A Novel Approach for In Vitro Testing and Hazard Evaluation of Nanoformulated RyR2-Targeting siRNA Drugs Using Human PBMCs
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of siRNA Loaded CaP NPs
2.3. Characterization of siRNA Loaded CaP NPs
2.4. PBMCs Isolation
2.5. Treatment with Positive Controls, RyR2 CaP-siRNA and CaP-Scramble Drugs
2.6. Cytotoxicity Evaluation
2.7. Cytokines Release
2.8. Toxicity Pathways Evaluation
2.9. Statistical Analysis
3. Results
3.1. Chemical-Physical Properties of siRNA-Loaded CaP NPs
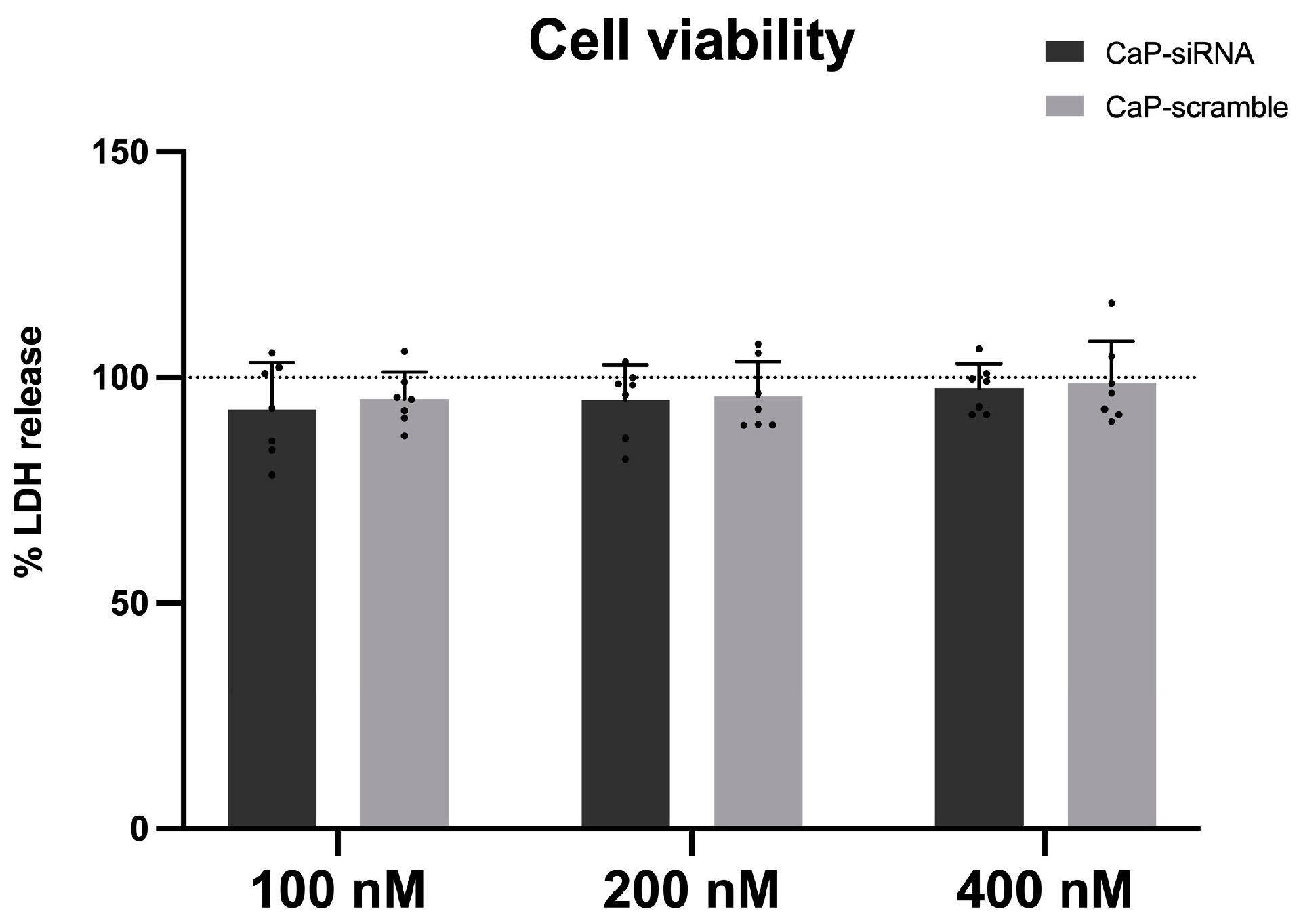
3.2. Cytotoxic Effects
3.3. Cytokines Release
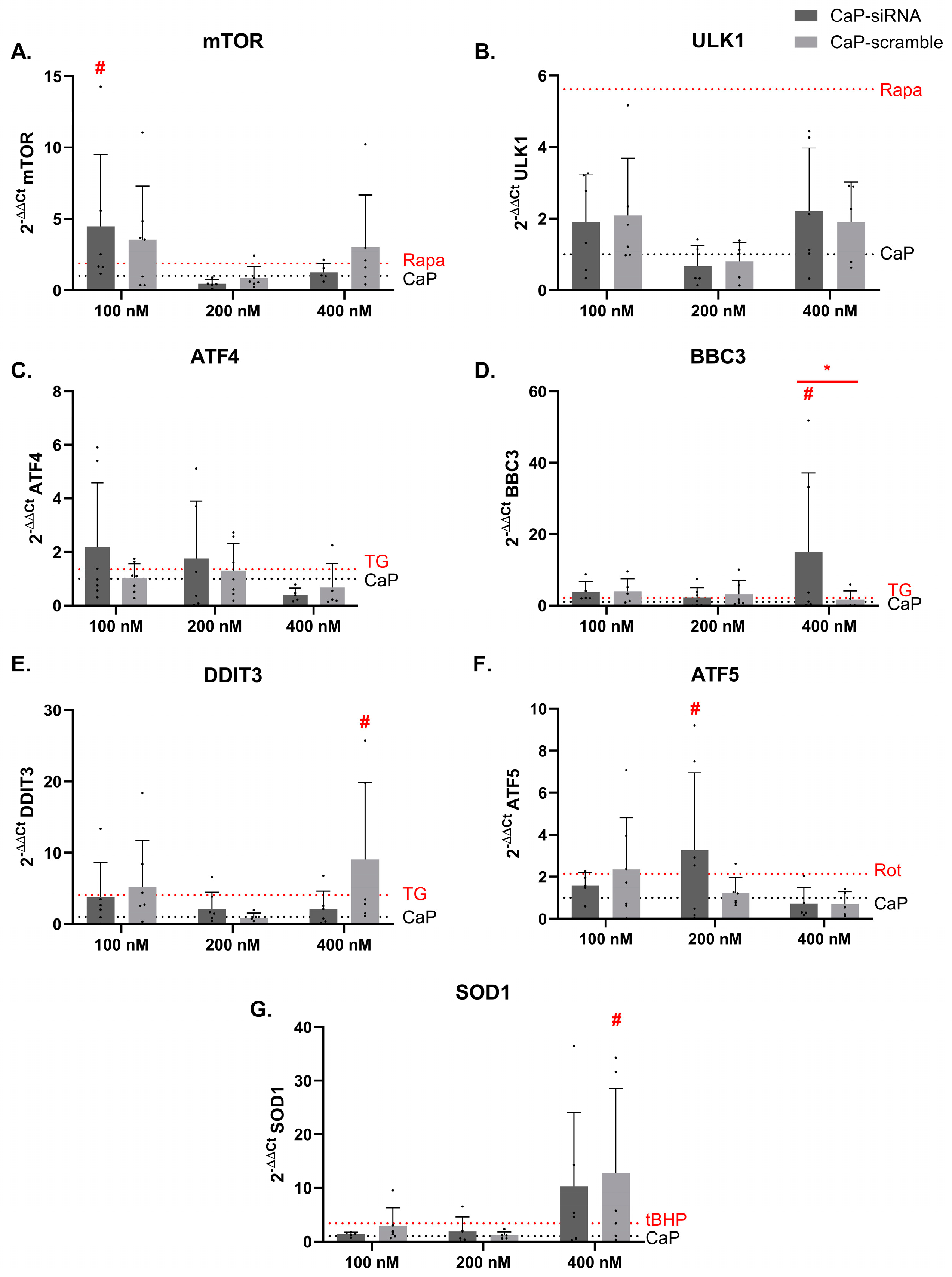
3.4. Effects on Toxicity Pathways
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. SiRNA Versus MiRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Witztum, J.L.; Bennett, C.F.; Baker, B.F. RNA-Targeted Therapeutics. Cell Metab. 2018, 27, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Arechavala-Gomeza, V.; Garanto, A. Antisense RNA Therapeutics: A Brief Overview. In Antisense RNA Design, Delivery, and Analysis; Springer: New York, NY, USA, 2022; pp. 33–49. [Google Scholar]
- Moreno-Montañés, J.; Sádaba, B.; Ruz, V.; Gómez-Guiu, A.; Zarranz, J.; González, M.V.; Pañeda, C.; Jimenez, A.I. Phase I Clinical Trial of SYL040012, a Small Interfering RNA Targeting β-Adrenergic Receptor 2, for Lowering Intraocular Pressure. Mol. Ther. 2014, 22, 226–232. [Google Scholar] [CrossRef]
- Benitez-Del-Castillo, J.M.; Moreno-Montañés, J.; Jiménez-Alfaro, I.; Muñoz-Negrete, F.J.; Turman, K.; Palumaa, K.; Sádaba, B.; González, M.V.; Ruz, V.; Vargas, B.; et al. Safety and Efficacy Clinical Trials for SYL1001, a Novel Short Interfering RNA for the Treatment of Dry Eye Disease. Investig. Opthalmology Vis. Sci. 2016, 57, 6447. [Google Scholar] [CrossRef]
- Nguyen, Q.D.; Schachar, R.A.; Nduaka, C.I.; Sperling, M.; Klamerus, K.J.; Chi-Burris, K.; Yan, E.; Paggiarino, D.A.; Rosenblatt, I.; Aitchison, R.; et al. Evaluation of the SiRNA PF-04523655 versus Ranibizumab for the Treatment of Neovascular Age-Related Macular Degeneration (MONET Study). Ophthalmology 2012, 119, 1867–1873. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Raal, F.J.; Kallend, D.; Ray, K.K.; Turner, T.; Koenig, W.; Wright, R.S.; Wijngaard, P.L.J.; Curcio, D.; Jaros, M.J.; Leiter, L.A.; et al. Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2020, 382, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Tanna, S.; Doshi, G.; Godad, A. SiRNA as Potential Therapeutic Strategy for Hypertension. Eur. J. Pharmacol. 2024, 969, 176467. [Google Scholar] [CrossRef]
- Desai, A.S.; Webb, D.J.; Taubel, J.; Casey, S.; Cheng, Y.; Robbie, G.J.; Foster, D.; Huang, S.A.; Rhyee, S.; Sweetser, M.T.; et al. Zilebesiran, an RNA Interference Therapeutic Agent for Hypertension. N. Engl. J. Med. 2023, 389, 228–238. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-Based Therapeutics: An Overview and Prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef] [PubMed]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug Delivery Systems for RNA Therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. SiRNA: Mechanism of Action, Challenges, and Therapeutic Approaches. Eur. J. Pharmacol. 2021, 905, 174178. [Google Scholar] [CrossRef]
- Yu, A.-M.; Choi, Y.H.; Tu, M.-J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef]
- Andersson, P. Preclinical Safety Assessment of Therapeutic Oligonucleotides. In Antisense RNA Design, Delivery, and Analysis; Springer: New York, NY, USA, 2022; pp. 355–370. [Google Scholar]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I Study of MRX34, a Liposomal MiR-34a Mimic, Administered Twice Weekly in Patients with Advanced Solid Tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of Pseudouridine into MRNA Enhances Translation by Diminishing PKR Activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef]
- Tanji, H.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Isobe, T.; Miyake, K.; Shimizu, T. Toll-like Receptor 8 Senses Degradation Products of Single-Stranded RNA. Nat. Struct. Mol. Biol. 2015, 22, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Janas, M.M.; Schlegel, M.K.; Harbison, C.E.; Yilmaz, V.O.; Jiang, Y.; Parmar, R.; Zlatev, I.; Castoreno, A.; Xu, H.; Shulga-Morskaya, S.; et al. Selection of GalNAc-Conjugated SiRNAs with Limited off-Target-Driven Rat Hepatotoxicity. Nat. Commun. 2018, 9, 723. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, X.-Y.; Lu, A.; Wang, X.-Y.; Jiang, L.-X.; Wang, J.-C. Non-Viral Vectors for RNA Delivery. J. Control. Release 2022, 342, 241–279. [Google Scholar] [CrossRef]
- Szebeni, J.; Simberg, D.; González-Fernández, Á.; Barenholz, Y.; Dobrovolskaia, M.A. Roadmap and Strategy for Overcoming Infusion Reactions to Nanomedicines. Nat. Nanotechnol. 2018, 13, 1100–1108. [Google Scholar] [CrossRef]
- Bitounis, D.; Jacquinet, E.; Rogers, M.A.; Amiji, M.M. Strategies to Reduce the Risks of MRNA Drug and Vaccine Toxicity. Nat. Rev. Drug Discov. 2024, 23, 281–300. [Google Scholar] [CrossRef] [PubMed]
- Priori, S.G.; Napolitano, C.; Tiso, N.; Memmi, M.; Vignati, G.; Bloise, R.; Sorrentino, V.; Danieli, G.A. Mutations in the Cardiac Ryanodine Receptor Gene (HRyR2) Underlie Catecholaminergic Polymorphic Ventricular Tachycardia. Circulation 2001, 103, 196–200. [Google Scholar] [CrossRef]
- Abdullah, N.M.; Ali, A. RYR2 Receptor Gene Mutation Associated with Catecholaminergic Polymorphic Ventricular Tachycardia in Children: A Case Report & Literature Review. Transl. Pediatr. 2024, 13, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Bers, D.M. Cardiac Excitation–Contraction Coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Colombi, B.; Memmi, M.; Zissimopoulos, S.; Rizzi, N.; Negri, S.; Imbriani, M.; Napolitano, C.; Lai, F.A.; Priori, S.G. Arrhythmogenesis in Catecholaminergic Polymorphic Ventricular Tachycardia. Circ. Res. 2006, 99, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Bongianino, R.; Denegri, M.; Mazzanti, A.; Lodola, F.; Vollero, A.; Boncompagni, S.; Fasciano, S.; Rizzo, G.; Mangione, D.; Barbaro, S.; et al. Allele-Specific Silencing of Mutant MRNA Rescues Ultrastructural and Arrhythmic Phenotype in Mice Carriers of the R4496C Mutation in the Ryanodine Receptor Gene (RYR2). Circ. Res. 2017, 121, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, V.; Iafisco, M.; Salvarani, N.; Vacchiano, M.; Carullo, P.; Ramírez-Rodríguez, G.B.; Patrício, T.; Tampieri, A.; Miragoli, M.; Catalucci, D. Bioinspired Negatively Charged Calcium Phosphate Nanocarriers for Cardiac Delivery of MicroRNAs. Nanomedicine 2016, 11, 891–906. [Google Scholar] [CrossRef] [PubMed]
- Miragoli, M.; Ceriotti, P.; Iafisco, M.; Vacchiano, M.; Salvarani, N.; Alogna, A.; Carullo, P.; Ramirez-Rodríguez, G.B.; Patrício, T.; Esposti, L.D.; et al. Inhalation of Peptide-Loaded Nanoparticles Improves Heart Failure. Sci. Transl. Med. 2018, 10, eaan6205. [Google Scholar] [CrossRef] [PubMed]
- Modica, J.; Di Mauro, V.; Barandalla-Sobrados, M.; Chavez, S.E.P.; Carullo, P.; Nemska, S.; Anselmo, A.; Condorelli, G.; Iafisco, M.; Miragoli, M.; et al. Nano-MiR-133a Replacement Therapy Blunts Pressure Overload–Induced Heart Failure. Circulation 2021, 144, 1973–1976. [Google Scholar] [CrossRef]
- Alogna, A.; Berboth, L.; Faragli, A.; Ötvös, J.; lo Muzio, F.P.; di Mauro, V.; Modica, J.; Quarta, E.; Semmler, L.; Deißler, P.M.; et al. Lung-to-Heart Nano-in-Micro Peptide Promotes Cardiac Recovery in a Pig Model of Chronic Heart Failure. J. Am. Coll. Cardiol. 2024, 83, 47–59. [Google Scholar] [CrossRef]
- Savi, M.; Rossi, S.; Bocchi, L.; Gennaccaro, L.; Cacciani, F.; Perotti, A.; Amidani, D.; Alinovi, R.; Goldoni, M.; Aliatis, I.; et al. Titanium Dioxide Nanoparticles Promote Arrhythmias via a Direct Interaction with Rat Cardiac Tissue. Part. Fibre Toxicol. 2014, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Mills, N.L.; Donaldson, K.; Hadoke, P.W.; Boon, N.A.; MacNee, W.; Cassee, F.R.; Sandström, T.; Blomberg, A.; Newby, D.E. Adverse Cardiovascular Effects of Air Pollution. Nat. Clin. Pract. Cardiovasc. Med. 2009, 6, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Iafisco, M.; Alogna, A.; Miragoli, M.; Catalucci, D. Cardiovascular Nanomedicine: The Route Ahead. Nanomedicine 2019, 14, 2391–2394. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, G.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of Autophagy and Reformation of Lysosomes Regulated by MTOR. Nature 2010, 465, 942–946. [Google Scholar] [CrossRef]
- Qiu, X.; Zheng, L.; Liu, X.; Hong, D.; He, M.; Tang, Z.; Tian, C.; Tan, G.; Hwang, S.; Shi, Z.; et al. ULK1 Inhibition as a Targeted Therapeutic Strategy for Psoriasis by Regulating Keratinocytes and Their Crosstalk With Neutrophils. Front. Immunol. 2021, 12, 714274. [Google Scholar] [CrossRef]
- Verfaillie, T.; Salazar, M.; Velasco, G.; Agostinis, P. Linking ER Stress to Autophagy: Potential Implications for Cancer Therapy. Int. J. Cell Biol. 2010, 2010, 930509. [Google Scholar] [CrossRef] [PubMed]
- Corazzari, M.; Gagliardi, M.; Fimia, G.M.; Piacentini, M. Endoplasmic Reticulum Stress, Unfolded Protein Response, and Cancer Cell Fate. Front. Oncol. 2017, 7, 78. [Google Scholar] [CrossRef]
- Zhao, Q. A Mitochondrial Specific Stress Response in Mammalian Cells. EMBO J. 2002, 21, 4411–4419. [Google Scholar] [CrossRef]
- Zhou, D.; Palam, L.R.; Jiang, L.; Narasimhan, J.; Staschke, K.A.; Wek, R.C. Phosphorylation of EIF2 Directs ATF5 Translational Control in Response to Diverse Stress Conditions. J. Biol. Chem. 2008, 283, 7064–7073. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, A.; Sena, L.A.; Diebold, L.P.; Mazar, A.P.; Chandel, N.S. Targeting SOD1 Reduces Experimental Non–Small-Cell Lung Cancer. J. Clin. Investig. 2014, 124, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, Z.; Zhao, X.; Keen, L.; Kong, X. Calcium Phosphate Nanoparticles-Based Systems for SiRNA Delivery. Regen. Biomater. 2016, 3, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Degli Esposti, L.; Carella, F.; Adamiano, A.; Tampieri, A.; Iafisco, M. Calcium Phosphate-Based Nanosystems for Advanced Targeted Nanomedicine. Drug Dev. Ind. Pharm. 2018, 44, 1223–1238. [Google Scholar] [CrossRef]
- Lee, Y.; Jeong, M.; Park, J.; Jung, H.; Lee, H. Immunogenicity of Lipid Nanoparticles and Its Impact on the Efficacy of MRNA Vaccines and Therapeutics. Exp. Mol. Med. 2023, 55, 2085–2096. [Google Scholar] [CrossRef] [PubMed]
- Najahi-Missaoui, W.; Arnold, R.D.; Cummings, B.S. Safe Nanoparticles: Are We There Yet? Int. J. Mol. Sci. 2020, 22, 385. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Jing, Q.; Li, Y.; Han, J. RNA Modification: Mechanisms and Therapeutic Targets. Mol. Biomed. 2023, 4, 25. [Google Scholar] [CrossRef]
- Pauls, E.; Senserrich, J.; Bofill, M.; Clotet, B.; Esté, J.A. Induction of Interleukins IL-6 and IL-8 by SiRNA. Clin. Exp. Immunol. 2006, 147, 189–196. [Google Scholar] [CrossRef]
- Sioud, M. Induction of Inflammatory Cytokines and Interferon Responses by Double-Stranded and Single-Stranded SiRNAs Is Sequence-Dependent and Requires Endosomal Localization. J. Mol. Biol. 2005, 348, 1079–1090. [Google Scholar] [CrossRef]
- Meng, Z.; Lu, M. RNA Interference-Induced Innate Immunity, Off-Target Effect, or Immune Adjuvant? Front. Immunol. 2017, 8, 331. [Google Scholar] [CrossRef]
- Robbins, M.; Judge, A.; MacLachlan, I. SiRNA and Innate Immunity. Oligonucleotides 2009, 19, 89–102. [Google Scholar] [CrossRef]


| Genes | Pathway | Reference |
|---|---|---|
| mTOR | Autophagy | [37] |
| ULK1 | Autophagy | [38] |
| ATF4 | Endoplasmic reticulum stress | [39] |
| BBC3 | Endoplasmic reticulum stress | [40] |
| DDIT3 | Endoplasmic reticulum stress | [41] |
| ATF5 | Mitochondrial stress | [42] |
| SOD1 | Oxidative stress | [43] |
| Sample | Z-Average (nm) | PDI | ζ-Potential (mV) | CaP NPs Concentration (mg/mL) | siRNA Loading (wt.%) |
|---|---|---|---|---|---|
| CaP NPs | 65 ± 6 | 0.30 ± 0.03 | −34 ± 3 | 0.6 ± 0.2 | - |
| CaP-siRNA | 40 ± 5 | 0.31 ± 0.02 | −35 ± 4 | 0.6 ± 0.1 | 2.7 ± 0.3 |
| CaP-scramble | 50 ± 5 | 0.32 ± 0.03 | −33 ± 5 | 0.6 ± 0.1 | 1.8 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bettinsoli, V.; Melzi, G.; Crea, A.; Degli Esposti, L.; Iafisco, M.; Catalucci, D.; Ciana, P.; Corsini, E. A Novel Approach for In Vitro Testing and Hazard Evaluation of Nanoformulated RyR2-Targeting siRNA Drugs Using Human PBMCs. Life 2025, 15, 95. https://doi.org/10.3390/life15010095
Bettinsoli V, Melzi G, Crea A, Degli Esposti L, Iafisco M, Catalucci D, Ciana P, Corsini E. A Novel Approach for In Vitro Testing and Hazard Evaluation of Nanoformulated RyR2-Targeting siRNA Drugs Using Human PBMCs. Life. 2025; 15(1):95. https://doi.org/10.3390/life15010095
Chicago/Turabian StyleBettinsoli, Valeria, Gloria Melzi, Angelica Crea, Lorenzo Degli Esposti, Michele Iafisco, Daniele Catalucci, Paolo Ciana, and Emanuela Corsini. 2025. "A Novel Approach for In Vitro Testing and Hazard Evaluation of Nanoformulated RyR2-Targeting siRNA Drugs Using Human PBMCs" Life 15, no. 1: 95. https://doi.org/10.3390/life15010095
APA StyleBettinsoli, V., Melzi, G., Crea, A., Degli Esposti, L., Iafisco, M., Catalucci, D., Ciana, P., & Corsini, E. (2025). A Novel Approach for In Vitro Testing and Hazard Evaluation of Nanoformulated RyR2-Targeting siRNA Drugs Using Human PBMCs. Life, 15(1), 95. https://doi.org/10.3390/life15010095








