Exploring the Effectiveness of Acyclovir against Ranaviral Thymidine Kinases: Molecular Docking and Experimental Validation in a Fish Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Protein Modeling
2.2. Acyclovir Docking
2.3. Cells and Viruses
2.4. Acyclovir Solutions
2.5. Cytotoxicity Assay
2.6. In Vitro Assays for Acyclovir and Virus Interactions
2.7. Quantitative PCRs
2.8. Statistical Data Analysis
3. Results
3.1. Acyclovir Docking
3.2. Cytotoxicity Assays
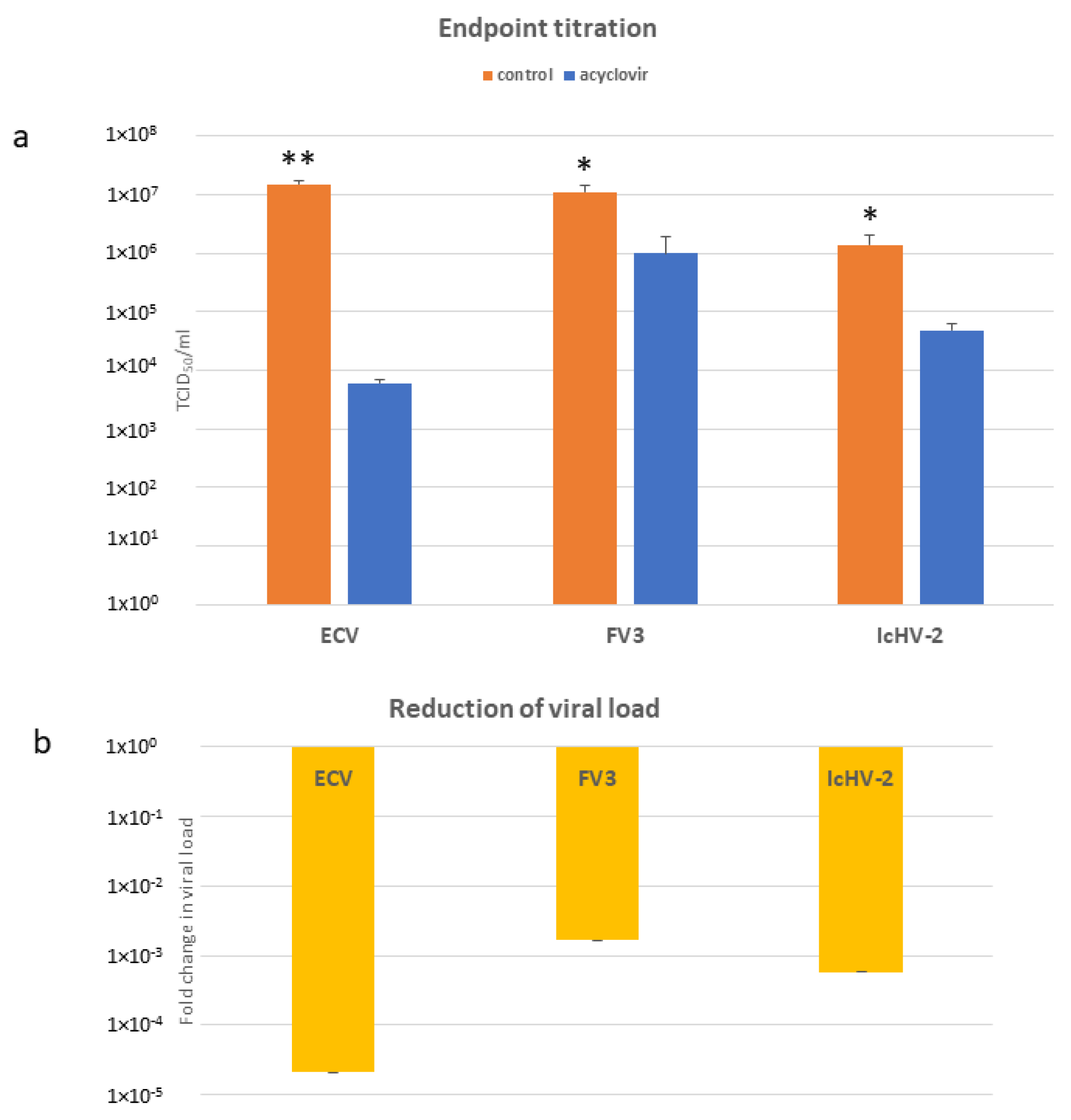
3.3. In Vitro Assays
3.4. qPCRs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zabrosky, C.A.; Milkovich, G. Acyclovir. Infect. Control 1986, 7, 228–229. [Google Scholar] [CrossRef]
- Laskin, O.L. Acyclovir. Pharmacology and clinical experience. Arch. Intern. Med. 1984, 144, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Maillard, M.; Gong, L.; Nishii, R.; Yang, J.J.; Whirl-Carrillo, M.; Klein, T.E. PharmGKB summary: Acyclovir/ganciclovir pathway. Pharmacogenet Genom. 2022, 32, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Hiraoka, W.; Kuwabara, M.; Sato, F.; Narita, T.; Niikura, M. Cytotoxic effect of acyclovir on cultured mammalian cells to which herpesvirus thymidine kinase gene was introduced. Jpn. J. Vet. Res. 1989, 37, 181–185. [Google Scholar]
- Benedetti, S.; Catalani, S.; Palma, F.; Canonico, B.; Luchetti, F.; Galati, R.; Papa, S.; Battistelli, S. Acyclovir induces cell cycle perturbation and apoptosis in Jurkat leukemia cells, and enhances chemotherapeutic drug cytotoxicity. Life Sci. 2018, 215, 80–85. [Google Scholar] [CrossRef]
- Doğan, H.H.; Karagöz, S.; Duman, R. In Vitro Evaluation of the Antiviral Activity of Some Mushrooms from Turkey. Int. J. Med. Mushrooms 2018, 20, 201–212. [Google Scholar] [CrossRef]
- Littler, E.; Stuart, A.D.; Chee, M.S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 1992, 358, 160–162. [Google Scholar] [CrossRef]
- Talarico, C.L.; Burnette, T.C.; Miller, W.H.; Smith, S.L.; Davis, M.G.; Stanat, S.C.; Ng, T.I.; He, Z.; Coen, D.M.; Roizman, B.; et al. Acyclovir is phosphorylated by the human cytomegalovirus UL97 protein. Antimicrob. Agents Chemother. 1999, 43, 1941–1946. [Google Scholar] [CrossRef]
- Elion, G.B. Mechanism of action and selectivity of acyclovir. Am. J. Med. 1982, 73, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.H.; Miller, R.L. Phosphorylation of acyclovir (acycloguanosine) monophosphate by GMP kinase. J. Biol. Chem. 1980, 255, 7204–7207. [Google Scholar] [CrossRef]
- Miller, W.H.; Miller, R.L. Phosphorylation of acyclovir diphosphate by cellular enzymes. Biochem. Pharmacol. 1982, 31, 3879–3884. [Google Scholar] [CrossRef] [PubMed]
- Elion, G.B. Acyclovir: Discovery, mechanism of action, and selectivity. J. Med. Virol. 1993, 41 (Suppl. S1), 2–6. [Google Scholar] [CrossRef]
- St Clair, M.H.; Furman, P.A.; Lubbers, C.M.; Elion, G.B. Inhibition of cellular alpha and virally induced deoxyribonucleic acid polymerases by the triphosphate of acyclovir. Antimicrob. Agents Chemother. 1980, 18, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Reardon, J.E. Herpes simplex virus type 1 and human DNA polymerase interactions with 2′-deoxyguanosine 5′-triphosphate analogues. Kinetics of incorporation into DNA and induction of inhibition. J. Biol. Chem. 1989, 264, 19039–19044. [Google Scholar]
- Allender, M.C.; Mitchell, M.A.; Yarborough, J.; Cox, S. Pharmacokinetics of a single oral dose of acyclovir and valacyclovir in North American box turtles (Terrapene sp.). J. Vet. Pharmacol. Ther. 2013, 36, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Marschang, R.E.; Gravendyck, M.; Kaleta, E.F. Herpesviruses in tortoises: Investigations into virus isolation and the treatment of viral stomatitis in Testudo hermanni and T. graeca. Zentralbl Vet. B 1997, 44, 385–394. [Google Scholar] [CrossRef]
- Troszok, A.; Kolek, L.; Szczygieł, J.; Wawrzeczko, J.; Borzym, E.; Reichert, M.; Kamińska, T.; Ostrowski, T.; Jurecka, P.; Adamek, M.; et al. Acyclovir inhibits Cyprinid herpesvirus 3 multiplication in vitro. J. Fish. Dis. 2018, 41, 1709–1718. [Google Scholar] [CrossRef]
- Hao, K.; Yuan, S.; Yu, F.; Chen, X.H.; Bian, W.J.; Feng, Y.H.; Zhao, Z. Acyclovir inhibits channel catfish virus replication and protects channel catfish ovary cells from apoptosis. Virus Res. 2021, 292, 198249. [Google Scholar] [CrossRef]
- Buck, C.C.; Loh, P.C. In vitro effect of acyclovir and other antiherpetic compounds on the replication of channel catfish virus. Antivir. Res. 1985, 5, 269–280. [Google Scholar] [CrossRef]
- Sosa-Higareda, M.; Yazdi, Z.; Littman, E.M.; Quijano Cardé, E.M.; Yun, S.; Soto, E. Efficacy of a multidose acyclovir protocol against cyprinid herpesvirus 3 infection in koi (Cyprinus carpio). Am. J. Vet. Res. 2022, 83. [Google Scholar] [CrossRef]
- Quijano Cardé, E.M.; Yazdi, Z.; Yun, S.; Hu, R.; Knych, H.; Imai, D.M.; Soto, E. Pharmacokinetic and Efficacy Study of Acyclovir Against Cyprinid Herpesvirus 3 in. Front. Vet. Sci. 2020, 7, 587952. [Google Scholar] [CrossRef] [PubMed]
- Troszok, A.; Kolek, L.; Szczygieł, J.; Ostrowski, T.; Adamek, M.; Irnazarow, I. Anti-CyHV-3 Effect of Fluorescent, Tricyclic Derivative of Acyclovir 6-(4-MeOPh)-TACV. J. Vet. Res. 2019, 63, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; et al. ICTV Virus Taxonomy Profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [CrossRef] [PubMed]
- Hanson, L.; Doszpoly, A.; van Beurden, S.; Viadanna, P.; Waltzek, T.; Kibenge, F.; Godoy, M. Alloherpesviruses of Fish. In Aquaculture Virology; Academic Press: Cambridge, MA, USA, 2016; pp. 153–172. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Ke, F.; Gui, L.; Zhao, Z. Recent insights into aquatic viruses: Emerging and reemerging pathogens, molecular features, biological effects, and novel investigative approaches. Water Biol. Secur. 2022, 1, 100062. [Google Scholar] [CrossRef]
- Abonyi, F.; Varga, A.; Sellyei, B.; Eszterbauer, E.; Doszpoly, A. Juvenile Wels Catfish (Silurus glanis) Display Age-Related Mortality to European Catfish Virus (ECV) under Experimental Conditions. Viruses 2022, 14, 1832. [Google Scholar] [CrossRef]
- Alborali, L.; Bovo, G.; Lavazza, A.; Capellaro, H.; Guadagnini, P.F. Isolation of a herpesvirus in breeding catfish (Ictalurus melas). Bull. Eur. Assoc. Fish. Pathol. 1996, 16, 134–137. [Google Scholar]
- Marschang, R.E. Viruses infecting reptiles. Viruses 2011, 3, 2087–2126. [Google Scholar] [CrossRef]
- Maniero, G.D.; Morales, H.; Gantress, J.; Robert, J. Generation of a long-lasting, protective, and neutralizing antibody response to the ranavirus FV3 by the frog Xenopus. Dev. Comp. Immunol. 2006, 30, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.A.; Garner, S.; Echaubard, P.; Lesbarrères, D.; Kyle, C.J.; Brunetti, C.R. Complete genome analysis of a frog virus 3 (FV3) isolate and sequence comparison with isolates of differing levels of virulence. Virol. J. 2014, 11, 46. [Google Scholar] [CrossRef]
- Doszpoly, A.; Shaalan, M.; El-Matbouli, M. Silver Nanoparticles Proved to Be Efficient Antivirals In Vitro against Three Highly Pathogenic Fish Viruses. Viruses 2023, 15, 1689. [Google Scholar] [CrossRef]
- Mugimba, K.K.; Byarugaba, D.K.; Mutoloki, S.; Evensen, Ø.; Munang’andu, H.M. Challenges and Solutions to Viral Diseases of Finfish in Marine Aquaculture. Pathogens 2021, 10, 673. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L. Schrödinger Suite; Schrödinger: New York, NY, USA, 2023. [Google Scholar]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, T.; Woynarovichne, L.; Csaba, G.; Farkas, L.; Dan, A. Isolation of ranavirus causing mass mortality in brown bullheads (Ameiurus nebulosus) in Hungary. Magy. Allatorvosok Lapja 2013, 135, 763–768. [Google Scholar]
- Granoff, A.; Came, P.E.; Breeze, D.C. Viruses and renal carcinoma of Rana pipiens. I. Isol. Prop. Virus Norm. Tumor Tissue. Virol. 1966, 29, 133–148. [Google Scholar] [CrossRef]
- Stöhr, A.C.; López-Bueno, A.; Blahak, S.; Caeiro, M.F.; Rosa, G.M.; Alves de Matos, A.P.; Martel, A.; Alejo, A.; Marschang, R.E. Phylogeny and differentiation of reptilian and amphibian ranaviruses detected in Europe. PLoS ONE 2015, 10, e0118633. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Killington, R.A. Virus isolation and quantitation. In Virology Methods Manual; Mahy, B.W., Kangro, H.O., Eds.; Academic Press: London, UK, 1996; pp. 25–46. [Google Scholar]
- Goodwin, A.E.; Marecaux, E. Validation of a qPCR assay for the detection of Ictalurid herpesvirus-2 (IcHV-2) in fish tissues and cell culture supernatants. J. Fish. Dis. 2010, 33, 341–346. [Google Scholar] [CrossRef]
- Bennett, M.S.; Wien, F.; Champness, J.N.; Batuwangala, T.; Rutherford, T.; Summers, W.C.; Sun, H.; Wright, G.; Sanderson, M.R. Structure to 1.9 A resolution of a complex with herpes simplex virus type-1 thymidine kinase of a novel, non-substrate inhibitor: X-ray crystallographic comparison with binding of aciclovir. FEBS Lett. 1999, 443, 121–125. [Google Scholar] [CrossRef]
- Shaalan, M.; Saleh, M.; El-Mahdy, M.; El-Matbouli, M. Recent progress in applications of nanoparticles in fish medicine: A review. Nanomedicine 2016, 12, 701–710. [Google Scholar] [CrossRef]
- Majewska, A.; Mlynarczyk-Bonikowska, B. 40 Years after the Registration of Acyclovir: Do We Need New Anti-Herpetic Drugs? Int. J. Mol. Sci. 2022, 23, 3431. [Google Scholar] [CrossRef] [PubMed]
- Szotowska, I.; Ledwoń, A. Antiviral Chemotherapy in Avian Medicine-A Review. Viruses 2024, 16, 593. [Google Scholar] [CrossRef]
- Maxwell, L.K. Antiherpetic Drugs in Equine Medicine. Vet. Clin. North. Am. Equine Pract. 2017, 33, 99–125. [Google Scholar] [CrossRef]
- Liang, C.S.; Chen, C.; Lin, Z.Y.; Shen, J.L.; Wang, T.; Jiang, H.F.; Wang, G.X. Acyclovir inhibits white spot syndrome virus replication in crayfish Procambarus clarkii. Virus Res. 2021, 305, 198570. [Google Scholar] [CrossRef]
- Fehér, E.; Doszpoly, A.; Horváth, B.; Marton, S.; Forró, B.; Farkas, S.L.; Bányai, K.; Juhász, T. Whole genome sequencing and phylogenetic characterization of brown bullhead (Ameiurus nebulosus) origin ranavirus strains from independent disease outbreaks. Infect. Genet. Evol. 2016, 45, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Borzym, E.; Karpińska, T.A.; Reichert, M. Outbreak of ranavirus infection in sheatfish, Silurus glanis (L.), in Poland. Pol. J. Vet. Sci. 2015, 18, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Borzák, R.; Haluk, T.; Bartha, D.; Doszpoly, A. Complete genome sequence and analysis of ictalurid herpesvirus 2. Arch Virol. 2018, 163, 1083–1085. [Google Scholar] [CrossRef]
- Troszok, A.; Kolek, L.; Szczygiel, J.; Wawrzeczko, J.; Jurecka, P.; Adamek, M.; Irnazarow, I. Acyclovir inhibits CyHV-3 replication in a dose-dependent manner. An in vitro study. Aquac. Res. 2022, 53, 1127–1130. [Google Scholar] [CrossRef]
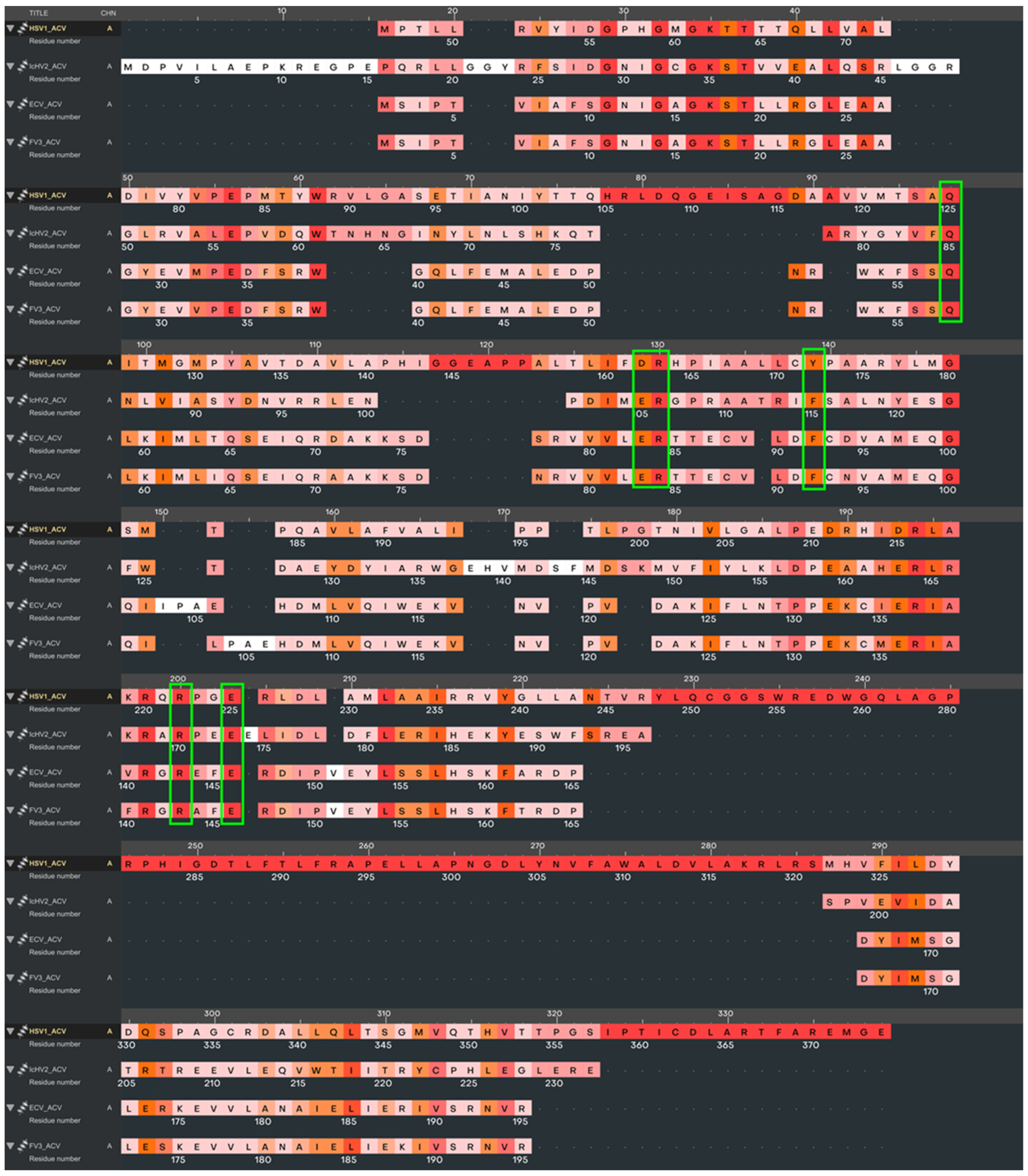
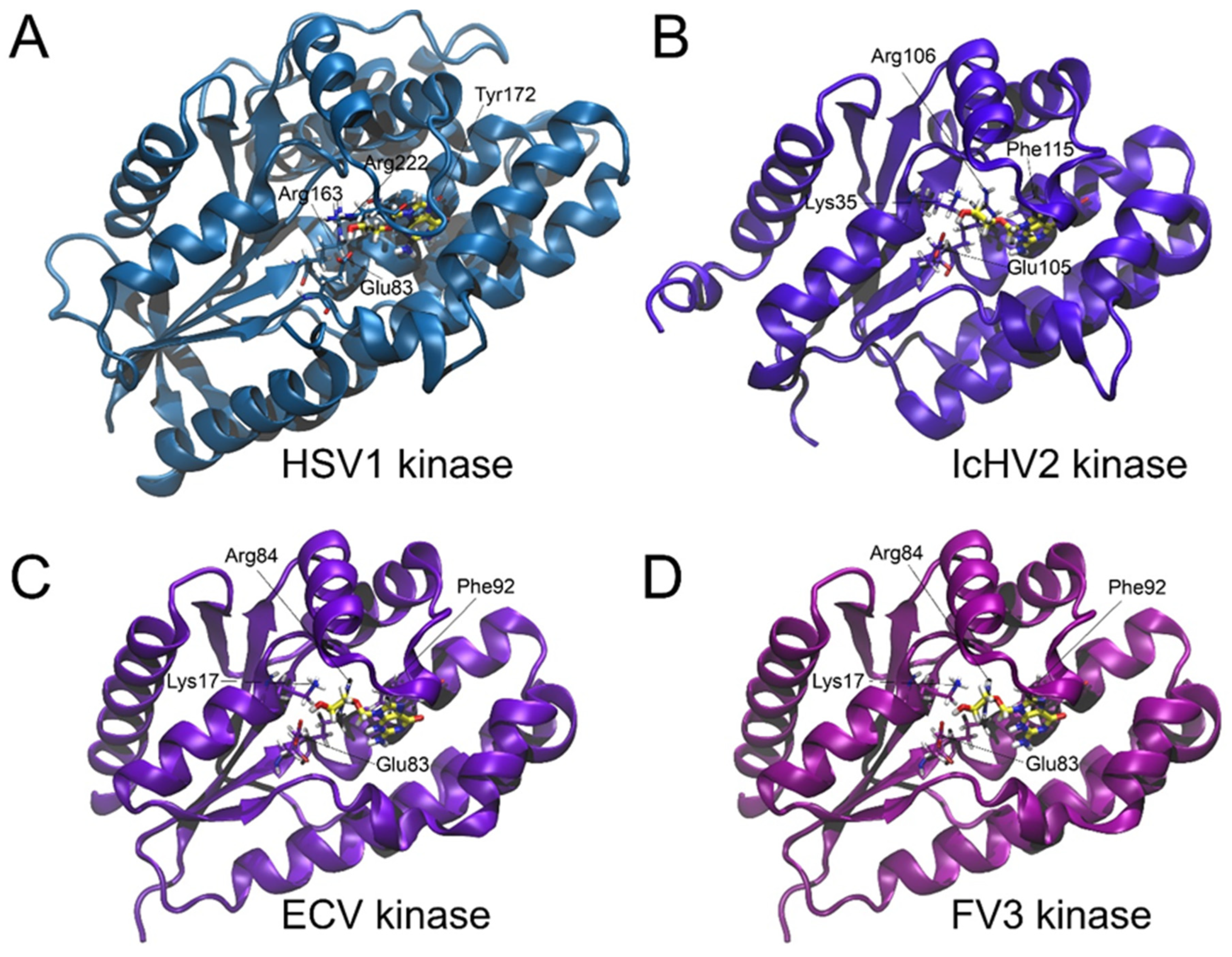

| Reference | HSV-1 | IcHV-2 | ECV | FV3 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Query | Identity % | Similarity % | Identity % | Similarity % | Identity % | Similarity % | Identity % | Similarity % | |
| HSV-1 | 16.6 | 24.0 | 12.5 | 19.9 | 11.2 | 16.9 | |||
| IcHV-2 | 16.1 | 22.4 | 21.5 | 37.2 | 21.1 | 36.4 | |||
| ECV | 12.5 | 19.9 | 21.5 | 37.2 | 93.8 | 96.9 | |||
| FV3 | 11.2 | 16.9 | 21.1 | 36.4 | 93.8 | 96.9 | |||
| Glide Score | Glide Emodel/(kcal/mol) | Phosphorylation Feasibility | |
|---|---|---|---|
| HSV-1 kinase (2KI5) | −8.730 | −71.527 | Yes |
| IcHV-2 kinase | −6.932 | −64.247 | Yes |
| ECV kinase | −7.239 | −65.365 | Yes |
| FV3 kinase | −7.299 | −65.376 | Yes |
| Concentration of Acyclovir (µM) | Cell Viability ± SEM (%) |
|---|---|
| 3200 | 103.373 ± 1.58 |
| 1600 | 94.35 ± 1.47 |
| 800 | 102.70 ± 1.41 |
| 400 | 110.17 ± 3.45 |
| 200 | 110.68 ± 2.27 |
| 100 | 104.99 ± 1.48 |
| 50 | 113.00 ± 1.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rácz, R.; Gellért, Á.; Papp, T.; Doszpoly, A. Exploring the Effectiveness of Acyclovir against Ranaviral Thymidine Kinases: Molecular Docking and Experimental Validation in a Fish Cell Line. Life 2024, 14, 1050. https://doi.org/10.3390/life14091050
Rácz R, Gellért Á, Papp T, Doszpoly A. Exploring the Effectiveness of Acyclovir against Ranaviral Thymidine Kinases: Molecular Docking and Experimental Validation in a Fish Cell Line. Life. 2024; 14(9):1050. https://doi.org/10.3390/life14091050
Chicago/Turabian StyleRácz, Richárd, Ákos Gellért, Tibor Papp, and Andor Doszpoly. 2024. "Exploring the Effectiveness of Acyclovir against Ranaviral Thymidine Kinases: Molecular Docking and Experimental Validation in a Fish Cell Line" Life 14, no. 9: 1050. https://doi.org/10.3390/life14091050





