TMEM9B Regulates Endosomal ClC-3 and ClC-4 Transporters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. HEK Cell Culture and Transfection
2.3. In Vitro cRNA Synthesis
2.4. Oocyte Expression
2.5. Two-Electrode Voltage-Clamp
2.6. Whole-Cell Patch Clamp
2.7. FLIM-FRET and Fluorescence Confocal Imaging
2.8. Data Representation and Statistics
3. Results
3.1. Identification of TMEM9B as a Candidate Interactor of CLC Proteins
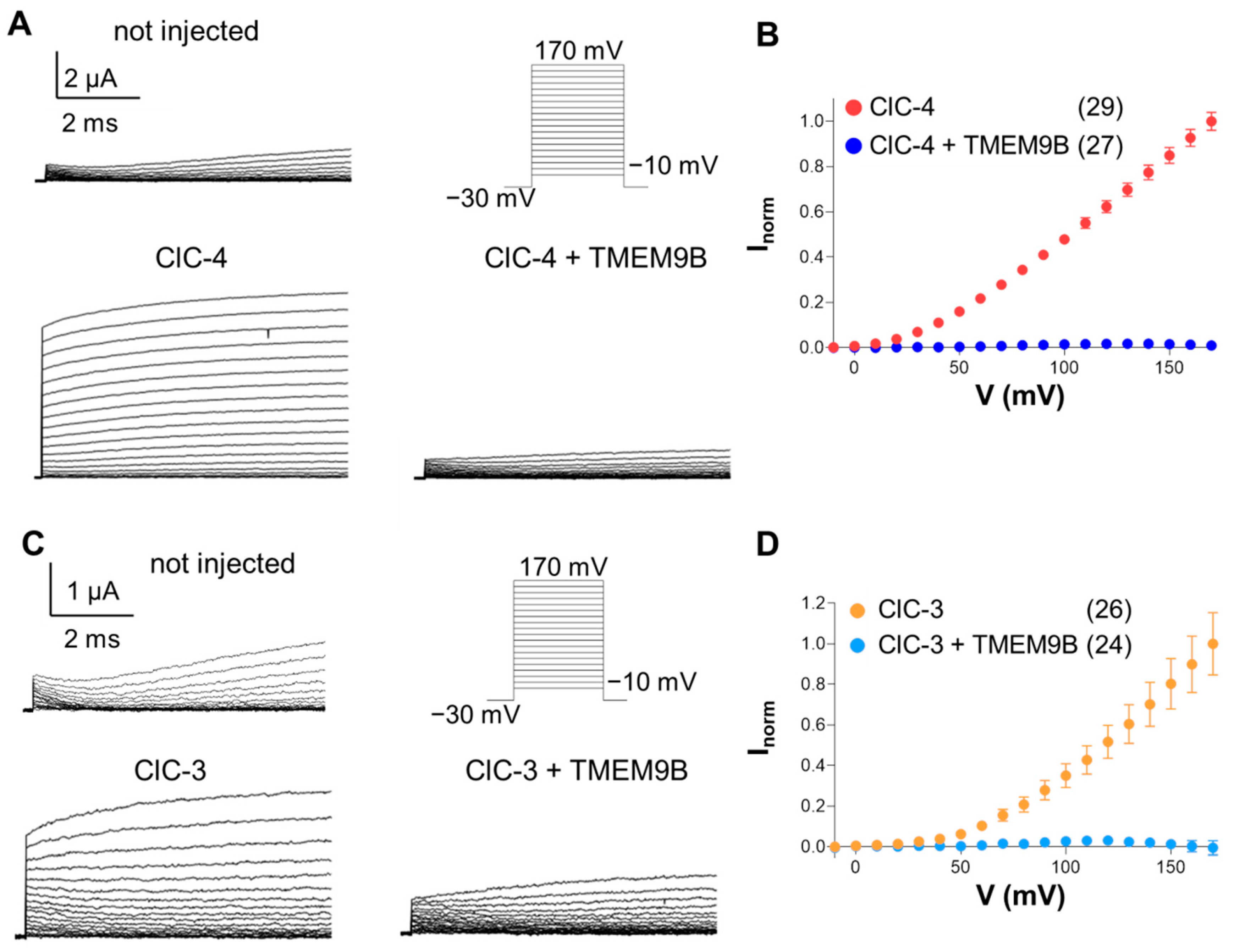
3.2. Co-Expression of CLC Proteins with TMEM9B in Xenopus Oocytes
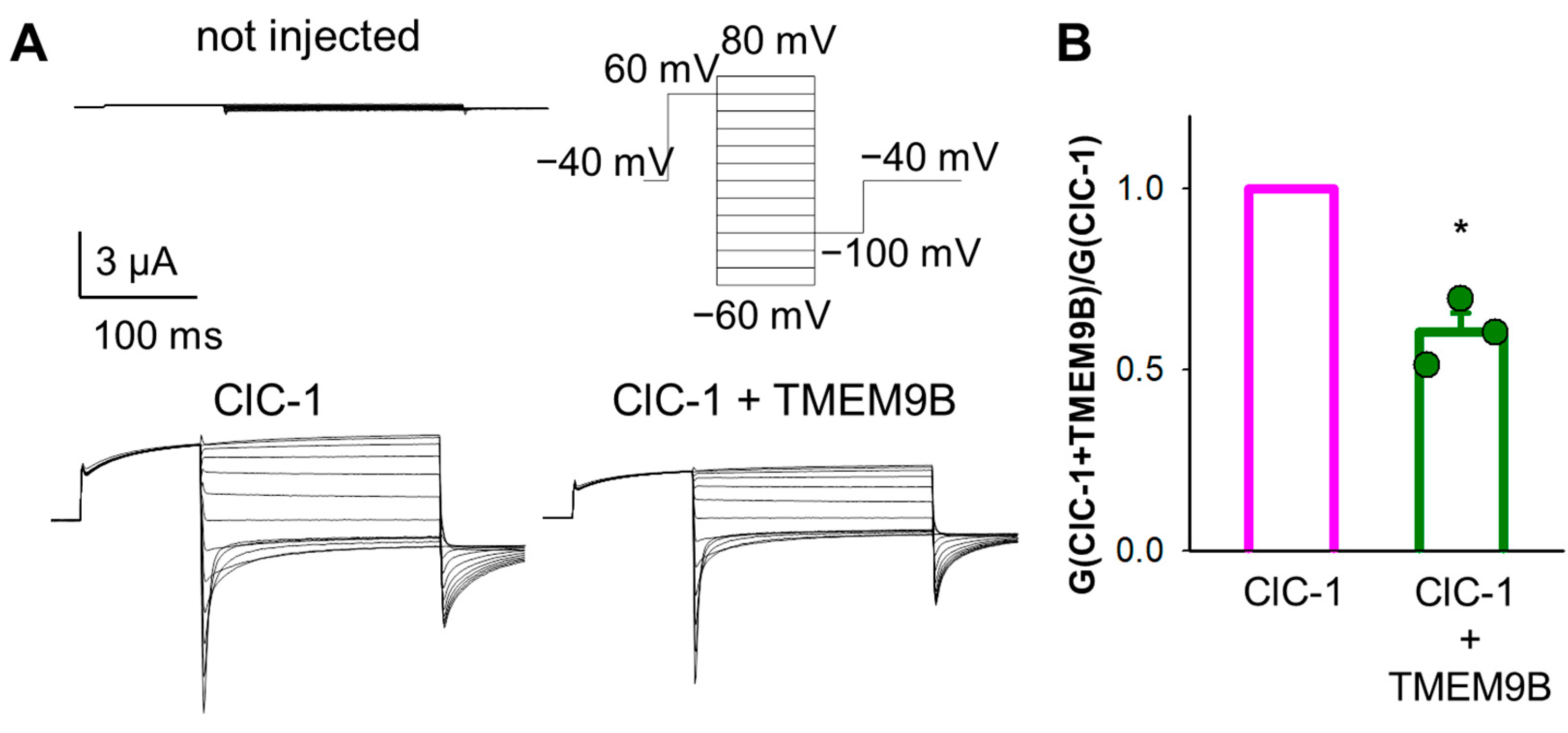
3.3. Co-Expression of CLC Proteins with TMEM9B in HEK Cells
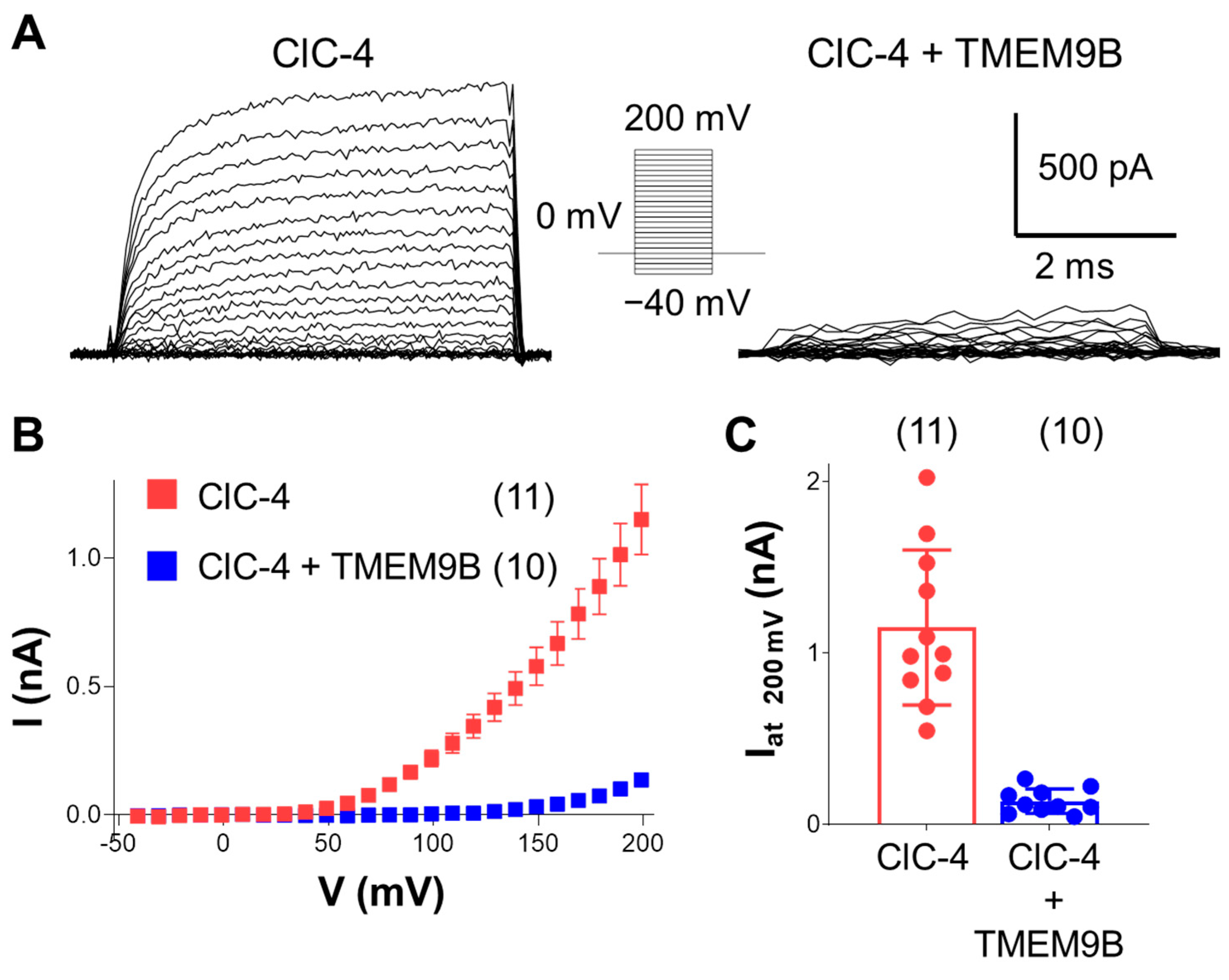
3.3.1. Co-Expression of ClC-4 with TMEM9B in HEK Cells
3.3.2. Co-Expression of ClC-3 with TMEM9B in HEK Cells
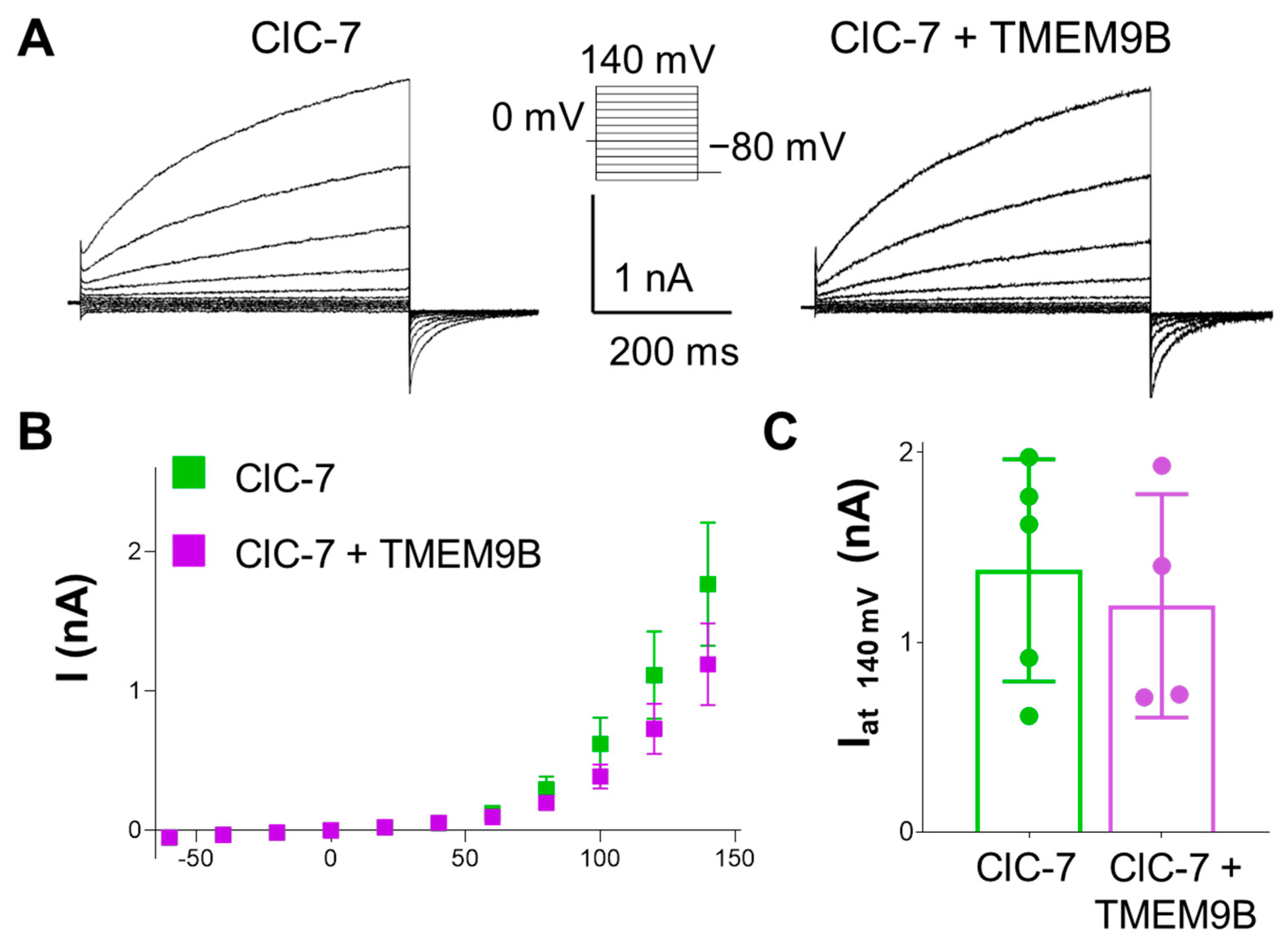
3.3.3. TMEM9B Does Not Affect the Lysosomal ClC-7 Antiporter
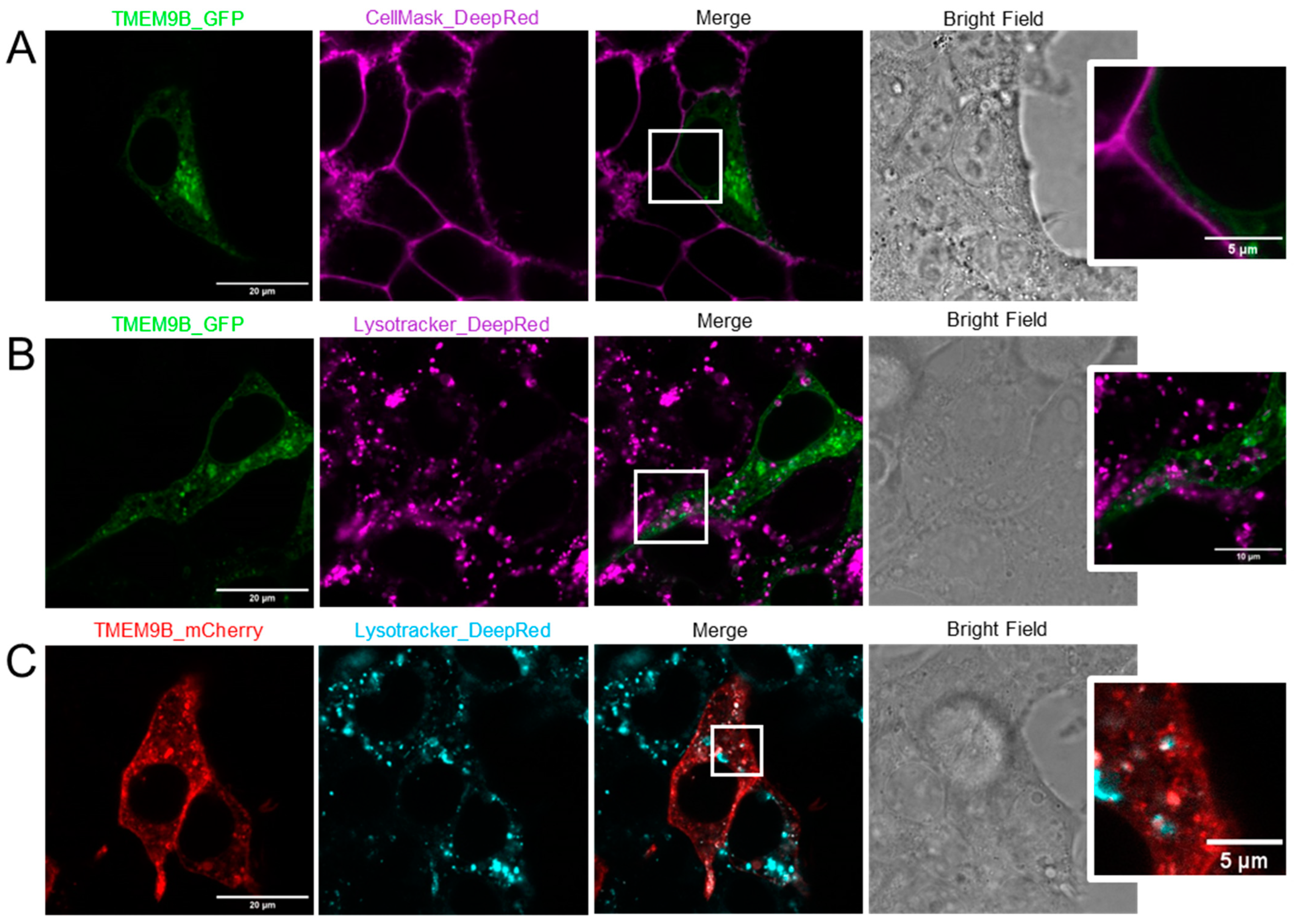
3.4. Subcellular Localization
3.5. FLIM-FRET Analysis of TMEM9B with CLC Proteins
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jentsch, T.J.; Pusch, M. CLC chloride channels and transporters: Structure, function, physiology, and disease. Physiol. Rev. 2018, 98, 1493–1590. [Google Scholar] [CrossRef]
- Miller, C. Open-state substructure of single chloride channels from Torpedo electroplax. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982, 299, 401–411. [Google Scholar]
- Middleton, R.E.; Pheasant, D.J.; Miller, C. Homodimeric architecture of a ClC-type chloride ion channel. Nature 1996, 383, 337–340. [Google Scholar] [CrossRef]
- Ludewig, U.; Pusch, M.; Jentsch, T.J. Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature 1996, 383, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Dutzler, R.; Campbell, E.B.; Cadene, M.; Chait, B.T.; MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 2002, 415, 287–294. [Google Scholar] [CrossRef]
- Polovitskaya, M.M.; Barbini, C.; Martinelli, D.; Harms, F.L.; Cole, F.S.; Calligari, P.; Bocchinfuso, G.; Stella, L.; Ciolfi, A.; Niceta, M.; et al. A Recurrent Gain-of-Function Mutation in CLCN6, Encoding the ClC-6 Cl(−)/H(+)-Exchanger, Causes Early-Onset Neurodegeneration. Am. J. Hum. Genet. 2020, 107, 1062–1077. [Google Scholar] [CrossRef]
- Duncan, A.R.; Polovitskaya, M.M.; Gaitan-Penas, H.; Bertelli, S.; VanNoy, G.E.; Grant, P.E.; O’Donnell-Luria, A.; Valivullah, Z.; Lovgren, A.K.; England, E.M.; et al. Unique variants in CLCN3, encoding an endosomal anion/proton exchanger, underlie a spectrum of neurodevelopmental disorders. Am. J. Hum. Genet. 2021, 108, 1450–1465. [Google Scholar] [CrossRef]
- Coppola, M.A.; Tettey-Matey, A.; Imbrici, P.; Gavazzo, P.; Liantonio, A.; Pusch, M. Biophysical Aspects of Neurodegenerative and Neurodevelopmental Disorders Involving Endo-/Lysosomal CLC Cl(−)/H(+) Antiporters. Life 2023, 13, 1317. [Google Scholar] [CrossRef]
- Piwon, N.; Günther, W.; Schwake, M.; Bösl, M.R.; Jentsch, T.J. ClC-5 Cl−-channel disruption impairs endocytosis in a mouse model for Dent’s disease. Nature 2000, 408, 369–373. [Google Scholar] [CrossRef]
- Guzman, R.E.; Sierra-Marquez, J.; Bungert-Plumke, S.; Franzen, A.; Fahlke, C. Functional Characterization of CLCN4 Variants Associated With X-Linked Intellectual Disability and Epilepsy. Front. Mol. Neurosci. 2022, 15, 872407. [Google Scholar] [CrossRef]
- He, H.; Guzman, R.E.; Cao, D.; Sierra-Marquez, J.; Yin, F.; Fahlke, C.; Peng, J.; Stauber, T. The molecular and phenotypic spectrum of CLCN4-related epilepsy. Epilepsia 2021, 62, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.E.; Pusch, M.; Picollo, A.; Forwood, C.; Nguyen, M.H.; Suckow, V.; Gibbons, J.; Hoff, A.; Sigfrid, L.; Megarbane, A.; et al. Functional and clinical studies reveal pathophysiological complexity of CLCN4-related neurodevelopmental condition. Mol. Psychiatry 2023, 28, 668–697. [Google Scholar] [CrossRef]
- Palmer, E.E.; Stuhlmann, T.; Weinert, S.; Haan, E.; Van Esch, H.; Holvoet, M.; Boyle, J.; Leffler, M.; Raynaud, M.; Moraine, C.; et al. De novo and inherited mutations in the X-linked gene CLCN4 are associated with syndromic intellectual disability and behavior and seizure disorders in males and females. Mol. Psychiatry 2016, 23, 222–230. [Google Scholar] [CrossRef]
- Estévez, R.; Boettger, T.; Stein, V.; Birkenhäger, R.; Otto, E.; Hildebrandt, F.; Jentsch, T.J. Barttin is a Cl− channel beta-subunit crucial for renal Cl− reabsorption and inner ear K+ secretion. Nature 2001, 414, 558–561. [Google Scholar] [CrossRef]
- Lange, P.F.; Wartosch, L.; Jentsch, T.J.; Fuhrmann, J.C. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature 2006, 440, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Jeworutzki, E.; López-Hernández, T.; Capdevila-Nortes, X.; Sirisi, S.; Bengtsson, L.; Montolio, M.; Zifarelli, G.; Arnedo, T.; Müller, C.S.; Schulte, U.; et al. GlialCAM, a protein defective in a leukodystrophy, serves as a ClC-2 Cl− channel auxiliary subunit. Neuron 2012, 73, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Günther, W.; Lüchow, A.; Cluzeaud, F.; Vandewalle, A.; Jentsch, T.J. ClC-5, the chloride channel mutated in Dent’s disease, colocalizes with the proton pump in endocytotically active kidney cells. Proc. Natl. Acad. Sci. USA 1998, 95, 8075–8080. [Google Scholar] [CrossRef]
- Lorenz, C.; Pusch, M.; Jentsch, T.J. Heteromultimeric CLC chloride channels with novel properties. Proc. Natl. Acad. Sci. USA 1996, 93, 13362–13366. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Vindas-Smith, R.; Fiore, M.; Vasquez, M.; Cuenca, P.; Del Valle, G.; Lagostena, L.; Gaitan-Penas, H.; Estevez, R.; Pusch, M.; Morales, F. Identification and functional characterization of CLCN1 mutationsg found in nondystrophic myotonia patients. Hum. Mutat. 2016, 37, 74–83. [Google Scholar] [CrossRef]
- Zanardi, I.; Zifarelli, G.; Pusch, M. An optical assay of the transport activity of ClC-7. Sci. Rep. 2013, 3, 1231. [Google Scholar] [CrossRef]
- Picollo, A. Vesicular CLC chloride/proton exchangers in health and diseases. Front. Pharmacol. 2023, 14, 1295068. [Google Scholar] [CrossRef]
- Coppola, M.A.; Gavazzo, P.; Zanardi, I.; Tettey-Matey, A.; Liantonio, A.; Fong, P.; Pusch, M. Distinct ClC-6 and ClC-7 Cl(−) sensitivities provide insight into ClC-7’s role in lysosomal Cl(−) homeostasis. J. Physiol. 2023, 601, 5635–5653. [Google Scholar] [CrossRef]
- Zifarelli, G.; Pusch, M.; Fong, P. Altered voltage-dependence of slowly activating chloride-proton antiport by late endosomal ClC-6 explains distinct neurological disorders. J. Physiol. 2022, 600, 2147–2164. [Google Scholar] [CrossRef]
- Ranjit, S.; Malacrida, L.; Jameson, D.M.; Gratton, E. Fit-free analysis of fluorescence lifetime imaging data using the phasor approach. Nat. Protoc. 2018, 13, 1979–2004. [Google Scholar] [CrossRef]
- Dodeller, F.; Gottar, M.; Huesken, D.; Iourgenko, V.; Cenni, B. The lysosomal transmembrane protein 9B regulates the activity of inflammatory signaling pathways. J. Biol. Chem. 2008, 283, 21487–21494. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef]
- Jung, Y.S.; Jun, S.; Kim, M.J.; Lee, S.H.; Suh, H.N.; Lien, E.M.; Jung, H.Y.; Lee, S.; Zhang, J.; Yang, J.I.; et al. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/beta-catenin signalling. Nat. Cell Biol. 2018, 20, 1421–1433. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, P.; Tian, K.; Qiao, Z.; Dong, H.; Li, J.; Guan, Z.; Su, H.; Song, Y.; Ma, X. TMEM9 promotes lung adenocarcinoma progression via activating the MEK/ERK/STAT3 pathway to induce VEGF expression. Cell Death Dis. 2024, 15, 295. [Google Scholar] [CrossRef]
- Guzman, R.E.; Miranda-Laferte, E.; Franzen, A.; Fahlke, C. Neuronal ClC-3 splice variants differ in subcellular localizations, but mediate identical transport functions. J. Biol. Chem. 2015, 290, 25851–25862. [Google Scholar] [CrossRef]
- Albertazzi, L.; Arosio, D.; Marchetti, L.; Ricci, F.; Beltram, F. Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem. Photobiol. 2009, 85, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Schrecker, M.; Korobenko, J.; Hite, R.K. Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1. eLife 2020, 9, e59555. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Y.; Zhang, B.; Zhou, J.; Li, T.; Liu, Z.; Li, Y.; Yang, M. Molecular insights into the human CLC-7/Ostm1 transporter. Sci. Adv. 2020, 6, eabb4747. [Google Scholar] [CrossRef] [PubMed]
- Steinmeyer, K.; Schwappach, B.; Bens, M.; Vandewalle, A.; Jentsch, T.J. Cloning and functional expression of rat CLC-5, a chloride channel related to kidney disease. J. Biol. Chem. 1995, 270, 31172–31177. [Google Scholar] [CrossRef]
- Lloyd, S.E.; Pearce, S.H.; Fisher, S.E.; Steinmeyer, K.; Schwappach, B.; Scheinman, S.J.; Harding, B.; Bolino, A.; Devoto, M.; Goodyer, P.; et al. A common molecular basis for three inherited kidney stone diseases. Nature 1996, 379, 445–449. [Google Scholar] [CrossRef]
- Zifarelli, G.; Pusch, M. CLC chloride channels and transporters: A biophysical and physiological perspective. Rev. Physiol. Biochem. Pharmacol. 2007, 158, 23–76. [Google Scholar]
- Kveine, M.; Tenstad, E.; Dosen, G.; Funderud, S.; Rian, E. Characterization of the novel human transmembrane protein 9 (TMEM9) that localizes to lysosomes and late endosomes. Biochem. Biophys. Res. Commun. 2002, 297, 912–917. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Festa, M.; Coppola, M.A.; Angeli, E.; Tettey-Matey, A.; Giusto, A.; Mazza, I.; Gatta, E.; Barbieri, R.; Picollo, A.; Gavazzo, P.; et al. TMEM9B Regulates Endosomal ClC-3 and ClC-4 Transporters. Life 2024, 14, 1034. https://doi.org/10.3390/life14081034
Festa M, Coppola MA, Angeli E, Tettey-Matey A, Giusto A, Mazza I, Gatta E, Barbieri R, Picollo A, Gavazzo P, et al. TMEM9B Regulates Endosomal ClC-3 and ClC-4 Transporters. Life. 2024; 14(8):1034. https://doi.org/10.3390/life14081034
Chicago/Turabian StyleFesta, Margherita, Maria Antonietta Coppola, Elena Angeli, Abraham Tettey-Matey, Alice Giusto, Irene Mazza, Elena Gatta, Raffaella Barbieri, Alessandra Picollo, Paola Gavazzo, and et al. 2024. "TMEM9B Regulates Endosomal ClC-3 and ClC-4 Transporters" Life 14, no. 8: 1034. https://doi.org/10.3390/life14081034
APA StyleFesta, M., Coppola, M. A., Angeli, E., Tettey-Matey, A., Giusto, A., Mazza, I., Gatta, E., Barbieri, R., Picollo, A., Gavazzo, P., Pusch, M., Picco, C., & Sbrana, F. (2024). TMEM9B Regulates Endosomal ClC-3 and ClC-4 Transporters. Life, 14(8), 1034. https://doi.org/10.3390/life14081034







