobacillus kunkeei | 11.7 | 24.4 | 6.3 | 3.0 | 4.6 |
| A0A087EPI7 | DNA-directed RNA polymerase subunit beta | Apilactobacillus kunkeei | 11.4 | 27.8 | 6.1 | 3.2 | 10.3 |
| A0A087EQ84 | Probable manganese-dependent inorganic pyrophosphatase | Apilactobacillus kunkeei | 9.2 | 18.0 | 4.9 | 3.9 | 2.9 |
| A0A0C3AFU8 | Nitroreductase | Apilactobacillus kunkeei | 9.0 | 14.7 | 3.8 | 2.6 | 5.7 |
| A0A087EMN0 | Glycine/betaine ABC transporter ATP-binding protein | Apilactobacillus kunkeei | 8.6 | 13.6 | 3.2 | 3.1 | 6.0 |
| A0A0M9D308 | Catalase | Apilactobacillus kunkeei | 6.5 | 18.3 | 4.3 | 2.7 | 3.8 |
| A0A087EPK8 | 30S ribosomal protein S8 | Apilactobacillus kunkeei | 6.2 | 8.1 | 2.1 | 1.5 | 4.8 |
| A0A0N0UVX1 | Beta sliding clamp | Apilactobacillus kunkeei | 5.5 | 8.3 | 2.1 | 2.0 | 3.0 |
| A0A087ENA2 | Aldo/keto reductase | Apilactobacillus kunkeei | 5.4 | 14.6 | 2.7 | 1.1 | 3.8 |
| A0A087EPK1 | 50S ribosomal protein L16 | Apilactobacillus kunkeei | 5.2 | 9.5 | 1.9 | 1.4 | 2.6 |
| A0A0N0CRP1 | DUF5776 domain-containing protein | Apilactobacillus kunkeei | 4.9 | 14.0 | 6.9 | 9.0 | 0.0 |
| A0A087EQ04 | 50S ribosomal protein L27 | Apilactobacillus kunkeei | 4.1 | 13.7 | 2.4 | 1.2 | 3.2 |
| A0A0N0CQ41 | 6-phosphogluconate dehydrogenase, decarboxylating | Apilactobacillus apinorum | 17.6 | 39.9 | 9.5 | 4.0 | 17.3 |
| A0A0M9D5F1 | L-lactate dehydrogenase | Apilactobacillus apinorum | 12.5 | 41.2 | 6.3 | 2.1 | 8.0 |
| A0A0M9D658 | Glutathione reductase | Apilactobacillus apinorum | 9.9 | 24.3 | 5.7 | 2.6 | 6.7 |
| A0A0N0CQC3 | Glutathione reductase | Apilactobacillus apinorum | 6.8 | 19.4 | 2.5 | 1.2 | 5.3 |
| A0A7M7RC42 | Uncharacterized protein | Apis mellifera | 43.6 | 15.5 | 11.2 | 48.2 | 10.4 |
| A0A7M7IFB4 | Glucosylceramidase | Apis mellifera | 56.0 | 27.0 | 15.2 | 45.4 | 16.5 |
| FBP | APN | LABN | LABC | |
|---|---|---|---|---|
| Number of faba bean pollen (FBP) | 1 | 0.415 | 0.943 | 0.935 |
| Aphid protein number (APN) | 0.415 | 1 | 0.693 | 0.065 |
| Lactic acid bacteria number (LABN) | 0.943 | 0.693 | 1 | 0.764 |
| Lactic acid bacteria content (LABC) | 0.935 | 0.065 | 0.764 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čeksterytė, V.; Kaupinis, A.; Aleliūnas, A.; Navakauskienė, R.; Jaškūnė, K. Composition of Proteins Associated with Red Clover (Trifolium pratense) and the Microbiota Identified in Honey. Life 2024, 14, 862. https://doi.org/10.3390/life14070862
Čeksterytė V, Kaupinis A, Aleliūnas A, Navakauskienė R, Jaškūnė K. Composition of Proteins Associated with Red Clover (Trifolium pratense) and the Microbiota Identified in Honey. Life. 2024; 14(7):862. https://doi.org/10.3390/life14070862
Chicago/Turabian StyleČeksterytė, Violeta, Algirdas Kaupinis, Andrius Aleliūnas, Rūta Navakauskienė, and Kristina Jaškūnė. 2024. "Composition of Proteins Associated with Red Clover (Trifolium pratense) and the Microbiota Identified in Honey" Life 14, no. 7: 862. https://doi.org/10.3390/life14070862
APA StyleČeksterytė, V., Kaupinis, A., Aleliūnas, A., Navakauskienė, R., & Jaškūnė, K. (2024). Composition of Proteins Associated with Red Clover (Trifolium pratense) and the Microbiota Identified in Honey. Life, 14(7), 862. https://doi.org/10.3390/life14070862






