Xenobiology for the Biocontainment of Synthetic Organisms: Opportunities and Challenges
Abstract
1. Background
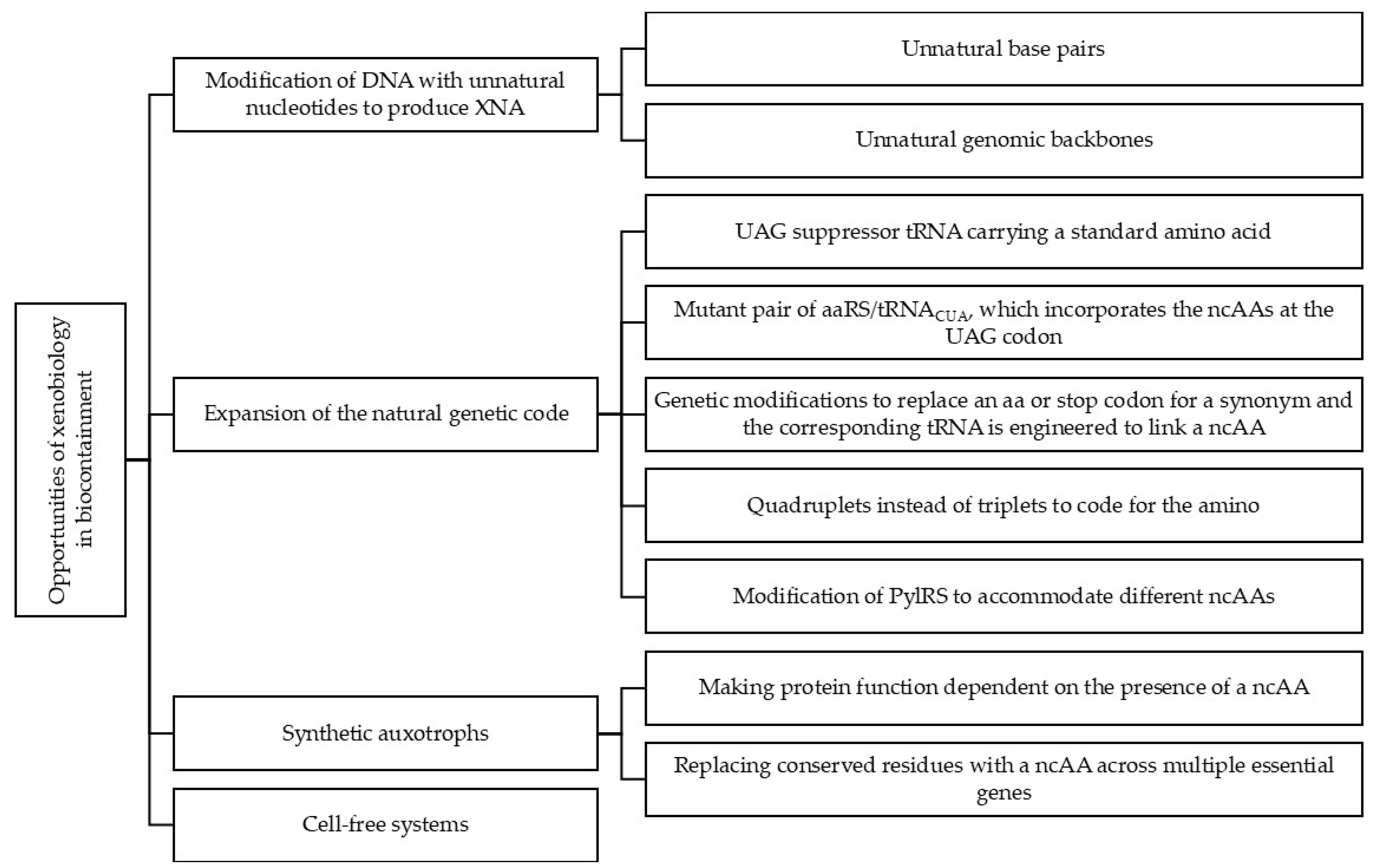
2. Opportunities of Xenobiology in Biocontainment
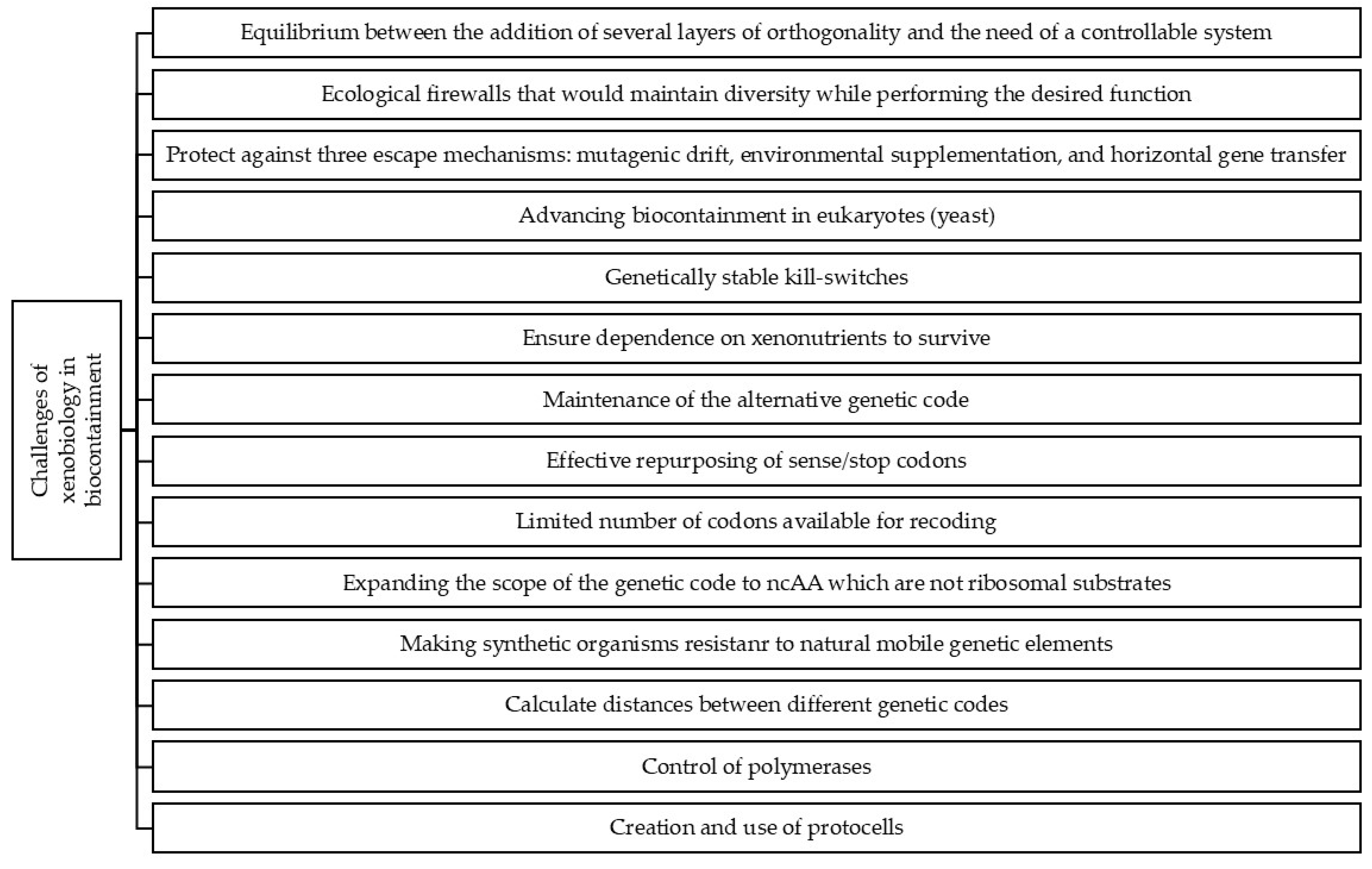
3. Challenges of Xenobiology in Biocontainment
4. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wooster, H. Xenobiology. Science 1961, 134, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Pei, L. Synthetic Toxicology: Where Engineering Meets Biology and Toxicology. Toxicol. Sci. 2011, 120 (Suppl. S1), S204–S224. [Google Scholar] [CrossRef] [PubMed]
- Deplazes, A. Piecing together a puzzle. An exposition of synthetic biology. EMBO Rep. 2009, 10, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Synthetic Biology II—Risk Assessment Methodologies and Safety Aspects. Available online: https://health.ec.europa.eu/publications/synthetic-biology-ii-risk-assessment-methodologies-and-safety-aspects_en (accessed on 23 February 2024).
- Schmidt, M.; Pei, L.; Budisa, N. Xenobiology: State-of-the-art, ethics, and philosophy of new-to-nature organisms. Adv. Biochem. Eng. Biotechnol. 2018, 162, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Budisa, N. The Alanine World Model for the Development of the Amino Acid Repertoire in Protein Biosynthesis. Int. J. Mol. Sci. 2019, 20, 5507. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Budisa, N. Anticipating alien cells with alternative genetic codes: Away from the alanine world! Curr. Opin. Biotechnol. 2019, 60, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Berg, P.; Baltimore, D.; Brenner, S.; Roblin, R.O.; Singer, M.F. Summary Statement of the Asilomar Conference on Recombinant DNA Molecules. Proc. Natl. Acad. Sci. USA 1975, 72, 1981–1984. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Rocha, C.G. The Synthetic Nature of Biology. In Ambivalences of Creating Life: Societal and Philosophical Dimensions of Synthetic Biology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 45, pp. 9–53. [Google Scholar] [CrossRef]
- Rovner, A.J.; Haimovich, A.D.; Katz, S.R.; Li, Z.; Grome, M.W.; Gassaway, B.M.; Amiram, M.; Patel, J.R.; Gallagher, R.R.; Rinehart, J.; et al. Recoded Organisms Engineered to Depend on Synthetic Amino Acids. Nature 2015, 518, 89–93. [Google Scholar] [CrossRef]
- Chaput, J.C.; Herdewijn, P.; Hollenstein, M. Orthogonal Genetic Systems. Chembiochem 2020, 21, 1408–1411. [Google Scholar] [CrossRef]
- Kubyshkin, V.; Budisa, N. Synthetic Alienation of Microbial Organisms by Using Genetic Code Engineering: Why and How? Biotechnol. J. 2017, 12, 1600097. [Google Scholar] [CrossRef]
- Schmidt, M. A Metric Space for Semantic Containment: Towards the Implementation of Genetic Firewalls. Biosystems 2019, 185, 104015. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M. Xenobiology: A New Form of Life as the Ultimate Biosafety Tool. Bioessays 2010, 32, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Torres, L.; Krüger, A.; Csibra, E.; Gianni, E.; Pinheiro, V.B. Synthetic biology approaches to biological containment: Pre-emptively tackling potential risks. Essays. Biochem. 2016, 60, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.J. NIH Guidelines for Research Involving Recombinant DNA Molecules. Account. Res. 1993, 3, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Diwo, C.; Budisa, N. Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation. Genes 2018, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.A.; Diggans, J.; Densmore, D.; Dai, J.; Knight, T.; Leproust, E.; Boeke, J.D.; Wheeler, N.; Cai, Y. Safety by design: Biosafety and biosecurity in the age of synthetic genomics. iScience 2023, 26, 106165. [Google Scholar] [CrossRef] [PubMed]
- Vladilo, G.; Hassanali, A. Hydrogen Bonds and Life in the Universe. Life 2018, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Beiranvand, N.; Freindorf, M.; Kraka, E. Hydrogen Bonding in Natural and Unnatural Base Pairs-A Local Vibrational Mode Study. Molecules 2021, 26, 2268. [Google Scholar] [CrossRef] [PubMed]
- Herdewijn, P.; Marlière, P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009, 6, 791–808. [Google Scholar] [CrossRef]
- Benner, S.A.; Karalkar, N.B.; Hoshika, S.; Laos, R.; Shaw, R.W.; Matsuura, M.; Fajardo, D.; Moussatche, P. Alternative Watson-Crick Synthetic Genetic Systems. Cold Spring Harb. Perspect. Biol. 2016, 8, a023770. [Google Scholar] [CrossRef]
- Kim, H.J.; Leal, N.A.; Hoshika, S.; Benner, S.A. Ribonucleosides for an artificially expanded genetic information system. J. Org. Chem. 2014, 79, 3194–3199. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jiménez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D.; et al. In Vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef]
- Zhang, Y.; Ptacin, J.L.; Fischer, E.C.; Aerni, H.R.; Caffaro, C.E.; San Jose, K.; Feldman, A.W.; Turner, C.R.; Romesberg, F.E. A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551, 644–647. [Google Scholar] [CrossRef]
- Hoshika, S.; Leal, N.A.; Kim, M.J.; Kim, M.S.; Karalkar, N.B.; Kim, H.J.; Bates, A.M.; Watkins, N.E., Jr.; SantaLucia, H.A.; Meyer, A.J.; et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 2019, 363, 884–887. [Google Scholar] [CrossRef]
- Futami, K.; Kimoto, M.; Lim, Y.W.S.; Hirao, I. Genetic Alphabet Expansion Provides Versatile Specificities and Activities of Unnatural-Base DNA Aptamers Targeting Cancer Cells. Mol. Ther. Nucleic Acids 2019, 14, 158–170. [Google Scholar] [CrossRef]
- Malyshev, D.A.; Dhami, K.; Lavergne, T.; Chen, T.; Dai, N.; Foster, J.M.; Corrêa, I.R., Jr.; Romesberg, F.E. A semi-synthetic organism with an expanded genetic alphabet. Nature 2014, 509, 385–388. [Google Scholar] [CrossRef]
- Dien, V.T.; Morris, S.E.; Karadeema, R.J.; Romesberg, F.E. Expansion of the genetic code via expansion of the genetic alphabet. Curr. Opin. Chem. Biol. 2018, 46, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Dien, V.T.; Holcomb, M.; Feldman, A.W.; Fischer, E.C.; Dwyer, T.J.; Romesberg, F.E. Progress Toward a Semi-Synthetic Organism with an Unrestricted Expanded Genetic Alphabet. J. Am. Chem. Soc. 2018, 140, 16115–16123. [Google Scholar] [CrossRef]
- Budisa, N. Expanded genetic code for the engineering of ribosomally synthetized and post-translationally modified peptide natural products (RiPPs). Curr. Opin. Biotechnol. 2013, 24, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y. Translational Control Using an Expanded Genetic Code. Int. J. Mol. Sci. 2019, 20, 887. [Google Scholar] [CrossRef]
- Fredens, J.; Wang, K.; de la Torre, D.; Funke, L.F.H.; Robertson, W.E.; Christova, Y.; Chia, T.; Schmied, W.H.; Dunkelmann, D.L.; Beránek, V.; et al. Total Synthesis of Escherichia Coli with a Recoded Genome. Nature 2019, 569, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Lajoie, M.J.; Xiao, H.; Church, G.M.; Schultz, P.G. A bacterial strain with a unique quadruplet codon specifying non-native amino acids. ChemBioChem 2014, 15, 1782–1786. [Google Scholar] [CrossRef]
- Fan, C.; Xiong, H.; Reynolds, N.M.; Söll, D. Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic. Acids. Res. 2015, 43, e156. [Google Scholar] [CrossRef] [PubMed]
- Crnković, A.; Suzuki, T.; Söll, D.; Reynolds, N.M. Pyrrolysyl-tRNA Synthetase, an Aminoacyl-tRNA Synthetase for Genetic Code Expansion. Croat. Chem. Acta 2016, 89, 163–174. [Google Scholar] [CrossRef]
- Koch, N.G.; Baumann, T.; Budisa, N. Efficient Unnatural Protein Production by Pyrrolysyl-tRNA Synthetase With Genetically Fused Solubility Tags. Front. Bioeng. Biotechnol. 2021, 9, 807438. [Google Scholar] [CrossRef]
- Mayer, C. Selection, Addiction and Catalysis: Emerging Trends for the Incorporation of Noncanonical Amino Acids into Peptides and Proteins In Vivo. ChemBioChem 2019, 20, 1357–1364. [Google Scholar] [CrossRef]
- Lee, J.W.; Chan, C.T.Y.; Slomovic, S.; Collins, J.J. Next-Generation Biocontainment Systems for Engineered Organisms. Nat. Chem. Biol. 2018, 14, 530–537. [Google Scholar] [CrossRef]
- Chang, T.; Ding, W.; Yan, S.; Wang, Y.; Zhang, H.; Zhang, Y.; Ping, Z.; Zhang, H.; Huang, Y.; Zhang, J.; et al. A Robust Yeast Biocontainment System with Two-Layered Regulation Switch Dependent on Unnatural Amino Acid. Nat. Commun. 2023, 14, 6487. [Google Scholar] [CrossRef] [PubMed]
- Orelle, C.; Carlson, E.D.; Szal, T.; Florin, T.; Jewett, M.C.; Mankin, A.S. Protein Synthesis by Ribosomes with Tethered Subunits. Nature 2015, 524, 119–124. [Google Scholar] [CrossRef]
- Jia, B.; Qi, H.; Li, B.-Z.; Pan, S.; Liu, D.; Liu, H.; Cai, Y.; Yuan, Y.-J. Orthogonal Ribosome Biofirewall. ACS Synth. Biol. 2017, 6, 2108–2117. [Google Scholar] [CrossRef]
- Kato, Y. An Engineered Bacterium Auxotrophic for an Unnatural Amino Acid: A Novel Biological Containment System. PeerJ 2015, 3, e1247. [Google Scholar] [CrossRef] [PubMed]
- Mandell, D.J.; Lajoie, M.J.; Mee, M.T.; Takeuchi, R.; Kuznetsov, G.; Norville, J.E.; Gregg, C.J.; Stoddard, B.L.; Church, G.M. Biocontainment of Genetically Modified Organisms by Synthetic Protein Design. Nature 2015, 518, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Tack, D.S.; Ellefson, J.W.; Thyer, R.; Wang, B.; Gollihar, J.; Forster, M.T.; Ellington, A.D. Addicting Diverse Bacteria to a Noncanonical Amino Acid. Nat. Chem. Biol. 2016, 12, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Schultz, P.G. A Strategy for Creating Organisms Dependent on Noncanonical Amino Acids. Angew. Chem. Weinh. Bergstr. Ger. Weinh. 2017, 129, 9298–9301. [Google Scholar] [CrossRef]
- Koh, M.; Yao, A.; Gleason, P.R.; Mills, J.H.; Schultz, P.G. A General Strategy for Engineering Noncanonical Amino Acid Dependent Bacterial Growth. J. Am. Chem. Soc. 2019, 141, 16213–16216. [Google Scholar] [CrossRef] [PubMed]
- Saccharomyces Cerevisiae 2.0. Available online: https://syntheticyeast.github.io/ (accessed on 23 February 2024).
- Kato, Y. Tight Translational Control Using Site-Specific Unnatural Amino Acid Incorporation with Positive Feedback Gene Circuits. ACS Synth. Biol. 2018, 7, 1956–1963. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yuan, Z.; Niu, W.; Li, Q.; Guo, J. Synthetic Biology Approach for the Development of Conditionally Replicating HIV-1 Vaccine. J. Chem. Technol. Biotechnol. 2017, 92, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Lai, Y.; Fuerte-Stone, J.; Mimee, M.; Lu, T.K. Cas9-assisted biological containment of a genetically engineered human commensal bacterium and genetic elements. Nat. Commun. 2024, 15, 2096. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Tozaki, M.; Murakami, H. An Amino Acid-Swapped Genetic Code. ACS Synth. Biol. 2020, 9, 2703–2713. [Google Scholar] [CrossRef]
- Whitford, C.M.; Dymek, S.; Kerkhoff, D.; März, C.; Schmidt, O.; Edich, M.; Droste, J.; Pucker, B.; Rückert, C.; Kalinowski, J. Auxotrophy to Xeno-DNA: An Exploration of Combinatorial Mechanisms for a High-Fidelity Biosafety System for Synthetic Biology Applications. J. Biol. Eng. 2018, 12, 13. [Google Scholar] [CrossRef]
- Chemla, Y.; Ozer, E.; Schlesinger, O.; Noireaux, V.; Alfonta, L. Genetically Expanded Cell-Free Protein Synthesis Using Endogenous Pyrrolysyl Orthogonal Translation System: Genetically Expanded Cell-Free Protein Synthesis. Biotechnol. Bioeng. 2015, 112, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Des Soye, B.J.; Patel, J.R.; Isaacs, F.J.; Jewett, M.C. Repurposing the Translation Apparatus for Synthetic Biology. Curr. Opin. Chem. Biol. 2015, 28, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Ntai, I.; Haimovich, A.D.; Kelleher, N.L.; Isaacs, F.J.; Jewett, M.C. Cell-Free Protein Synthesis from a Release Factor 1 Deficient Escherichia Coli Activates Efficient and Multiple Site-Specific Nonstandard Amino Acid Incorporation. ACS Synth. Biol. 2014, 3, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Khambhati, K.; Bhattacharjee, G.; Gohil, N.; Braddick, D.; Kulkarni, V.; Singh, V. Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems. Front. Bioeng. Biotechnol. 2019, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- Copeland, C.E.; Langlois, A.; Kim, J.; Kwon, Y.-C. The Cell-Free System: A New Apparatus for Affordable, Sensitive, and Portable Healthcare. Biochem. Eng. J. 2021, 175, 108124. [Google Scholar] [CrossRef]
- Karig, D.K. Cell-free synthetic biology for environmental sensing and remediation. Curr. Opin. Biotechnol. 2017, 45, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Lee, J.A.; Biondo, J.R.; Lux, J.E.; Raig, R.M.; Berger, P.N.; Bernhards, C.B.; Kuhn, D.L.; Gupta, M.K.; Lux, M.W. Cell-Free Protein Expression in Polymer Materials. ACS Synth. Biol. 2024, 13, 1152–1164. [Google Scholar] [CrossRef]
- Lee, M.S.; Raig, R.M.; Gupta, M.K.; Lux, M.W. Lyophilized Cell-Free Systems Display Tolerance to Organic Solvent Exposure. ACS Synth. Biol. 2020, 9, 1951–1957. [Google Scholar] [CrossRef]
- Wright, O.; Stan, G.-B.; Ellis, T. Building-in Biosafety for Synthetic Biology. Microbiology 2013, 159, 1221–1235. [Google Scholar] [CrossRef]
- Vidiella, B.; Solé, R. Ecological firewalls for synthetic biology. iScience 2022, 25, 104658. [Google Scholar] [CrossRef]
- Pavão, G.; Sfalcin, I.; Bonatto, D. Biocontainment Techniques and Applications for Yeast Biotechnology. Fermentation 2023, 9, 341. [Google Scholar] [CrossRef]
- Richardson, S.M.; Mitchell, L.A.; Stracquadanio, G.; Yang, K.; Dymond, J.S.; DiCarlo, J.E.; Lee, D.; Huang, C.L.; Chandrasegaran, S.; Cai, Y.; et al. Design of a synthetic yeast genome. Science 2017, 355, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Chari, R.; Church, G.M. Beyond editing to writing large genomes. Nat. Rev. Genet. 2017, 18, 749–760. [Google Scholar] [CrossRef]
- Kato, Y.; Mori, H. Genetically stable kill-switch using “demon and angel” expression construct of essential genes. Front. Bioeng. Biotechnol. 2024, 12, 1365870. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Kubyshkin, V. How to Quantify a Genetic Firewall? A Polarity-Based Metric for Genetic Code Engineering. ChemBioChem 2021, 22, 1268–1284. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Nakano, Y. Breaking the deadlock in genetic code expansion. Nat. Chem. Biol. 2024, 20, 406–407. [Google Scholar] [CrossRef] [PubMed]
- Hartman, H.; Smith, T.F. The Evolution of the Ribosome and the Genetic Code. Life 2014, 4, 227–249. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Hartman, H. The Evolution of Class II Aminoacyl-tRNA Synthetases and the First Code. FEBS Lett. 2015, 589, 3499–3507. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.M.; Reynolds, N.M.; Rivera, K.; Connolly, M.; Guo, L.-T.; Ling, J.; Pappin, D.J.; Church, G.M.; Söll, D. Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia Coli. ACS Synth. Biol. 2016, 5, 163–171. [Google Scholar] [CrossRef]
- Dunkelmann, D.L.; Piedrafita, C.; Dickson, A.; Liu, K.C.; Elliott, T.S.; Fiedler, M.; Bellini, D.; Zhou, A.; Cervettini, D.; Chin, J.W. Adding α, α-disubstituted and β-linked monomers to the genetic code of an organism. Nature 2024, 625, 603–610. [Google Scholar] [CrossRef]
- Zürcher, J.F.; Robertson, W.E.; Kappes, T.; Petris, G.; Elliott, T.S.; Salmond, G.P.C.; Chin, J.W. Refactored Genetic Codes Enable Bidirectional Genetic Isolation. Science 2022, 378, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Nyerges, A.; Vinke, S.; Flynn, R.; Owen, S.V.; Rand, E.A.; Budnik, B.; Keen, E.; Narasimhan, K.; Marchand, J.A.; Baas-Thomas, M.; et al. A Swapped Genetic Code Prevents Viral Infections and Gene Transfer. Nature 2023, 615, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Ghadessy, F.J.; Holliger, P. Compartmentalized Self-Replication: A Novel Method for the Directed Evolution of Polymerases and Other Enzymes. Methods Mol. Biol. 2007, 352, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sismour, A.M.; Sheng, P.; Puskar, N.L.; Benner, S.A. Enzymatic Incorporation of a Third Nucleobase Pair. Nucleic Acids Res. 2007, 35, 4238–4249. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.; Chen, L.; Nilsson, M.; Abe, S. Bridging Nonliving and Living Matter. Artif. Life 2003, 9, 269–316. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, W.; Wang, Z.; Zhao, H.; Shi, S. Development of Host-Orthogonal Genetic Systems for Synthetic Biology. Adv. Biol. 2021, 5, e2000252. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Tatay, L.; Hernández-Andreu, J.M. Xenobiology for the Biocontainment of Synthetic Organisms: Opportunities and Challenges. Life 2024, 14, 996. https://doi.org/10.3390/life14080996
Gómez-Tatay L, Hernández-Andreu JM. Xenobiology for the Biocontainment of Synthetic Organisms: Opportunities and Challenges. Life. 2024; 14(8):996. https://doi.org/10.3390/life14080996
Chicago/Turabian StyleGómez-Tatay, Lucía, and José Miguel Hernández-Andreu. 2024. "Xenobiology for the Biocontainment of Synthetic Organisms: Opportunities and Challenges" Life 14, no. 8: 996. https://doi.org/10.3390/life14080996
APA StyleGómez-Tatay, L., & Hernández-Andreu, J. M. (2024). Xenobiology for the Biocontainment of Synthetic Organisms: Opportunities and Challenges. Life, 14(8), 996. https://doi.org/10.3390/life14080996






