Molecular Characterization, Evolution and Expression Analysis of Ammonium Transporter from Four Closely Related Bactrocera Species (Tephritidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Sample Collection
2.2. RNA Isolation and cDNA Synthesis
2.3. Homology Search and Molecular Cloning
2.4. Bioinformatic Analyses
2.5. The Phylogenetic Analysis of the Amt Family
2.6. Analysis of Expression Patterns of Amt in Different Peripheral Tissues
2.7. Statistical Analyses
3. Results
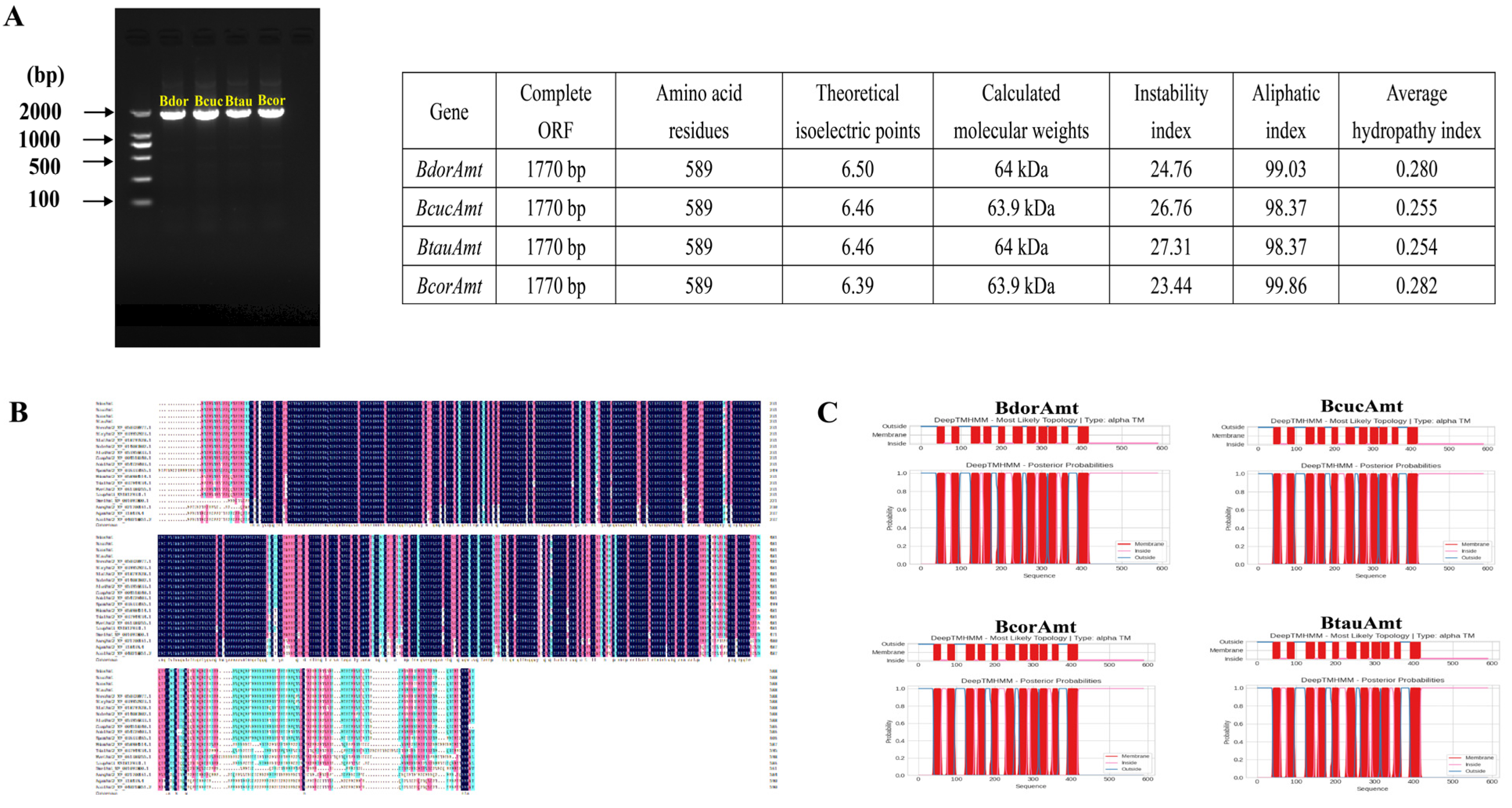
3.1. Identification and Sequence Characteristics of Amts
3.2. Phylogenetic Analysis of Amts
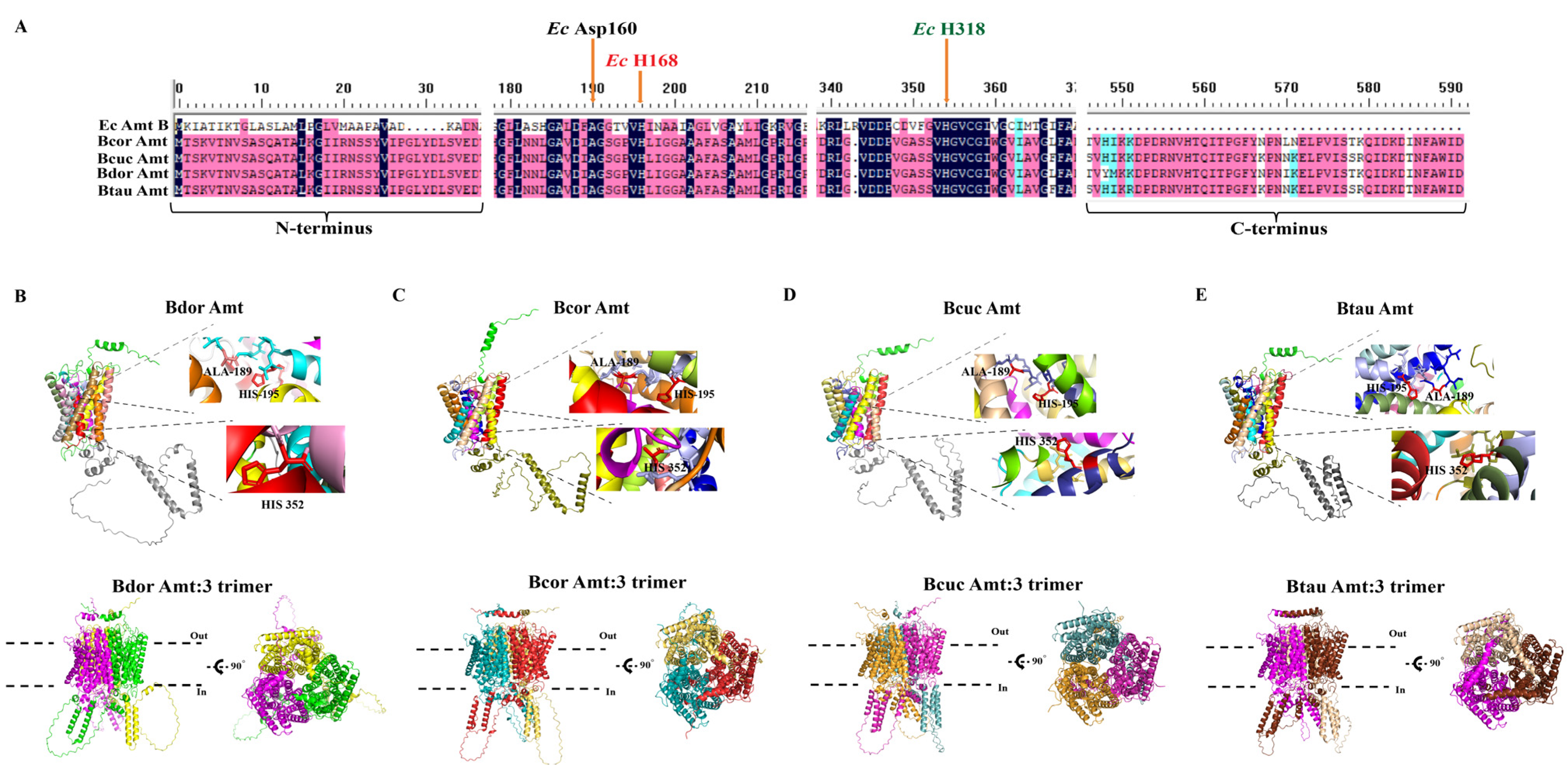
3.3. Structural Prediction of Amt and Analysis of Conserved Functional Sites
3.4. Expression Profiles of Amts in Different Peripheral Tissues
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| PCR Primers | PCR Conditions | |
|---|---|---|
| BdorAmt | BdorAmt-CDS-F: ATGACTTCAAAAGTGACAAATGTCT BdorAmt-CDS-R: TCAATCGATCCATGCAAAATTGATAT | 98 °C for 3 min; then 35 cycles of 98 °C for 10 s, annealing at 60 °C for 15 s, extension at 72 °C for 2 min; followed by a final extension of 72 °C for 10 min; stored at 10 °C |
| BcorAmt | BcorAmt-CDS-F: ATGACTTCAAAAGTGACAAATGTCTC BcorAmt-CDS-R: TCAATCGATCCATGCAAAATTGATAT | 98 °C for 3 min; then 35 cycles of 98 °C for 10 s, annealing at 60 °C for 15 s, extension at 72 °C for 2 min; followed by a final extension of 72 °C for 10 min; stored at 10 °C |
| BcucAmt | BcucAmt-CDS-F: ATGACTTCAAAAGTGACAAATGTCTCAG BcucAmt-CDS-R: TCAATCGATCCATGCGAAATTGGTAT | 98 °C for 3 min; then 35 cycles of 98 °C for 10 s, annealing at 60 °C for 15 s, extension at 72 °C for 2 min; followed by a final extension of 72 °C for 10 min; stored at 10 °C |
| BtauAmt | BtauAmt-CDS-F: ATGACTTCAAAAGTGACAAATGTCT BtauAmt-CDS-R: TCAATCGATCCATGCGAAATTGGTAT | 98 °C for 3 min; then 35 cycles of 98 °C for 10 s, annealing at 60 °C for 15 s, extension at 72 °C for 2 min; followed by a final extension of 72 °C for 10 min; stored at 10 °C |
| BdorAmt(q-PCR) | BdorAmt-q-F: GAACACCGGATTCGTCACT BdorAmt-q-R: CGCGTAGCTGTTCAATGCTC | 95 °C for 3 min; then 39 cycles of 95 °C for 10 s, annealing at 55 °C for 30 s, +Plate Read; followed Melt Curve 65 °C to 95 °C, increment 0.5 °C, for 5 s; Plate Read |
| BcorAmt(q-PCR) | BcorAmt-q-F: CTTTGTGCTCTGGTGGGGAT BcorAmt-q-R: CATCATGACGCCAAAGCGAG | |
| BcucAmt(q-PCR) | BcucAmt-q-F: CGCCATGATGGACGTATGGA BcucAmt-q-R: CCCCTAACCGGTCGAACAAA | |
| BtauAmt(q-PCR) | BtauAmt-q-F: GCTCGGTTCACTGGTCTCAA BtauAmt-q-R: CCCCTAACCGGTCGAACAAA | |
| BdorActin-2(q-PCR) | BdorActin-2-F: GTGTGATGGTTGGTATGGGA BdorActin-2-R: GGCTGGGGAGTTGAAGGTTT | |
| Bcor_GAPDH2(q-PCR) | Bcor-F: CTTCAACTGGTGCCGCTAAG Bcor-R: ACCCTTCATTGGACCCTCTG | |
| Bcuc_GAPDH2(q-PCR) | Bcuc-F: TGCTACTACTGCCACCCAAA Bcuc-R: ACGGAAAGCCATACCAGTCA | |
| Btau_GAPDH2(q-PCR) | Btau-F: TGCTACTACTGCCACCCAAA Btau-R: ACGGAAAGCCATACCAGTCA |
Appendix B
| Gene Name | Accession Number |
|---|---|
| Bactrocera neohumeralis Amt2 (BneoAmt2) | XP_050328977.1 |
| Bactrocera tryoni Amt2 (BtryAmt2) | XP_039952923.1 |
| Bactrocera latifrons Amt2 (BlatAmt2) | XP_018797628.1 |
| Bactrocera oleae Amt2 (BoleAmt2) | XP_014087802.1 |
| Anastrepha ludens Amt2 (AludAmt2) | XP_053956833.1 |
| Ceratitis capitata Amt2 (CcapAmt2) | XP_004518340.1 |
| Anastrepha obliqua Amt2 (AoblAmt2) | XP_054726403.1 |
| Rhagoletis pomonella Amt2 (RpomAmt2) | XP_036333465.1 |
| Musca domestica Amt2 (MdomAmt2) | XP_058984514.1 |
| Teleopsis dalmanni Amt2 (TdalAmt2) | XP_037949734.1 |
| Musca vetustissima Amt2 (MvetAmt2) | XP_061386255.1 |
| Lucilia cuprina Amt3 (LcupAmt3) | KAI8129331.1 |
| Drosophila melanogaster Amt (DmelAmt) | NP_001097800.1 |
| Aedes aegypti Amt2 (AaegAmt2) | XP_021700161.1 |
| Anopheles gambiae Amt2 (AgamAmt2) | XP_318439.4 |
| Anopheles coluzzii Amt2 (AcolAmt2) | XP_040218851.2 |
Appendix C
Appendix D

References
- Shelly, T.; Epsky, N.; Jang, E.B.; Reyes-Flores, J.; Vargas, R. Trapping and the Detection, Control, and Regulation of Tephritid Fruit Flies; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Christenson, L.D.; Foote, R.H. Biology of fruit flies. Annu. Rev. Entomol. 1960, 5, 171–192. [Google Scholar] [CrossRef]
- Ono, H.; Hee, A.K.W.; Jiang, H.B. Recent advancements in studies on chemosensory mechanisms underlying detection of semiochemicals in Dacini fruit flies of economic importance (Diptera: Tephritidae). Insects 2021, 12, 106. [Google Scholar] [CrossRef]
- Smith, P.T.; Kambhampati, S.; Armstrong, K.S. Phylogenetic relationships among Bactrocera species (Diptera: Tephritidae) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2003, 26, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Bateman, M.A.; Morton, T.C. The importance of ammonia in proteinaceous attractants for fruit flies (Diptera: Tephritidae). Aust. J. Agric. Res. 1981, 32, 883–903. [Google Scholar] [CrossRef]
- Mazor, M.; Peysakhis, A.; Reuven, G. Release rate of ammonia-a key component in the attraction of female mediterranean fruit fly to protein-based food lures. IOBC-WPRS Bull. 2002, 25, 1–6. [Google Scholar]
- Epsky, N.D.; Heath, R.R. Exploiting the interactions of chemical and visual cues in behavioral control measures for pest Tephritid fruit flies. Flo. Entomol. 1998, 81, 273–282. [Google Scholar] [CrossRef]
- Robacker, D.; Flath, R. Attractants from Staphylococcus aureus cultures for mexican fruit fly, Anastrepha ludens. J. Chem. Ecol. 1995, 21, 1861–1874. [Google Scholar] [CrossRef] [PubMed]
- Robacker, D.C.; Heath, R.R. Attraction of mexican fruit flies (Diptera: Tephritidae) to lures emitting host-fruit volatiles in a citrus orchard. Flo. Entomol. 1996, 79, 600–602. [Google Scholar] [CrossRef]
- Hull, C.; Cribb, B. Olfaction in the Queensland Fruit Fly, Bactrocera tryoni. I: Identification of olfactory receptor neuron types responding to environmental odors. J. Chem. Ecol. 2001, 27, 871–887. [Google Scholar] [CrossRef]
- Phyoe, A.; Nwet, T.T.; Maung, K.L. Effectiveness of protein bait on the attraction and reproduction of Bactrocera dorsalis and Bactrocera correcta (Diptera: Tephritidae). Univ. J. Sci. Eng. Res. 2020, 2, 10–15. [Google Scholar]
- Drew, R.; Courtice, A.C.; Teakle, A. Bacteria as a natural source of food for adult fruit flies (Diptera: Tephritidae). Oecologia 1983, 60, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Hagen, K.S.; Finney, G.L. A food supplement for effectively increasing the fecundity of certain tephritid species. J. Econ. Entomol. 1950, 43, 735. [Google Scholar] [CrossRef]
- Biasazin, T.D.; Chernet, H.T.; Herrera, S.L.; Bengtsson, M.; Karlsson, M.F.; Lemmen-Lechelt, J.K.; Dekker, T. Detection of volatile constituents from food lures by Tephritid fruit flies. Insects 2018, 9, 119. [Google Scholar] [CrossRef]
- Piñero, J.C.; Mau, R.F.L.; Vargas, R.I. A comparative assessment of the response of three fruit fly species (Diptera: Tephritidae) to a spinosad-based bait: Effect of ammonium acetate, female age, and protein hunger. Bull. Entomol. Res. 2011, 101, 373–381. [Google Scholar] [CrossRef]
- Piñero, J.C.; Souder, S.K.; Smith, T.R.; Vargas, R.I. Attraction of Bactrocera cucurbitae and Bactrocera dorsalis (Diptera: Tephritidae) to beer waste and other protein sources laced with ammonium acetate. Fla. Entomol. 2017, 100, 70–76. [Google Scholar] [CrossRef]
- Bruce, T.J.A.; Wadhams, L.J.; Woodcock, C.M. Insect host location: A volatile situation. Trends Plant Sci. 2005, 10, 269–274. [Google Scholar] [CrossRef]
- Montagné, N.; de Fouchier, A.; Newcomb, R.D.; Jacquin-Joly, E. Advances in the identification and characterization of olfactory receptors in insects. Prog. Mol. Biol. Transl. Sci. 2015, 130, 55–80. [Google Scholar] [PubMed]
- Menuz, K.; Larter, N.K.; Park, J.; Carlson, J.R. An RNA-seq screen of the Drosophila antenna identifies a transporter necessary for ammonia detection. PLoS Genet. 2014, 10, e1004810. [Google Scholar] [CrossRef]
- Pitts, R.J.; Derryberry, S.L., Jr.; Pulous, F.E.; Zwiebel, L.J. Antennal-expressed ammonium transporters in the malaria vector mosquito Anopheles gambiae. PLoS ONE 2014, 9, e111858. [Google Scholar] [CrossRef]
- Vulpe, A.; Kim, H.S.; Ballou, S.; Wu, S.T.; Grabe, V.; Nava Gonzales, C.; Liang, T.; Sachse, S.; Jeanne, J.M.; Su, C.Y.; et al. An ammonium transporter is a non-canonical olfactory receptor for ammonia. Curr. Biol. 2021, 31, 3382–3390.e7. [Google Scholar] [CrossRef]
- Khademi, S.; Stroud, R.M. The Amt/MEP/Rh family: Structure of AmtB and the mechanism of ammonia gas conduction. Physiology 2006, 21, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Kostrewa, D.; Bernèche, S.; Winkler, F.K.; Li, X.D. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 2004, 101, 17090–17095. [Google Scholar] [CrossRef] [PubMed]
- Khademi, S.; O’Connell, J., III; Remis, J.; Robles-Colmenares, Y.; Miercke, L.J.; Stroud, R.M. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science 2004, 305, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Maniero, R.A.; Koltun, A.; Vitti, M.; Factor, B.G.; de Setta, N.; Câmara, A.S.; Lima, J.E.; Figueira, A. Identification and functional characterization of the sugarcane (Saccharum spp.) AMT2-type ammonium transporter ScAMT3;3 revealed a presumed role in shoot ammonium remobilization. Front. Plant Sci. 2023, 14, 1299025. [Google Scholar] [CrossRef] [PubMed]
- Delventhal, R.; Menuz, K.; Joseph, R.; Park, J.; Sun, J.S.; Carlson, J.R. The taste response to ammonia in Drosophila. Sci. Rep. 2017, 7, 43754. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome. Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.N.; Videvall, E.; Walden, K.K.O.; Harris, M.O.; Robertson, H.M.; Lofstedt, C. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae). BMC Genom. 2014, 15, 501. [Google Scholar] [CrossRef]
- Leitch, O.; Papanicolaou, A.; Lennard, C.; Kirkbride, K.P.; Anderson, A. Chemosensory genes identified in the antennal transcriptome of the blowfly Calliphora stygia. BMC Genom. 2015, 16, 255. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gruswitz, F.; Chaudhary, S.; Ho, J.D.; Schlessinger, A.; Pezeshki, B.; Ho, C.M.; Sali, A.; Westhoff, C.M.; Stroud, R.M. Function of human Rh based on structure of RhCG at 2.1 A. Proc. Natl. Acad. Sci. USA 2010, 107, 9638–9643. [Google Scholar] [CrossRef]
- Schlessinger, A.; Yee, S.W.; Sali, A.; Giacomini, K.M. SLC classification: An update. Clin. Pharmacol. Ther. 2013, 94, 19–23. [Google Scholar] [CrossRef]
- Zhang, X.X.; Liu, Y.; Guo, M.B.; Sun, D.D.; Zhang, M.J.; Chu, X.; Berg, B.G.; Wang, G.R. A female-specific odorant receptor mediates oviposition deterrence in the moth Helicoverpa armigera. Curr. Biol. 2024, 34, 1–11. [Google Scholar] [CrossRef]
- Zhang, R.B.; Wang, B.; Grossi, G.; Falabella, P.; Liu, Y.; Yan, S.C.; Lu, J.; Xi, J.H.; Wang, G.R. Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 2017, 27, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Pitts, R.J.; Fox, A.N.; Zwiebel, L.J. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 5058–5063. [Google Scholar] [CrossRef] [PubMed]
- Brigaud, I.; Montagné, N.; Monsempes, C.; François, M.C.; Jacquin-Joly, E. Identification of an atypical insect olfactory receptor subtype highly conserved within noctuids. FEBS J. 2009, 276, 6537–6547. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Q.; Zhou, Y.L.; Shan, S.; Cui, H.H.; Xiao, Y.; Dong, K.; Khashaveh, A.; Sun, L.; Zhang, Y.J. Characterization and comparative analysis of olfactory receptor co-receptor Orco orthologs among five mirid bug species. Front. Physiol. 2018, 9, 158. [Google Scholar] [CrossRef]
- Hallem, E.A.; Ho, M.G.; Carlson, J.R. The molecular basis of odor coding in the Drosophila antenna. Cell 2004, 117, 965–979. [Google Scholar] [CrossRef]
- Chang, H.T.; Cassau, S.; Krieger, J.; Guo, X.J.; Knaden, M.; Kang, L.; Hansson, B.S. A chemical defense deters cannibalism in migratory locusts. Science 2023, 380, 537–543. [Google Scholar] [CrossRef]


| Gene Name | Blastx Best Hit (Reference/Name/Species) | E-Value | Score |
|---|---|---|---|
| BdorAmt | XP_049304235.1 putative ammonium transporter 2 [Bactrocera dorsalis] | 0 | 1211 |
| BcorAmt | XP_049304235.1 putative ammonium transporter 2 [Bactrocera dorsalis] | 0 | 1204 |
| BcucAmt | XP_054081809.1 putative ammonium transporter 2 [Zeugodacus cucurbitae] | 0 | 1209 |
| BtauAmt | XP_054081809.1 putative ammonium transporter 2 [Zeugodacus cucurbitae] | 0 | 1097 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, Q.; Liu, C.; Liu, J.; Qian, Q.; Ru, C.; Liu, L.; Yan, S.; Liu, W.; Wang, G. Molecular Characterization, Evolution and Expression Analysis of Ammonium Transporter from Four Closely Related Bactrocera Species (Tephritidae). Life 2024, 14, 1114. https://doi.org/10.3390/life14091114
Zhang J, Wang Q, Liu C, Liu J, Qian Q, Ru C, Liu L, Yan S, Liu W, Wang G. Molecular Characterization, Evolution and Expression Analysis of Ammonium Transporter from Four Closely Related Bactrocera Species (Tephritidae). Life. 2024; 14(9):1114. https://doi.org/10.3390/life14091114
Chicago/Turabian StyleZhang, Jie, Qi Wang, Chenhao Liu, Jiaying Liu, Qian Qian, Chuanjian Ru, Leyuan Liu, Shanchun Yan, Wei Liu, and Guirong Wang. 2024. "Molecular Characterization, Evolution and Expression Analysis of Ammonium Transporter from Four Closely Related Bactrocera Species (Tephritidae)" Life 14, no. 9: 1114. https://doi.org/10.3390/life14091114
APA StyleZhang, J., Wang, Q., Liu, C., Liu, J., Qian, Q., Ru, C., Liu, L., Yan, S., Liu, W., & Wang, G. (2024). Molecular Characterization, Evolution and Expression Analysis of Ammonium Transporter from Four Closely Related Bactrocera Species (Tephritidae). Life, 14(9), 1114. https://doi.org/10.3390/life14091114





