Canine Distemper Virus: Origins, Mutations, Diagnosis, and Epidemiology in Mexico
Abstract
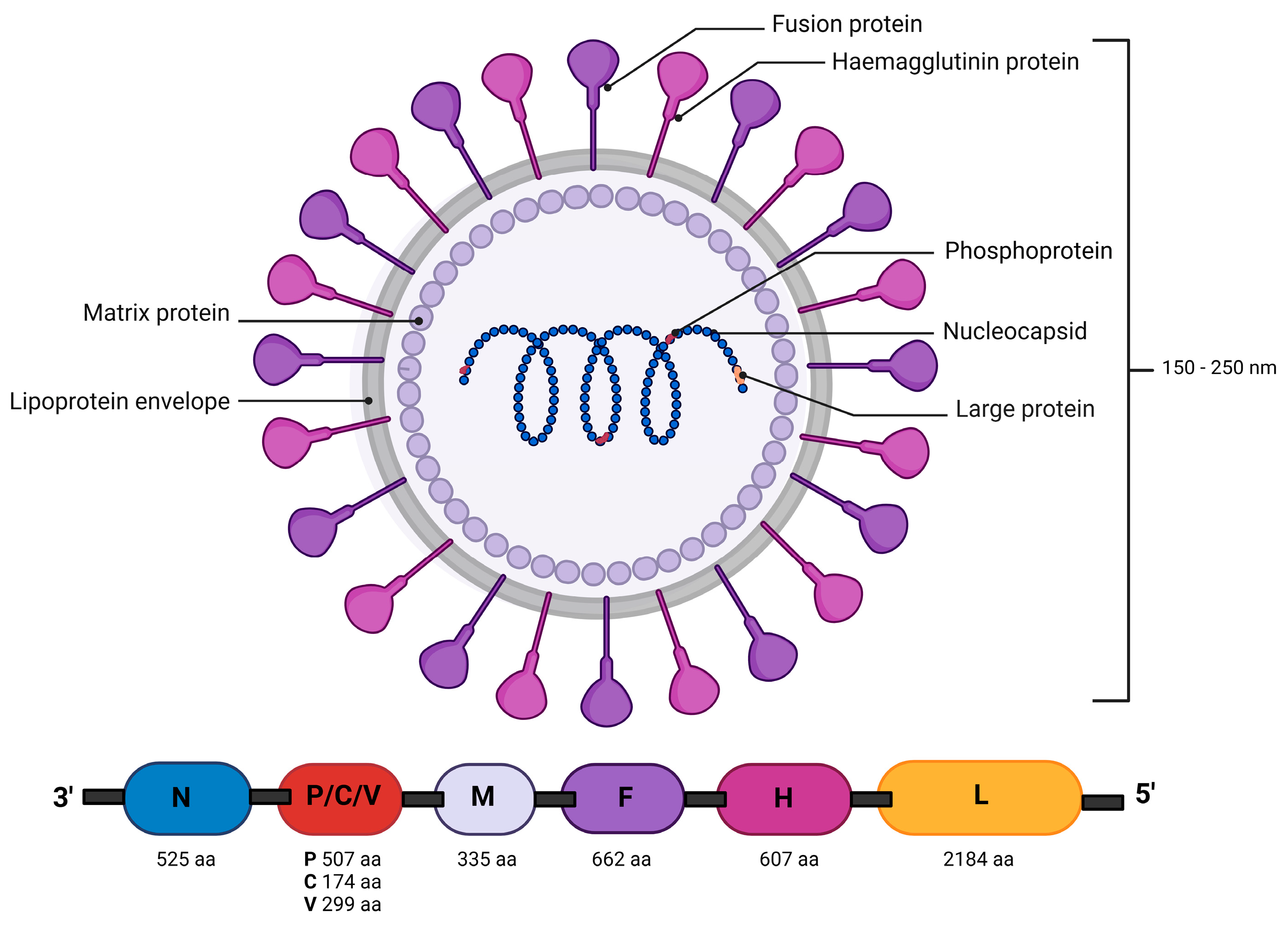
1. Introduction
2. Materials and Methods
3. Results Overview: Insights into Canine Distemper Virus
3.1. Hypothesis on CDV’s Origin
3.2. Epidemiology of CDV in Mexico
3.3. Genomic Mutations of CDV
3.4. Advances in the Diagnosis of CDV
| Diagnostic Method | Detection Method | Type of Diagnostic Method | Target | Notes on the Test | Reference |
|---|---|---|---|---|---|
| MDCK, MV1 Lu, cells Vero-SLAM B95a | Virus isolation | Cell culture | Virus | Gold-standard test, but currently infrequently used | [57] |
| Direct ELISA | Antigen detection | Serological | CDV antigen | Detects antigen in serum | [58] |
| Sandwich ELISA | Antigen detection | Serological | Protein H antigen | High specificity Detection and quantification | [59] |
| Protein F antigen | Efficient in field application with fecal and serum samples | [60] | |||
| Sandwich dot ELISA | Antigen detection | Serological | Virus | Rapid test, used in epidemiological surveillance | [61] |
| LFA | Antigen detection | Serological | Protein F antigen | Rapid test | [62] |
| Immunofluorescence | Detection of fluorescently labeled antibodies | Immunofluorescence | Protein F/H antigen | Laborious test, a fluorescence microscope is needed, highly sensitive and specific | [40] |
| RT-PCR | RNA detection | Genomic | Gen N | Standard laboratory test, currently the most used | [63] |
| One-step nested RT-PCR | Antigen detection | Genomic | Gen N | 100 times greater sensitivity than RT-PCR and nested PCR | [38] |
| Double-step real-time RT-PCR | RNA detection | Genomic | Gen N | Highly sensitive and specific Quantify the viral load in clinical samples | [7] |
| One-step real-time RT-PCR | RNA detection | Genomic | Gen N | Allows the study of viral replication and the kinetics of the viral load of viral RNA in infection | [64] |
| RT-LAMP assay | RNA detection | Genomic | Gen H | 100 times more sensitive than RT-PCR Only 1 h of reaction | [65] |
| RT-qPCR | RNA detection | Genomic | Protein M gene and MF intergenic region | Uses the TaqMan probe based on the CDV-N and P genes and is highly sensitive and specific compared to other tests | [66] |
| RT-PCR-RFLP | RNA detection plus restriction enzymes | Genomic | N gene amplification plus BamHI Restriction enzyme (RE) digestion Gen N amplification plus enzyme digestion using MspI | It results in a different number of fragments in both strains | [67] |
| ELISA | Detection of specific antibodies against the virus | Serological | Antibody IgG | Detect within 6 days of infection | [68] |
| Dot blot assay | Detection of specific antibodies against the virus | Serological | N protein-specific IgM | Detect recent infections | [69] |
| Capture sandwich ELISA | Detection of specific antibodies against the virus | Serological | N protein-specific IgG and IgM antibody | No cross-reactivity with other morbilliviruses | [70] |
4. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blancou, J. Dog distemper: Imported into Europe from South America? Hist. Med. Vet. 2004, 29, 35–41. [Google Scholar] [PubMed]
- Calzada Nova, L.A.; Vázquez Manríquez, L. Origen e historia del Moquillo canino. Vanguard. Vet. 2020, 98, 22–26. [Google Scholar]
- Carré, H. Sur la maladie des jeunes chiens. C. R. Acad. Sci. 1905, 140 (689–690), 1489–1491. [Google Scholar]
- Bolio González, M.E.; Rodríguez Vivas, R.I.; Rosado Aguilar, J.A.; Gutiérrez Ruiz, E.J.; Gutiérrez Calzada Nova, L.A.; Vázquez Manríquez, L. Distemper canino, infección viral multisistémica, que produce síndromes clínicos neurológicos: Una revisión actualizada. Vanguard. Vet. 2020, 102, 6–12. [Google Scholar]
- Zipperle, L.; Langedijk, J.P.; Orvell, C.; Vandevelde, M.; Zurbriggen, A.; Plattet, P. Identification of key residues in virulent canine distemper virus hemagglutinin that control CD150/SLAM-binding activity. J. Virol. 2010, 84, 9618–9624. [Google Scholar] [CrossRef] [PubMed]
- Loots, A.K.; Mitchell, E.; Dalton, D.L.; Kotzé, A.; Venter, E.H. Advances in canine distemper virus pathogenesis research: A wildlife perspective. J. Gen. Virol. 2017, 98, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Duque-Valencia, J.; Sarute, N.; Olarte-Castillo, X.A.; Ruíz-Sáenz, J. Evolution and Interspecies Transmission of Canine Distemper Virus—An Outlook of the Diverse Evolutionary Landscapes of a Multi-Host Virus. Viruses 2019, 11, 582. [Google Scholar] [CrossRef] [PubMed]
- Uhl, E.W.; Thomas, R. Uncovering tales of transmission: An integrated palaeopathological perspective on the evolution of shared human and animal pathogens. In Palaeopathology and Evolutionary Medicine: An Integrated Approach; Plomp, K.A., Roberts, C.A., Elton, S., Bentley, G.R., Eds.; Oxford Academic: Oxford, UK, 2022; pp. 317–349. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Cordero, R.; Lee, B.; Rota, P.; et al. Perfil taxonómico del virus ICTV: Paramyxoviridae. J. Gen. Virol. Rev. 2019, 100, 1593–1594. [Google Scholar] [CrossRef]
- Da Fontoura Budaszewski, R.; von Messling, V. Morbillivirus Experimental Animal Models: Measles Virus Pathogenesis Insights from Canine Distemper Virus. Viruses 2016, 8, 274. [Google Scholar] [CrossRef]
- Uhl, E.W.; Kelderhouse, C.; Buikstra, J.; Blick, J.P.; Bolon, B.; Hogan, R.J. New world origin of canine distemper: Interdisciplinary insights. Int. J. Paleopathol. 2019, 24, 266–278. [Google Scholar] [CrossRef]
- Wilkes, R.P. Canine distemper virus in endangered species: Species jump, clinical variations, and vaccination. Pathogens 2022, 12, 57. [Google Scholar] [CrossRef]
- Rebollar-Zamorano, M.; Morales-Ubaldo, A.L.; González-Alamilla, E.N.; Ángeles-Rodríguez, A.; Valladares-Carranza, B.; Velásquez-Ordoñez, V.; Rivero-Pérez, N.; Zaragoza-Bastida, A. Análisis epidemiológico retrospectivo de Distemper Canino en la ciudad de Pachuca de Soto, Estado de Hidalgo. J. Selva Andin. Anim. Sci. 2020, 7, 40–46. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, N.; Sun, Y.; Martella, V.; Nikolin, V.; Zhu, C.; Zhang, H.; Hu, B.; Bai, X.; Yan, X. Pathogenesis of canine distemper virus in experimentally infected raccon dogs, foxes, and minks. J. Antiv Res. 2015, 122, 1–11. [Google Scholar] [CrossRef]
- Pillet, S.; Svitek, N.; Von Messling, V. Ferrets as a model for morbillivirus pathogenesis, complications, and vaccines. Measles Pathog. Control 2009, 330, 73–87. [Google Scholar]
- de Vries, R.D.; Ludlow, M.; Verburgh, R.J.; van Amerongen, G.; Yüksel, S.; Nguyen, D.T.; McQuaid, S.; Osterhaus, A.D.M.E.; Duprex, W.P.; de Swart, R.L. Measles vaccination of nonhuman primates provides partial protection against infection with canine distemper virus. J. Virol. 2014, 88, 4423–4433. [Google Scholar] [CrossRef] [PubMed]
- Nambulli, S.; Sharp, C.R.; Acciardo, A.S.; Drexler, J.F.; Duprex, W.P. Mapping the evolutionary trajectories of morbilliviruses: What, where and whither. Curr. Opin. Virol. 2016, 16, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Gil, C.; Rendon-Marin, S.; Martinez-Gutierrez, M.; Ruiz-Saenz, J. Origin of canine distemper virus. Consolidating evidence to understand potential zoonoses. Front. Microbiol. 2019, 10, 1–5. [Google Scholar] [CrossRef]
- Padilla, J. Signologia Clínica de la Enfermedad de Carré. In Memoria Foro sobre Enfermedad de Carré; Unidad de Congreso del Centro Medico Nacional: Ciudad de México, México, 1987; pp. 6–8. [Google Scholar]
- Roelke-Parker, M.E.; Munson, L.; Packer, C.; Kock, R.; Cleaveland, S.; Carpenter, M.; O’Brien, S.J.; Pospischil, A.; Hofmann-Lehmann, R.; Lutz, H.; et al. A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature 1996, 379, 441–445. [Google Scholar] [CrossRef]
- Kameo, Y.; Nagao, Y.; Nishio, Y.; Shimoda, H.; Nakano, H.; Suzuki, K.; Une, Y.; Sato, H.; Shimojima, M.; Maeda, K. Epizootic canine distemper virus infection among wild mammals. Vet. Microbiol. 2012, 154, 222–229. [Google Scholar] [CrossRef]
- Anis, E.; Needle, D.B.; Stevens, B.; Yan, L.; Wilkes, R.P. Genetic characteristics of canine distemper viruses circulating in wildlife in the United States. J. Zoo Wildl. Med. 2020, 50, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Timm, S.F.; Munson, L.; Summers, B.A.; Terio, K.A.; Dubovi, E.J.; Rupprecht, C.E.; Kapil, S.; Garcelon, D.K. A suspected canine distemper epidemic as the cause of a catastrophic decline in Santa Catalina island foxes (Urocyon littoralis catalinae). J. Wildl. Dis. 2009, 45, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cabo-Mercado, R.; Martínez-Hernández, F.; Aréchiga-Ceballos, N.; López-Díaz, O.; Muñoz-García, C.I.; Aguilar-Setién, A.; Villalobos, G.; Villanueva-García, C.; Verdugo-Rodríguez, A.; Iturbe-Ramírez, R.; et al. Canine distemper in neotropical procyonids: Molecular evidence, humoral immune response and epidemiology. Virus Res. 2020, 290, 198164. [Google Scholar] [CrossRef] [PubMed]
- Gámiz-Mejía, C.E.; Simón-Martínez, J.; Fajardo-Muñoz, R.C. Identification of new genovariants of canine distemper virus in dogs from the State of Mexico by analyzing the nucleocapsid gene. Arch. Med. Vet. 2012, 44, 53–58. [Google Scholar] [CrossRef]
- González Vallejo, M.V. Detección Molecular de Virus Asociados con el Complejo Respiratorio Canino en Perros del Área Metropolitana de Monterrey. Doctoral Dissertation, Universidad Autónoma de Nuevo León, San Nicolás de los Garza, Mexico, 2014. [Google Scholar]
- Garcia Vidaña, V.J. Diagnóstico de Distemper Canino por Medio de Prueba Rápida Para Detección de Antígeno en Perros. 2016. Available online: https://repositorio.uaaan.mx/handle/123456789/8015 (accessed on 26 June 2024).
- Martinez, J.S.; Arvizu, R.A.; Soriano, V.E.; Fajardo, R. Identification of a genetic variant of canine distemper virus from clinical cases in two vaccinated dog in México. Vet. J. 2018, 175, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Almuna, R.; López-Pérez, A.M.; Sarmiento, R.E.; Suzán, G. Drivers of canine distemper virus exposure in dogs at a wildlife interface in Janos, Mexico. Vet. Rec. Open 2021, 8, e7. [Google Scholar] [CrossRef] [PubMed]
- Tatsuo, H.; Yanagi, Y. El SLAM del receptor de morbillivirus (CD150). Microbiol. Inmunol 2002, 46, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Cirone, F.; Elia, G.; Lorusso, E.; Decaro, N.; Campolo, M.; Desario, C.; Lucente, M.S.; Bellacicco, A.L.; Blixenkrone-Moller, M.; et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet. Microbiol. 2006, 116, 301–309. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, H.; Bai, X.; Martella, V.; Hu, B.; Sun, Y.; Zhu, C.; Zhang, L.; Liu, H.; Xu, S.; et al. Emergence of canine distemper virus strains with two amino acid substitutions in the haemagglutinin protein, detected from vaccinated carnivores in North-Eastern China in 2012–2013. Vet. J. 2014, 200, 191–194. [Google Scholar] [CrossRef]
- Zhao, J.-J.; Yan, X.-J.; Chai, X.-L.; Martella, V.; Luo, G.-L.; Zhang, H.-L.; Gao, H.; Liu, Y.-X.; Bai, X.; Zhang, L.; et al. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet. Microbiol. 2010, 140, 34–42. [Google Scholar] [CrossRef]
- Demeter, Z.; Lakatos, B.; Palade, E.A.; Kozma, T.; Forgách, P.; Rusvai, M. Genetic diversity of Hungarian canine distemper virus strains. Vet. Microbiol. 2007, 122, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Karki, M.; Rajak, K.K.; Singh, P.; Fayaz, A.; Kiran Yadav, A.K.; Bhatt, M.; Rai, V.; Einstein, C.; Singh, R.P. Optimization of competitive lateral flow assay for detection of canine distemper virus antibody. Pharma Innov. J. 2022, SP-11, 1568–1572. [Google Scholar]
- Singh, R.P.; Sreenivasa, B.P.; Dhar, P.; Bandyopadhyay, S.K. A sandwich-ELISA for the diagnosis of Peste des petits ruminants (PPR) infection in small ruminants using anti-nucleocapsid protein monoclonal antibody. Arch. Virol. 2004, 149, 2155–2170. [Google Scholar] [CrossRef]
- Karki, M.; Rajak, K.K.; Singh, R.P. Canine morbillivirus (CDV): A review on current status, emergence and the diagnostics. Virusdisease 2022, 33, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Messling, V.V.; Harder, T.C.; Moening, V.; Rautemberg, P.; Nolte, I.; Haas, L. Rapid and sensitive detection of immunoglobulin M (IgM) and IgG against canine distemper virus by a new recombinant nucleocapsid Protein-based enzyme-linked immunosorbent assay. J. Clin. Microbiol. 1999, 37, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, M.; Namikawa, K.; Maruo, T.; Orito, K.; Lynch, J.; Sahara, H. Antibody titers for canine parvovirus type-2, canine distemper virus, and canine adenovirus type-1 in adult household dogs. Can. Vet. J. 2011, 52, 983. [Google Scholar]
- Greene, C.E. Infectious Diseases of the Dog and Cat, 3rd ed.; WB Saunders/Elsevier Science: Philadelphia, PA, USA, 2006. [Google Scholar]
- Li, Z.; Zhang, Y.; Wang, H.; Jin, J.; Li, W. Sandwich-dot enzyme-linked immunosorbent assay for the detection of canine distemper virus. Can. J. Vet. Res. 2013, 77, 303–308. [Google Scholar] [PubMed]
- Hassenin, A.S.H.; Durrani, A. Sensitivity & Specificity of ELISA Kit for Antibody Detection of Canine Parvovirus and Canine Distemper Infection in Dogs. Mathews J. Vet. Sci. 2023, 7, 34. [Google Scholar]
- Amude, A.M.; Alfieri, A.A.; y Alfieri, A.F. Antemorten Diagnosis of VDC Infection by RT-PCR in Distemper Dogs with Neurological Deficits Without the Typical Clinical Presentation. Vet. Rearc. 2006, 30, 679–687. [Google Scholar]
- Frisk, A.L.; Konig, M.; Moritz, A.; Baumgartner, W. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J. Clin. Microbiol. 1999, 37, 3634–3643. [Google Scholar] [CrossRef]
- Appel, M.J.; Summers, B.A. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet. Microbiol. 1995, 44, 187–191. [Google Scholar] [CrossRef]
- Martella, V.; Elia, G.; Buonavoglia, C. Canine distemper virus. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Sarchahi, A.A.; Arbabi, M.; Mohebalian, H. Detection of canine distemper virus in cerebrospinal fluid, whole blood and mucosal specimens of dogs with distemper using RT-PCR and immunochromatographic assays. Vet. Med. Sci. 2022, 8, 1390–1399. [Google Scholar] [CrossRef]
- Jóźwik, A.; Frymus, T. Comparison of the immunofluorescence assay with RT-PCR and nested PCR in the diagnosis of canine distemper. Vet. Res. Commun. 2005, 29, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Conceição-Neto, N.; Godinho, R.; Álvares, F.; Yinda, C.K.; Deboutte, W.; Zeller, M.; Laenen, L.; Heylen, E.; Roque, S.; Petrucci-Fonseca, F.; et al. Viral gut metagenomics of sympatric wild and domestic canids, and monitoring of viruses: Insights from an endangered wolf population. Ecol. Evol. 2017, 7, 4135–4146. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; He, Y.; Chen, X.; Kalim, U.; Wang, Y.; Yang, S.; Qi, H.; Cheng, H.; Lu, X.; Wang, X.; et al. Viral Metagenomics Reveals Diverse Viruses in the Feces Samples of Raccoon Dogs. Front. Vet. Sci. 2021, 8, 693564. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.; Li, C.; Ma, Y.; Sun, Q.; Zhang, H.; Niu, G.; Wei, J.; Yao, H.; Ma, Z. Viral Metagenomic Analysis of the Fecal Samples in Domestic Dogs (Canis lupus familiaris). Viruses 2023, 15, 685. [Google Scholar] [CrossRef]
- Lanszki, Z.; Tóth, G.E.; Schütz, É.; Zeghbib, S.; Rusvai, M.; Jakab, F.; Kemenesi, G. Complete genomic sequencing of canine distemper virus with nanopore technology during an epizootic event. Sci. Rep. 2022, 12, 4116. [Google Scholar] [CrossRef]
- Roingeard, P.; Raynal, P.I.; Eymieux, S.; Blanchard, E. Virus detection by transmission electron microscopy: Still useful for diagnosis and a plus for biosafety. Rev. Med. Virol. 2019, 29, e2019. [Google Scholar] [CrossRef]
- Stancu, A.C.; Voia, O.S.; Boldura, O.M.; Pasca, S.A.; Luca, I.; Hulea, A.S.; Ivan, O.R.; Dragoescu, A.A.; Lungu, B.C.; Hutu, I. Unusual Canine Distemper Virus Infection in Captive Raccoons (Procyon lotor). Viruses. 2023, 15, 1536. [Google Scholar] [CrossRef]
- Chludzinski, E.; Ciurkiewicz, M.; Stoff, M.; Klemens, J.; Krüger, J.; Shin, D.-L.; Herrler, G.; Beineke, A. Canine Distemper Virus Alters Defense Responses in an Ex Vivo Model of Pulmonary Infection. Viruses 2023, 15, 834. [Google Scholar] [CrossRef]
- Appel, M.J.G.; Gillespie, J.H. Canine Distemper Virus. In Virology Monographs; Hallauer, S.C.G., Meyer, K., Eds.; Springer: Vienna, NY, USA, 1972; pp. 1–96. [Google Scholar]
- Sindhu, N.; Borah, J.; Shah, S.; Rajput, N.; Jadav, K.K. Is canine distemper virus (CDV) a lurking threat to large carnivores? A case study from Ranthambhore landscape in Rajasthan. India JoTT 2019, 11, 14220–14223. [Google Scholar]
- Pan, Z.; Liu, J.; Ma, J.; Jin, Q.; Yao, H.; Osterrieder, N. The recombinant EHV-1 vector producing CDV hemagglutinin as potential vaccine against canine distemper. MicrobPathog 2017, 111, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Wostenberg, D.J.; Walker, N.; Fox, K.A.; Spraker, T.R.; Piaggio, A.J.; Gilbert, A. Evidence of two cocirculating canine distemper virus strains in mesocarnivores from northern Colorado, USA. J. Wildl. Dis. 2018, 54, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Latha, D.; Geetha, M.; Ramadass, P.; Narayanan, R.B. Evaluation of ELISA based on the conserved and functional middle region of nucleocapsid protein to detect distemper infection in dogs. Vet. Microbiol. 2017, 120, 251–260. [Google Scholar] [CrossRef] [PubMed]
- An, D.J.; Kim, T.Y.; Song, D.S.; Kang, B.K.; Park, B.K. An immunochromatography assay for rapid antemortem diagnosis of dogs suspected to have canine distemper. J. Virol. Methods 2008, 147, 244–249. [Google Scholar] [CrossRef]
- Freitas, L.A.; Leme, R.A.; Saporiti, V.; Alfieri, A.A.; Alfieri, A.F. Molecular analysis of the full-length F gene of Brazilian strains of canine distemper virus shows lineage co-circulation and variability between field and vaccine strains. Virus Res. 2019, 264, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Nagata, N.; Ami, Y.; Seki, F.; Suzaki, Y.; Iwata-Yoshikawa, N.; Suzuki, T.; Fukushi, S.; Mizutani, T.; Yoshikawa, T.; et al. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J. Virol. 2013, 87, 1105–1114. [Google Scholar] [CrossRef]
- Latha, D.; Srinivasan, S.R.; Thirunavukkarasu, P.S.; Gunaselan, L.; Ramadass, P.; Narayanan, R.B. Assessment of canine distemper virus infection in vaccinated and unvaccinated dogs. Indian J. Biotechnol. 2007, 6, 35–40. [Google Scholar]
- Wilkes, R.P.; Sanchez, E.; Riley, M.C.; Kennedy, M.A. Real-time reverse transcription polymerase chain reaction method for detection of canine distemper virus modified live vaccine shedding for differentiation from infection with wild-type strains. J. Vet. Diag. Invest. 2014, 26, 27–34. [Google Scholar] [CrossRef]
- Wang, F.; Yan, X.; Chai, X.; Zhang, H.; Zhao, J.; Wen, Y.; Wu, W. Differentiation of canine distemper virus isolates in fur animals from various vaccine strains by reverse transcription-polymerase chain reaction-restriction fragment length polymorphism according to phylogenetic relations in China. Virol. J. 2011, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Barben, G.; Stettler, M.; Jaggy, A.; Vandevelde, M.; Zurbriggen, A. Detection of IgM antibodies against a recombinant nucleocapsid protein of canine distemper virus in dog sera using a dot-blot assay. J. Vet. Med. 1999, 46, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ashmi, J.M.; Thangavelu, A.; Senthilkumar, T.M.A.; Manimaran, K. Molecular characterization of canine distemper virus from Tamil Nadu, India. Indian J. Anim. Sci. 2017, 87, 1062–1067. [Google Scholar] [CrossRef]
- Takenaka, A.; Yoneda, M.; Seki, T.; Uema, M.; Kooriyama, T.; Nishi, T.; Fujita, K.; Miura, R.; Tsukiyama-Kohara, K.; Sato, H.; et al. Characterization of two recent Japanese field isolates of canine distemper virus and examination of the avirulent strain utility as an attenuated vaccine. Vet. Microbial. 2014, 174, 372–381. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera-Martínez, A.; Rodríguez-Alarcón, C.A.; Adame-Gallegos, J.R.; Laredo-Tiscareño, S.V.; de Luna-Santillana, E.d.J.; Hernández-Triana, L.M.; Garza-Hernández, J.A. Canine Distemper Virus: Origins, Mutations, Diagnosis, and Epidemiology in Mexico. Life 2024, 14, 1002. https://doi.org/10.3390/life14081002
Rivera-Martínez A, Rodríguez-Alarcón CA, Adame-Gallegos JR, Laredo-Tiscareño SV, de Luna-Santillana EdJ, Hernández-Triana LM, Garza-Hernández JA. Canine Distemper Virus: Origins, Mutations, Diagnosis, and Epidemiology in Mexico. Life. 2024; 14(8):1002. https://doi.org/10.3390/life14081002
Chicago/Turabian StyleRivera-Martínez, Alejandra, Carlos A. Rodríguez-Alarcón, Jaime R. Adame-Gallegos, S. Viridiana Laredo-Tiscareño, Erick de Jesús de Luna-Santillana, Luis M. Hernández-Triana, and Javier A. Garza-Hernández. 2024. "Canine Distemper Virus: Origins, Mutations, Diagnosis, and Epidemiology in Mexico" Life 14, no. 8: 1002. https://doi.org/10.3390/life14081002
APA StyleRivera-Martínez, A., Rodríguez-Alarcón, C. A., Adame-Gallegos, J. R., Laredo-Tiscareño, S. V., de Luna-Santillana, E. d. J., Hernández-Triana, L. M., & Garza-Hernández, J. A. (2024). Canine Distemper Virus: Origins, Mutations, Diagnosis, and Epidemiology in Mexico. Life, 14(8), 1002. https://doi.org/10.3390/life14081002







