Adversarial Training Based Domain Adaptation of Skin Cancer Images
Abstract
1. Introduction
2. Related Work
- We present a deep learning-based methodology for unsupervised domain adaptation designed to tackle the drift and bias issues prevalent in skin lesion datasets.
- We compared the performance of AlexNet, VGG-11, and VGG-13 as feature extractors within two state-of-the-art domain adaptation frameworks, finding that VGG-13-based features yielded the best classification results.
- We compared the performance of our model with VGG-11, VGG-13, and AlexNet.
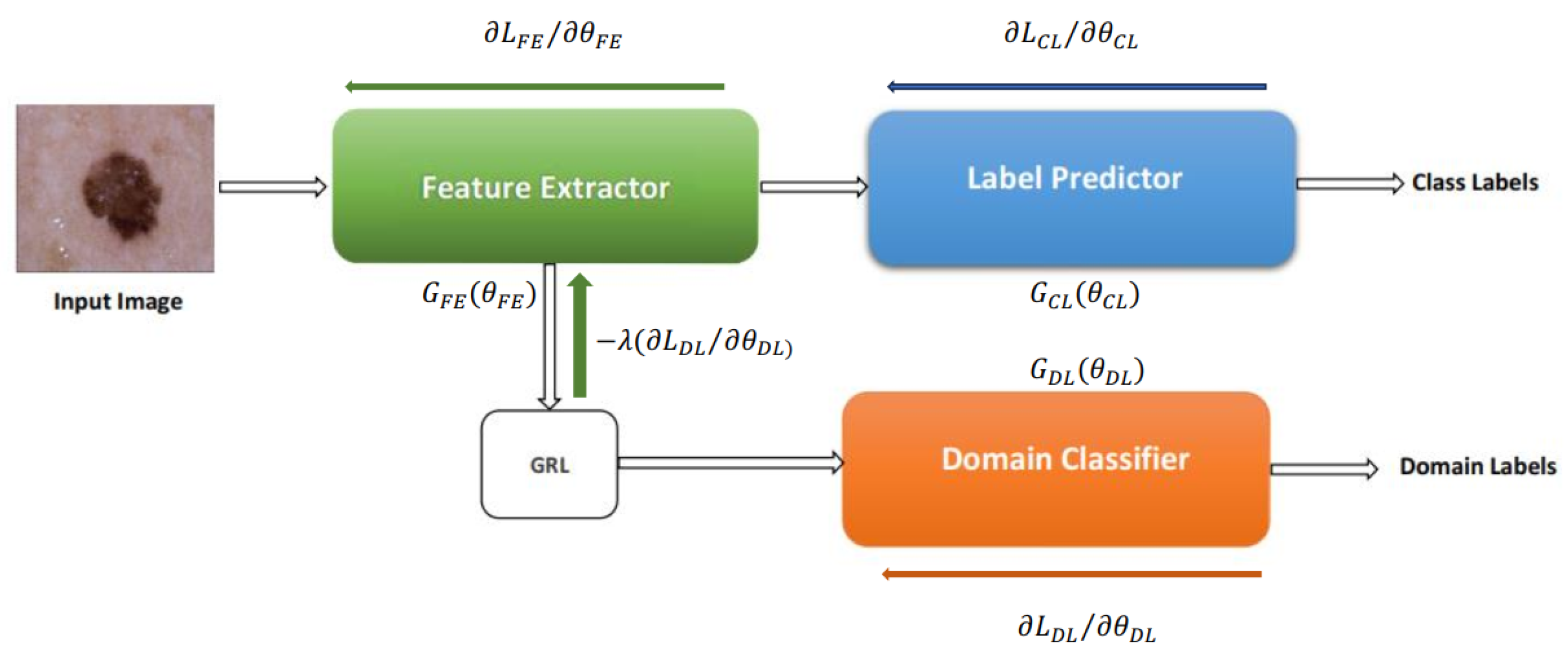
3. Overview of the Proposed Method
4. Methadology
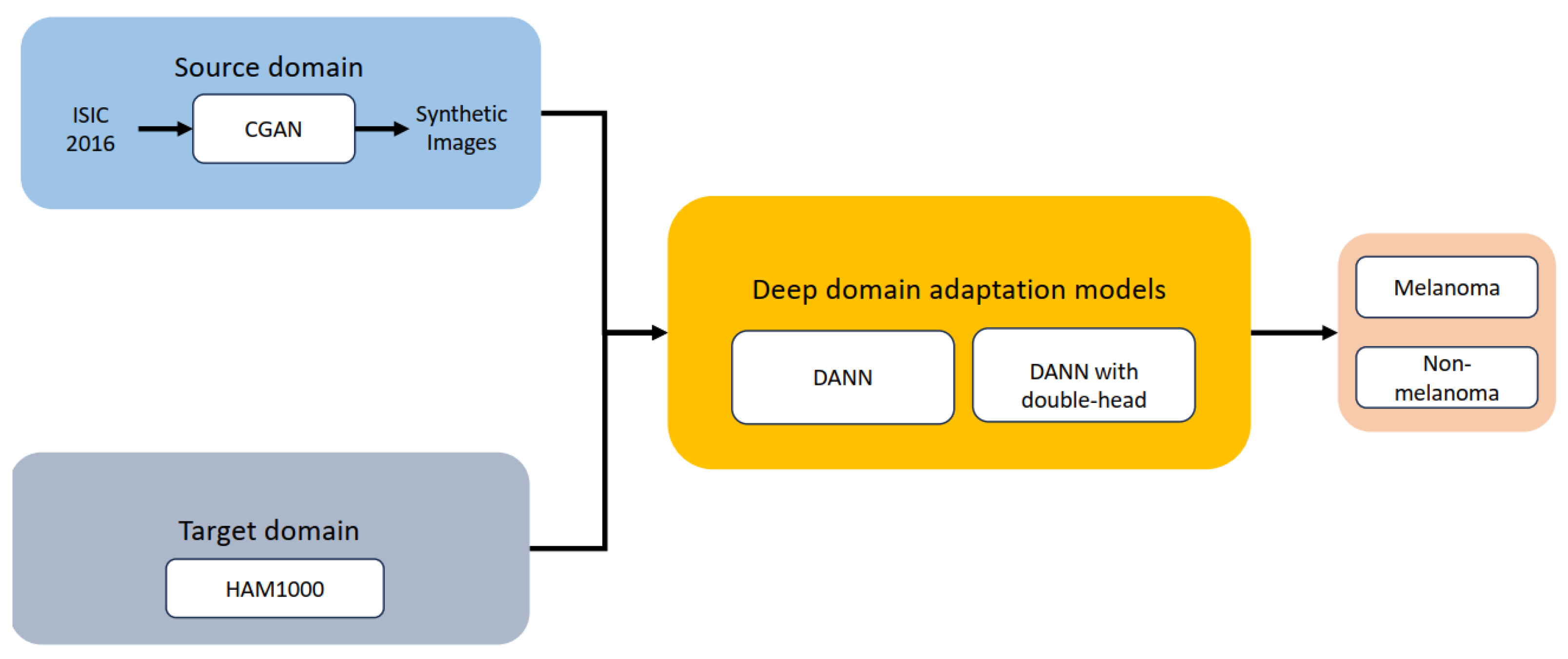
5. Experiments and Results
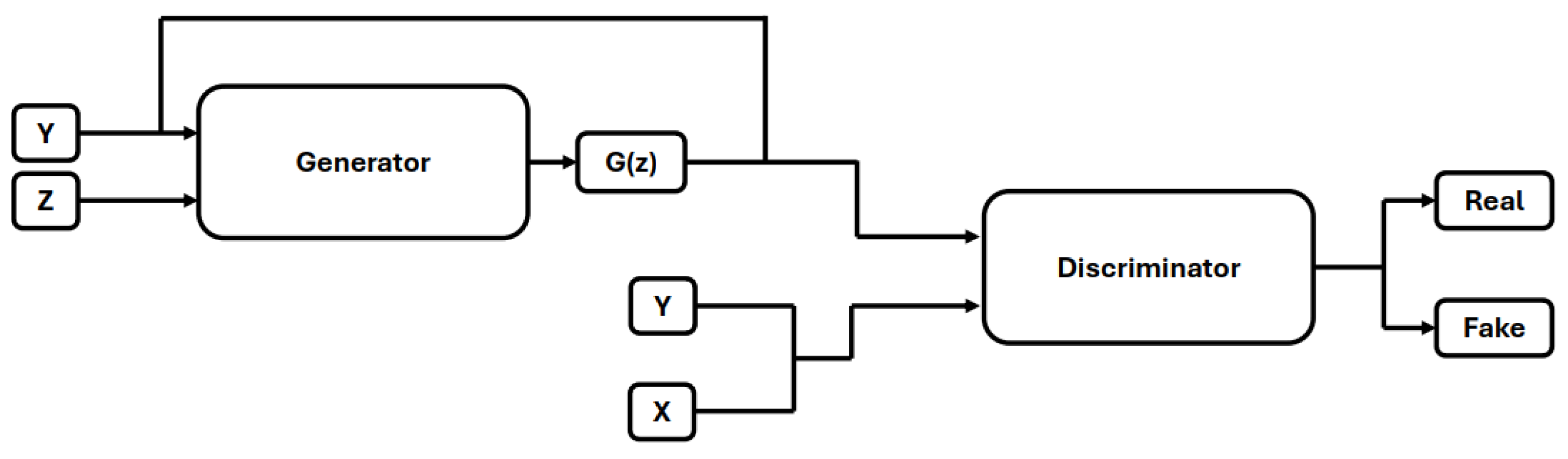
5.1. Image Generation
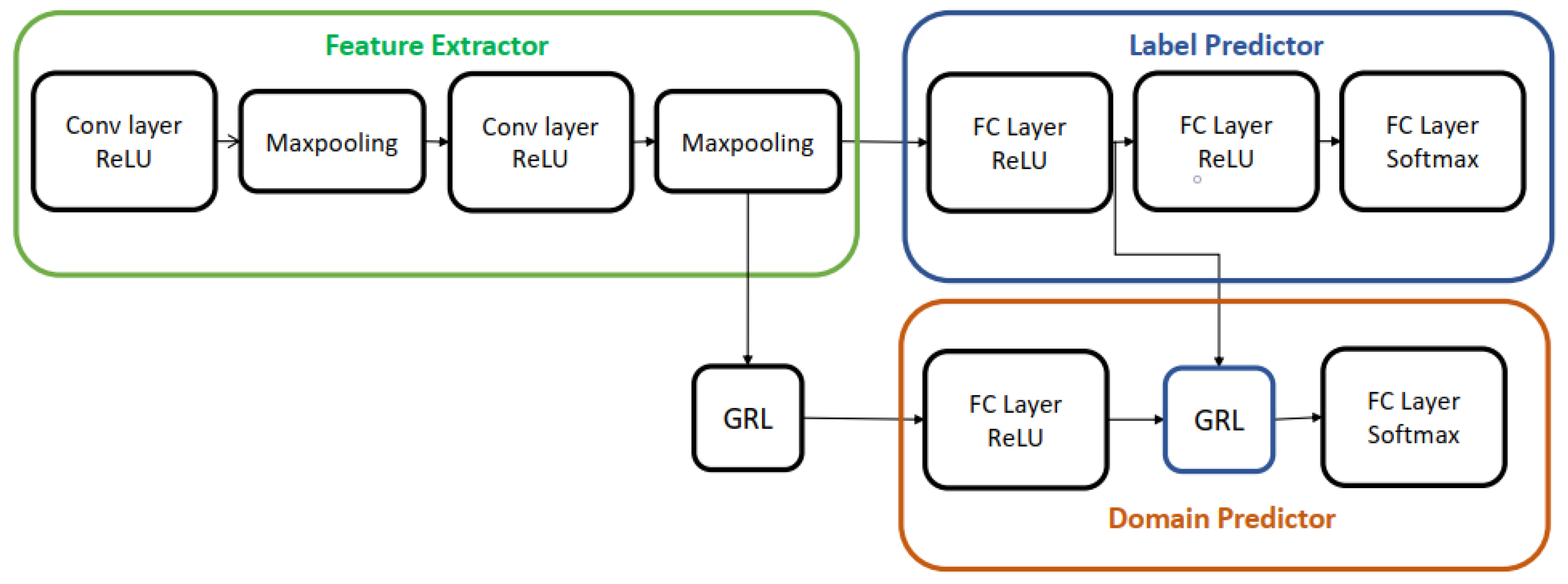
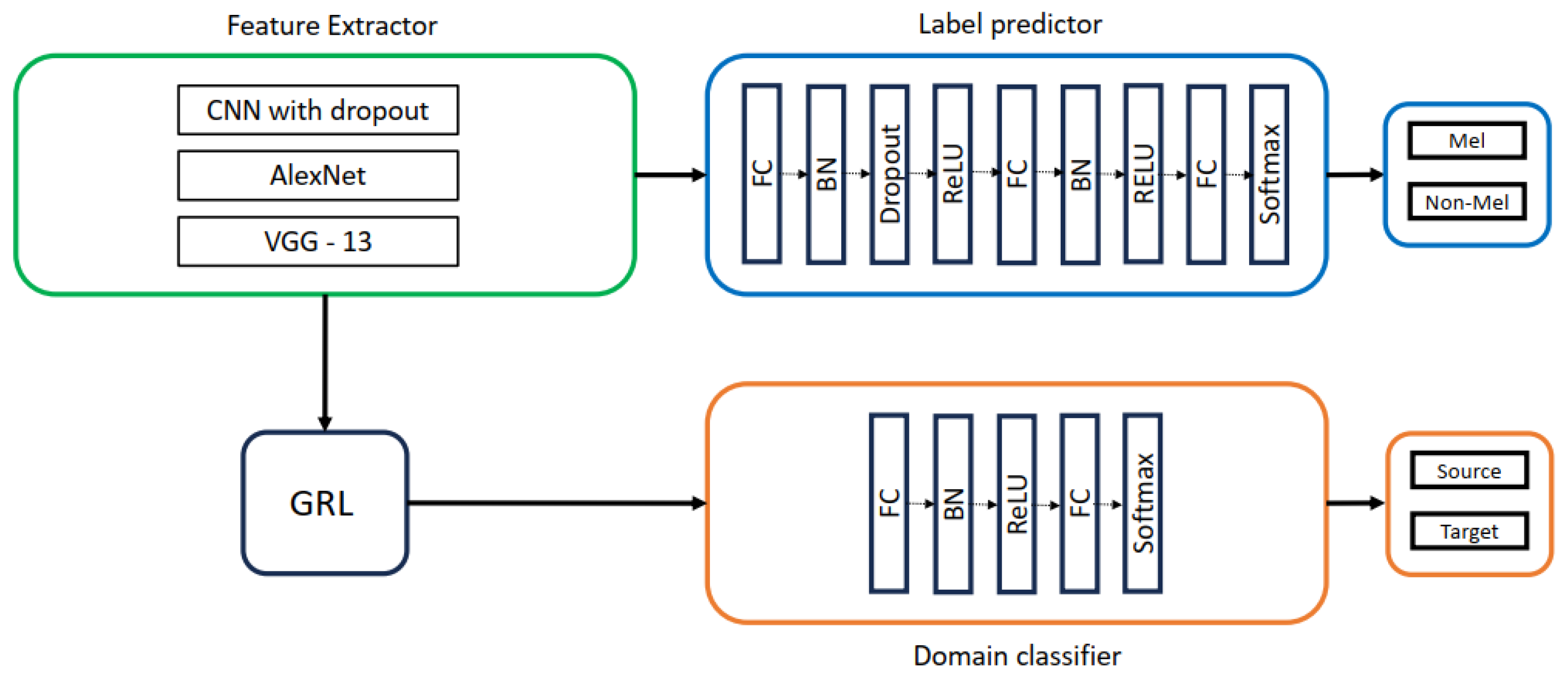
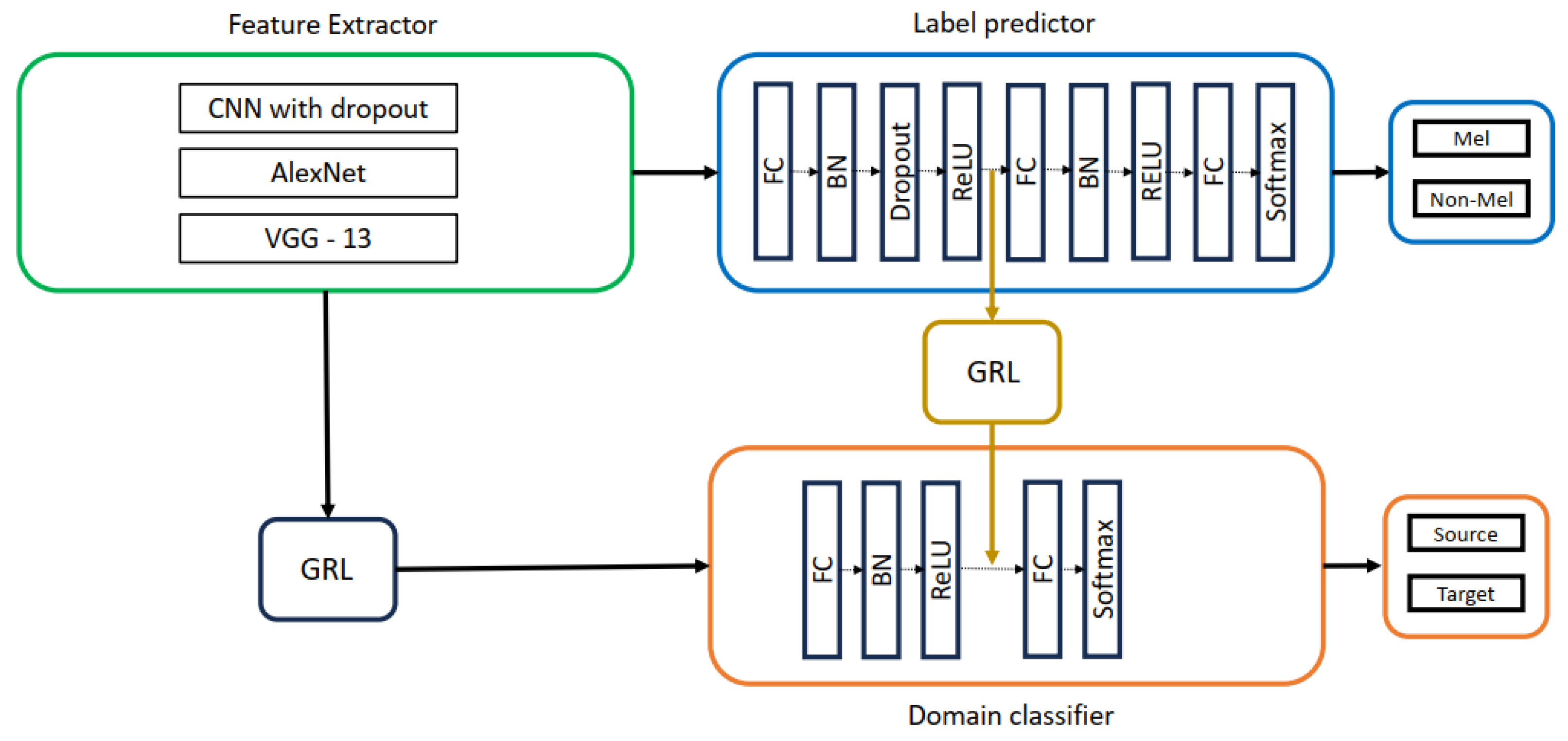
5.2. DANN Architectures Used in This Study
5.3. Dataset
5.3.1. Source Domain Dataset
5.3.2. Target Domain Dataset
5.4. Experimental Settings
5.5. Evaluation Metrics
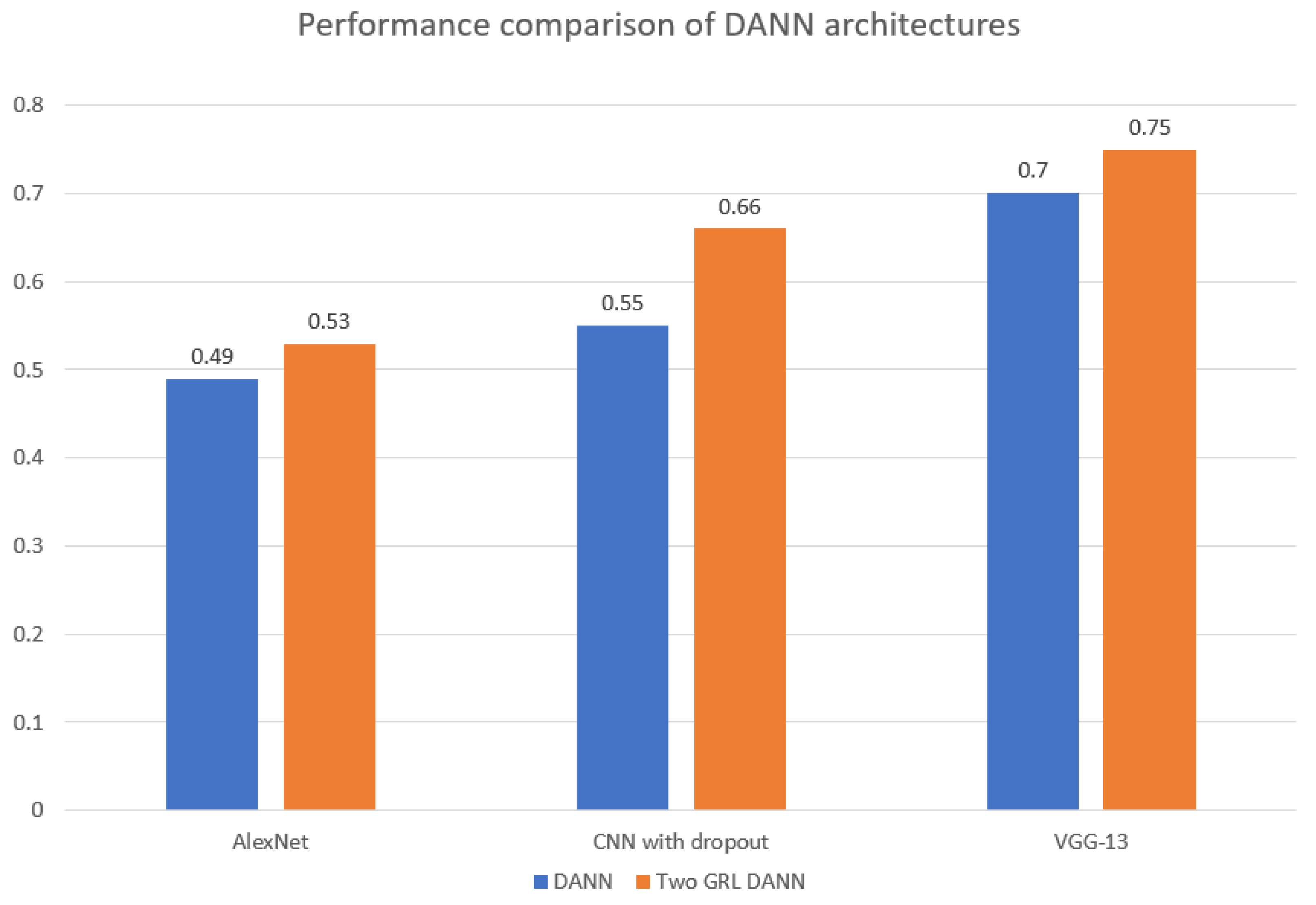
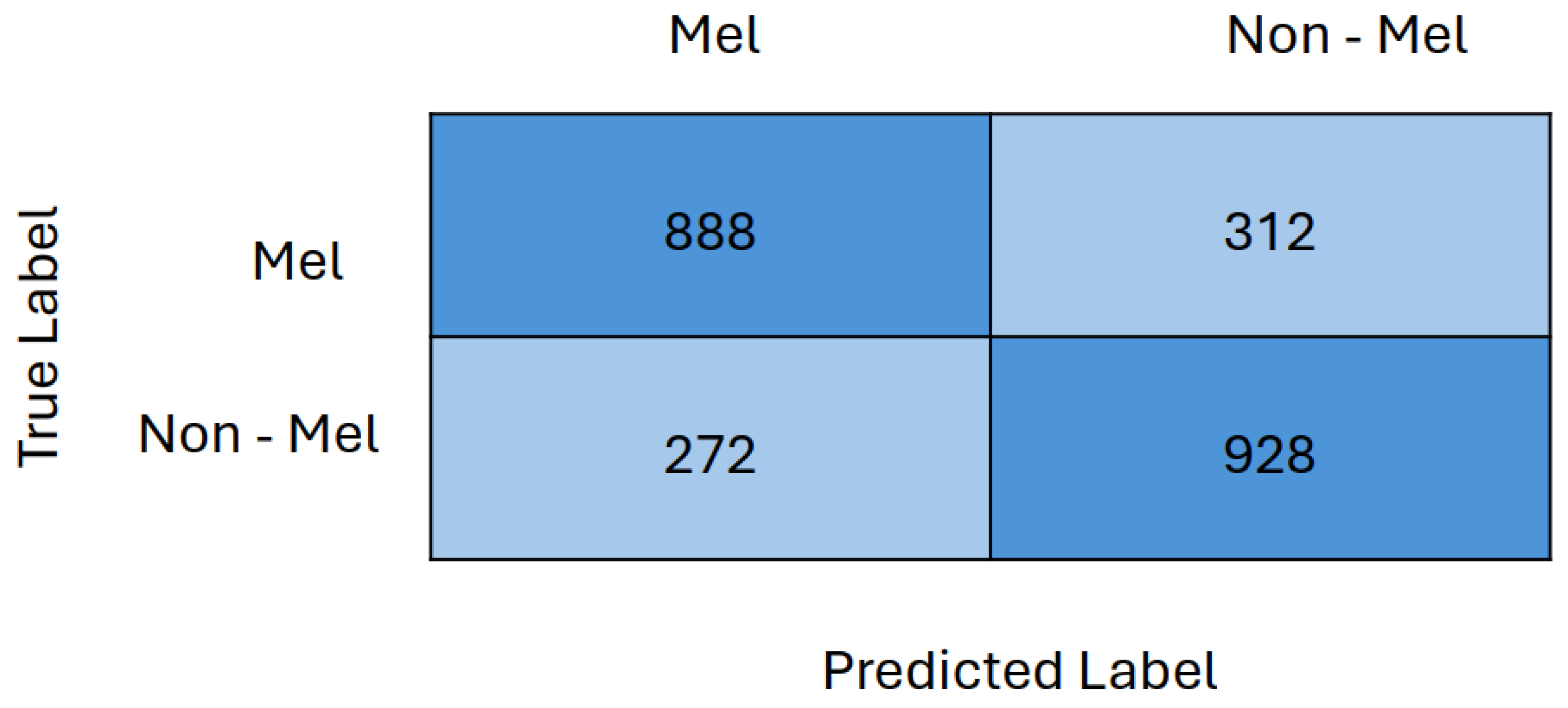
5.6. Results
5.7. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Available online: https://tinyurl.com/ptp97uzv (accessed on 12 March 2024).
- Rosendahl, C.; Tschandl, P.; Cameron, A.; Kittler, H. Diagnostic accuracy of dermatoscopy for melanocytic and nonmelanocytic pigmented lesions. J. Am. Acad. Dermatol. 2011, 64, 1068–1073. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, M.; Gilani, S.Q.; Syed, T.; Marques, O.; Kim, H.C. Skin Cancer Detection Using Deep Learning—A Review. Diagnostics 2023, 13, 1911. [Google Scholar] [CrossRef]
- Javanmardi, M.; Tasdizen, T. Domain adaptation for biomedical image segmentation using adversarial training. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 554–558. [Google Scholar]
- Available online: https://tinyurl.com/4avchh6j (accessed on 16 March 2014).
- Huang, J.; Gretton, A.; Borgwardt, K.; Schölkopf, B.; Smola, A. Correcting sample selection bias by unlabeled data. Adv. Neural Inf. Process. Syst. 2006, 19, 601–608. [Google Scholar]
- Muandet, K.; Balduzzi, D.; Schölkopf, B. Domain generalization via invariant feature representation. In Proceedings of the International Conference on Machine Learning, Atlanta, GE, USA, 17–19 June 2013; pp. 10–18. [Google Scholar]
- Pan, S.J.; Tsang, I.W.; Kwok, J.T.; Yang, Q. Domain adaptation via transfer component analysis. IEEE Trans. Neural Netw. 2010, 22, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Ghifary, M.; Balduzzi, D.; Kleijn, W.B.; Zhang, M. Scatter component analysis: A unified framework for domain adaptation and domain generalization. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1414–1430. [Google Scholar] [CrossRef] [PubMed]
- Bousmalis, K.; Silberman, N.; Dohan, D.; Erhan, D.; Krishnan, D. Unsupervised pixel-level domain adaptation with generative adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3722–3731. [Google Scholar]
- Hoffman, J.; Tzeng, E.; Park, T.; Zhu, J.Y.; Isola, P.; Saenko, K.; Efros, A.; Darrell, T. Cycada: Cycle-consistent adversarial domain adaptation. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 1989–1998. [Google Scholar]
- Li, H.; Wan, R.; Wang, S.; Kot, A.C. Unsupervised domain adaptation in the wild via disentangling representation learning. Int. J. Comput. Vis. 2021, 129, 267–283. [Google Scholar] [CrossRef]
- Long, M.; Cao, Y.; Wang, J.; Jordan, M. Learning transferable features with deep adaptation networks. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 97–105. [Google Scholar]
- Tzeng, E.; Hoffman, J.; Saenko, K.; Darrell, T. Adversarial discriminative domain adaptation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7167–7176. [Google Scholar]
- Ghifary, M.; Kleijn, W.B.; Zhang, M.; Balduzzi, D.; Li, W. Deep reconstruction-classification networks for unsupervised domain adaptation. In Proceedings of the Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Proceedings, Part IV 14. Springer: Amsterdam, The Netherlands, 2016; pp. 597–613. [Google Scholar]
- Tzeng, E.; Hoffman, J.; Darrell, T.; Saenko, K. Simultaneous deep transfer across domains and tasks. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 4068–4076. [Google Scholar]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Synergistic image and feature adaptation: Towards cross-modality domain adaptation for medical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 865–872. [Google Scholar]
- Li, H.; Wang, Y.; Wan, R.; Wang, S.; Li, T.Q.; Kot, A. Domain generalization for medical imaging classification with linear-dependency regularization. Adv. Neural Inf. Process. Syst. 2020, 33, 3118–3129. [Google Scholar]
- Ganin, Y.; Ustinova, E.; Ajakan, H.; Germain, P.; Larochelle, H.; Laviolette, F.; Marchand, M.; Lempitsky, V. Domain-adversarial training of neural networks. J. Mach. Learn. Res. 2016, 17, 2030–2096. [Google Scholar]
- Aubreville, M.; Bertram, C.A.; Jabari, S.; Marzahl, C.; Klopfleisch, R.; Maier, A. Learning New Tricks from Old Dogs–Inter-Species, Inter-Tissue Domain Adaptation for Mitotic Figure Assessment. arXiv 2019, arXiv:1911.10873. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.; Rozeira, J. PH 2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
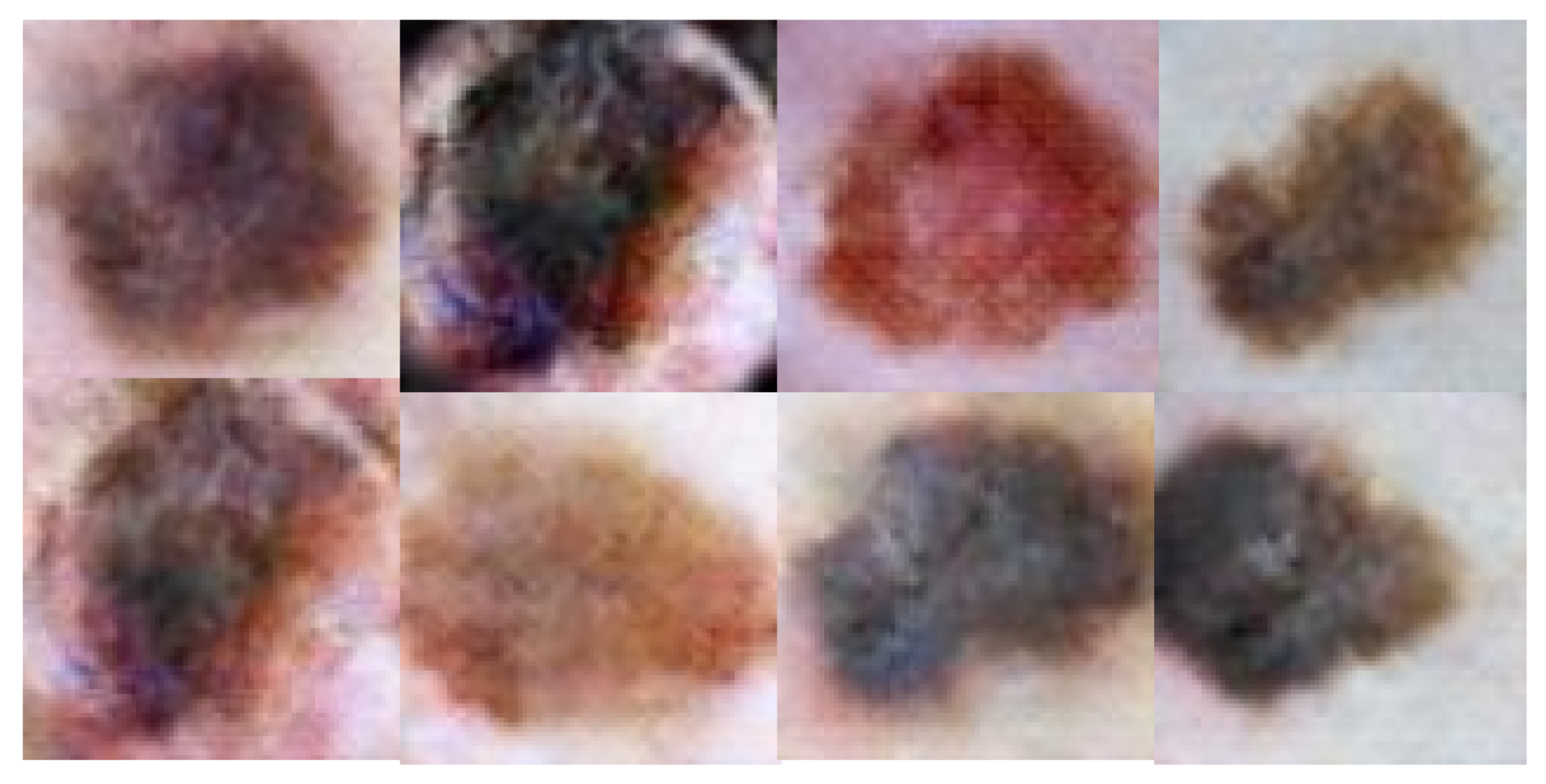

| Model | Feature Extractor | Accuracy |
|---|---|---|
| Baseline | CNN with dropout | 0.4783 |
| AlexNet | 0.5023 | |
| VGG13 | 0.5720 | |
| DANN | CNN with dropout | 0.6404 |
| AlexNet | 0.5421 | |
| VGG13 | 0.6650 | |
| Two GRL DANN | CNN with dropout | 0.6692 |
| AlexNet | 0.6332 | |
| VGG13 | 0.7567 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilani, S.Q.; Umair, M.; Naqvi, M.; Marques, O.; Kim, H.-C. Adversarial Training Based Domain Adaptation of Skin Cancer Images. Life 2024, 14, 1009. https://doi.org/10.3390/life14081009
Gilani SQ, Umair M, Naqvi M, Marques O, Kim H-C. Adversarial Training Based Domain Adaptation of Skin Cancer Images. Life. 2024; 14(8):1009. https://doi.org/10.3390/life14081009
Chicago/Turabian StyleGilani, Syed Qasim, Muhammad Umair, Maryam Naqvi, Oge Marques, and Hee-Cheol Kim. 2024. "Adversarial Training Based Domain Adaptation of Skin Cancer Images" Life 14, no. 8: 1009. https://doi.org/10.3390/life14081009
APA StyleGilani, S. Q., Umair, M., Naqvi, M., Marques, O., & Kim, H.-C. (2024). Adversarial Training Based Domain Adaptation of Skin Cancer Images. Life, 14(8), 1009. https://doi.org/10.3390/life14081009








