Innovative Diagnostic Approaches and Challenges in the Management of HIV: Bridging Basic Science and Clinical Practice
Abstract
1. Introduction
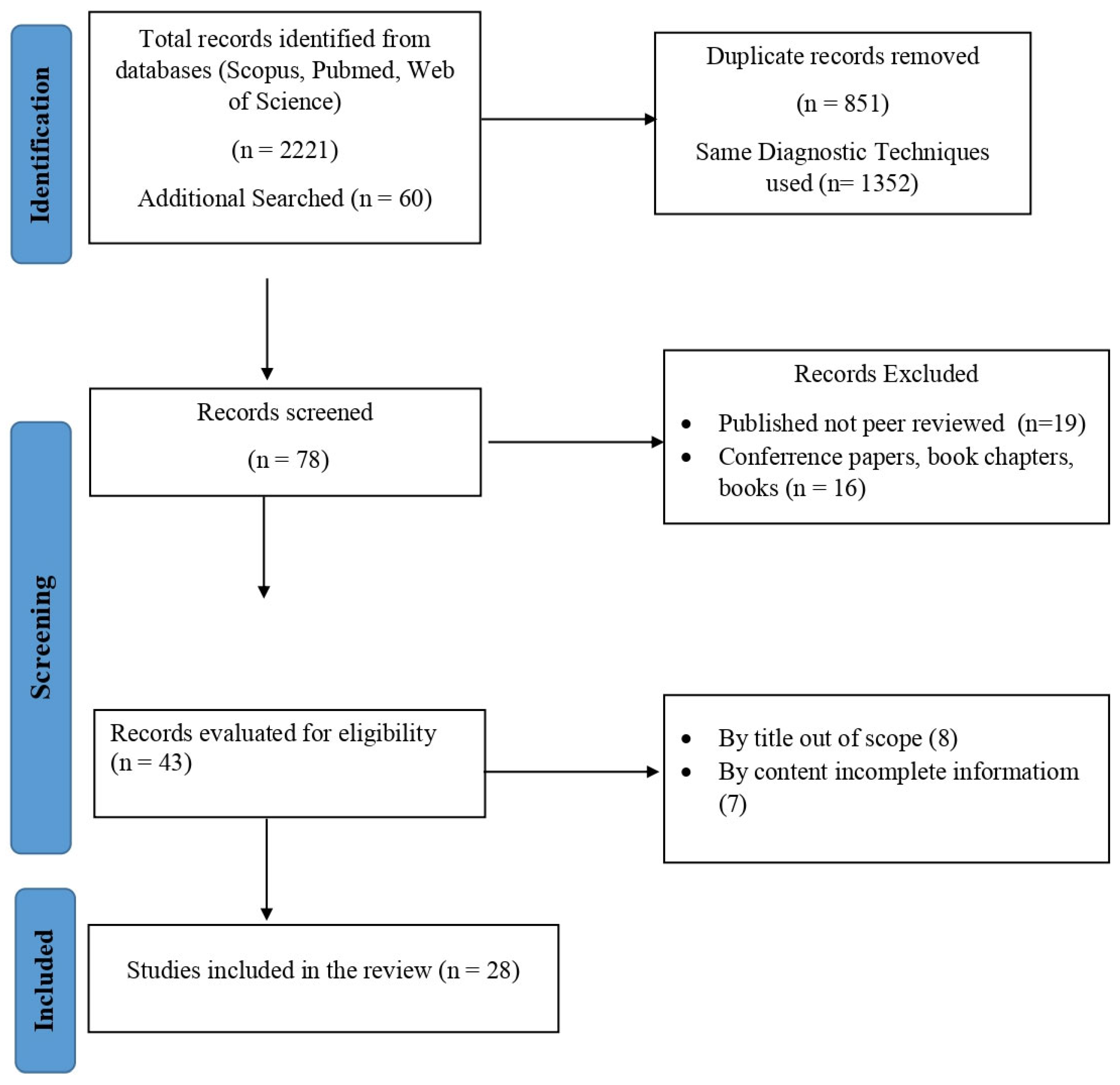
2. Methodology
2.1. Search Strategy
2.2. Study Selection
3. Diagnostic Innovations in HIV
| Diagnostic Technique | Description | References | Time Period |
|---|---|---|---|
| ELISA (Enzyme-Linked Immunosorbent Assay) | Detects HIV antibodies in blood samples, providing first lab-based serological test for HIV | [3] | Early 1980s |
| Western Blot | Confirmatory test for HIV, identifying specific HIV proteins via antibody binding | [4] | Mid-1980s |
| Rapid Antibody Tests | Quick detection of HIV antibodies using fingerstick blood or oral fluids, e.g., OraQuick HIV test | [5] | Early 2000s |
| NAT (Nucleic Acid Testing) | Directly detects HIV RNA in blood, useful for early detection and confirmation | [5] | 2000s |
| PCR (Polymerase Chain Reaction) | Identifies HIV DNA/RNA in blood, especially valuable in early detection and viral load assessment | [6] | 1990s |
| qPCR (Quantitative PCR) | Quantifies HIV viral load in blood to monitor treatment effectiveness and disease progression | [7] | Early 2000s |
| Multiplex Testing | Combines HIV antibody and antigen detection to increase sensitivity, identifying both acute and chronic infections | [8] | 2010s |
| Lab-on-a-Chip and Microfluidics | Miniaturized diagnostics integrating multiple assays for rapid POC HIV testing, e.g., CD4+ counts | [9] | 2010s–present |
| NGS (Next-Generation Sequencing) | High-throughput sequencing allowing detailed HIV genetic analysis, detecting drug resistance and viral diversity | [10] | Late 2000s–present |
| Biosensors | Detects HIV antigens/antibodies or nucleic acids with portable sensors for POC, enabling rapid results | [11] | 2010s–present |
| CRISPR-Based Diagnostics | Gene-editing technology adapted to detect HIV nucleic acids with high sensitivity, e.g., SHERLOCK assay | [12] | 2016–present |
| Machine Learning and AI | Analyzes large genomic datasets to predict HIV drug resistance patterns, optimizing treatment regimens | [13] | 2020s–present |
3.1. Molecular Assays
3.1.1. Polymerase Chain Reaction (PCR) Developments
3.1.2. Real-Time Quantitative PCR (qPCR)
3.1.3. Nucleic Acid Testing (NAT)
3.2. Biosensors and Lab-on-a-Chip Technologies
3.3. Next-Generation Sequencing
3.4. Point-of-Care Testing: Rapid Antigen and Antibody Testing
3.5. CRISPR-Based Diagnostics for HIV
3.6. Machine Learning and AI for HIV Diagnosis
3.6.1. Predictive Modeling for HIV Diagnosis and Progression
3.6.2. Diagnostic Accuracy and Rapid Screening
3.6.3. AI in HIV Screening and Early Detection
3.7. Limitations in HIV Diagnostic Approaches
3.7.1. Sensitivity Gaps
3.7.2. Latency Issues
3.7.3. Resource Constraints
3.7.4. Cost-Effective Diagnostic Innovations for Resource-Limited Settings
4. Challenges in Clinical Translation
4.1. Logistic Barriers
4.2. Economic and Infrastructural Constraints
4.3. Patient Accessibility and Acceptability Issues
4.4. Education and Training Gaps
5. Bridging the Gap: Strategies for Integration
| Strategy | Description | References |
|---|---|---|
| Interdisciplinary Collaboration | Collaboration among healthcare providers, researchers, and policymakers fosters innovation and translates research into clinical practice. | [44] |
| Infrastructure and Technological Investment | Building diagnostic infrastructure and investing in technologies like point-of-care testing and mobile health units expand reach and reliability. | [45] |
| Healthcare Professional Training and Education | Equipping healthcare workers with updated skills through training in diagnostics, patient handling, and emerging technologies enhances service delivery. | [46] |
| Public Health Policies and Supportive Frameworks | Establishing supportive policies, including funding for diagnostics and patient access programs, ensures sustainable health outcomes and continuity of care. | [47] |
5.1. Interdisciplinary Collaboration
5.2. Infrastructure and Technological Investment
5.3. Healthcare Professional Training and Education
5.4. Public Health Policies and Supportive Frameworks
6. Future Directions and Emerging Technologies
6.1. CRISPR-Based Diagnostics
6.2. AI-Powered Diagnostic Algorithms
6.3. Advancements in Biosensor Technology
6.4. Nanotechnology and Next-Generation Sequencing (NGS)
6.5. Integrative Platforms and Telemedicine
6.6. Future Directions: Emphasis on Management
6.7. Future Directions
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zaid, D.; Greenman, Y. Human Immunodeficiency Virus Infection and the Endocrine System. Endocrinol. Metab. 2019, 34, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S. Democratizing nucleic acid-based molecular diagnostic tests for infectious diseases at resource-limited settings–from point of care to extreme point of care. Sens. Diagn. 2024, 3, 536–561. [Google Scholar] [CrossRef]
- Iha, K.; Inada, M.; Kawada, N.; Nakaishi, K.; Watabe, S.; Tan, Y.H.; Shen, C.; Ke, L.Y.; Yoshimura, T.; Ito, E. Ultrasensitive ELISA Developed for Diagnosis. Diagnostics 2019, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.S. Human immunodeficiency virus diagnostic testing: 30 years of evolution. Clin. Vaccine Immunol. 2016, 23, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Parekh, B.S.; Ou, C.Y.; Fonjungo, P.N.; Kalou, M.B.; Rottinghaus, E.; Puren, A.; Alexander, H.; Hurlston Cox, M.; Nkengasong, J.N. Diagnosis of Human Immunodeficiency Virus Infection. Clin. Microbiol. Rev. 2018, 32, e00064-18. [Google Scholar] [CrossRef]
- Summah, H.; Zhu, Y.G.; Falagas, M.E.; Vouloumanou, E.K.; Qu, J.M. Use of real-time polymerase chain reaction for the diagnosis of Pneumocystis pneumonia in immunocompromised patients: A meta-analysis. Chin. Med. J. 2013, 126, 1965–1973. [Google Scholar] [CrossRef]
- Gaebler, C.; Lorenzi, J.C.C.; Oliveira, T.Y.; Nogueira, L.; Ramos, V.; Lu, C.L.; Pai, J.A.; Mendoza, P.; Jankovic, M.; Caskey, M.; et al. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J. Exp. Med. 2019, 216, 2253–2264. [Google Scholar] [CrossRef]
- Lee, J.H.; Seo, H.S.; Kwon, J.H.; Kim, H.T.; Kwon, K.C.; Sim, S.J.; Cha, Y.J.; Lee, J. Multiplex diagnosis of viral infectious diseases (AIDS, hepatitis C, and hepatitis A) based on point of care lateral flow assay using engineered proteinticles. Biosens. Bioelectron. 2015, 69, 213–225. [Google Scholar] [CrossRef]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef]
- Gibson, R.M.; Schmotzer, C.L.; Quiñones-Mateu, M.E. Next-Generation Sequencing to Help Monitor Patients Infected with HIV: Ready for Clinical Use? Curr. Infect. Dis. Rep. 2014, 16, 401. [Google Scholar] [CrossRef]
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. HIV biosensors for early diagnosis of infection: The intertwine of nanotechnology with sensing strategies. Talanta 2020, 206, 120201. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, A.; Jude Serpes, N.; Gupta, T.; James, A.; Sharma, A.; Kumar, D.; Nagraik, R.; Kumar, V.; Pandey, S. Advancements in CRISPR-Based Biosensing for Next-Gen Point of Care Diagnostic Application. Biosensors 2023, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Turbé, V.; Herbst, C.; Mngomezulu, T.; Meshkinfamfard, S.; Dlamini, N.; Mhlongo, T.; Smit, T.; Cherepanova, V.; Shimada, K.; Budd, J.; et al. Deep learning of HIV field-based rapid tests. Nat. Med. 2021, 27, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Pai, N.P.; Karellis, A.; Kim, J.; Peter, T. Modern diagnostic technologies for HIV. Lancet HIV 2020, 7, E574–E581. [Google Scholar] [CrossRef]
- Boyle, D.S.; Lehman, D.A.; Lillis, L.; Peterson, D.; Singhal, M.; Armes, N.; Parker, M.; Piepenburg, O.; Overbaugh, J. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio 2013, 4, e00135-13. [Google Scholar] [CrossRef]
- Rutsaert, S.; Bosman, K.; Trypsteen, W.; Nijhuis, M.; Vandekerckhove, L. Digital PCR as a tool to measure HIV persistence. Retrovirology 2018, 15, 16. [Google Scholar] [CrossRef]
- Zhao, J.; Chang, L.; Wang, L. Nucleic acid testing and molecular characterization of HIV infections. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 829–842. [Google Scholar] [CrossRef]
- Sengupta, J.; Adhikari, A.; Hussain, C.M. Graphene-based analytical lab-on-chip devices for detection of viruses: A review. Carbon. Trends 2021, 4, 100072. [Google Scholar] [CrossRef]
- Mauk, M.; Song, J.; Bau, H.H.; Gross, R.; Bushman, F.D.; Collman, R.G.; Liu, C. Miniaturized devices for point of care molecular detection of HIV. Lab Chip 2017, 17, 382–394. [Google Scholar] [CrossRef]
- Glynn, M.T.; Kinahan, D.J.; Ducrée, J. Rapid, low-cost and instrument-free CD4+ cell counting for HIV diagnostics in resource-poor settings. Lab Chip 2014, 14, 2844–2851. [Google Scholar] [CrossRef]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses—Current challenges and future perspectives. Biosens. Bioelectron. 2020, 163, 112291. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Ríos, S.; Parkin, N.; Swanstrom, R.; Paredes, R.; Shafer, R.; Ji, H.; Kantor, R. Next-Generation Sequencing for HIV Drug Resistance Testing: Laboratory, Clinical, and Implementation Considerations. Viruses 2020, 12, 617. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of care testing for infectious diseases. Clin. Chim. Acta 2019, 493, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef]
- Pant Pai, N.; Balram, B.; Shivkumar, S.; Martinez-Cajas, J.L.; Claessens, C.; Lambert, G.; Peeling, R.W.; Joseph, L. Head-to-head comparison of accuracy of a rapid point-of-care HIV test with oral versus whole-blood specimens: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 373–380. [Google Scholar]
- Luo, W.; Masciotra, S.; Delaney, K.P.; Charurat, M.; Croxton, T.; Constantine, N.; Blattner, W.; Wesolowski, L.; Owen, S.M. Comparison of HIV oral fluid and plasma antibody results during early infection in a longitudinal Nigerian cohort. J. Clin. Virol. 2013, 58 (Suppl. S1), e113–e118. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Huang, D.; Zhao, Y.; Fang, M.; Shen, P.; Xu, H.; He, Y.; Chen, S.; Si, Z.; Xu, Z. Magnetofluid-integrated biosensors based on DNase-dead Cas12a for visual point-of-care testing of HIV-1 by an up and down chip. Lab Chip 2023, 23, 4265–4275. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Steiner, M.C.; Gibson, K.M.; Crandall, K.A. Drug Resistance Prediction Using Deep Learning Techniques on HIV-1 Sequence Data. Viruses 2020, 12, 560. [Google Scholar] [CrossRef] [PubMed]
- Bhaiyya, M.; Panigrahi, D.; Rewatkar, P.; Haick, H. Role of Machine Learning Assisted Biosensors in Point-of-Care-Testing For Clinical Decisions. ACS Sens. 2024, 9, 4495–4519. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.A.; Zilouchian, H.; Caputi, M.; Asghar, W. Advances in HIV diagnosis and monitoring. Crit. Rev. Biotechnol. 2020, 40, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Bacon, A.; Wang, W.; Lee, H.; Umrao, S.; Sinawang, P.D.; Akin, D.; Khemtonglang, K.; Tan, A.; Hirshfield, S.; Demirci, U.; et al. Review of HIV Self Testing Technologies and Promising Approaches for the Next Generation. Biosensors 2023, 13, 298. [Google Scholar] [CrossRef]
- Wi, T.E.; Ndowa, F.J.; Ferreyra, C.; Kelly-Cirino, C.; Taylor, M.M.; Toskin, I.; Kiarie, J.; Santesso, N.; Unemo, M. Diagnosing sexually transmitted infections in resource-constrained settings: Challenges and ways forward. J. Int. AIDS Soc. 2019, 22 (Suppl. S6), e25343. [Google Scholar] [CrossRef]
- Heidt, B.; Siqueira, W.F.; Eersels, K.; Diliën, H.; van Grinsven, B.; Fujiwara, R.T.; Cleij, T.J. Point of care diagnostics in resource-limited settings: A review of the present and future of PoC in its most needed environment. Biosensors 2020, 10, 133. [Google Scholar] [CrossRef]
- Younis, M.A.; Tawfeek, H.M.; Abdellatif, A.A.H.; Abdel-Aleem, J.A.; Harashima, H. Clinical translation of nanomedicines: Challenges, opportunities, and keys. Adv. Drug Deliv. Rev. 2022, 181, 114083. [Google Scholar] [CrossRef]
- Zhang, Y.; Guy, R.; Camara, H.; Applegate, T.L.; Wiseman, V.; Treloar, C.; Lafferty, L. Barriers and facilitators to HIV and syphilis rapid diagnostic testing in antenatal care settings in low-income and middle-income countries: A systematic review. BMJ Glob. Health 2022, 7, e009408. [Google Scholar] [CrossRef]
- Roberts, T.; Cohn, J.; Bonner, K.; Hargreaves, S. Scale-up of Routine Viral Load Testing in Resource-Poor Settings: Current and Future Implementation Challenges. Clin. Infect. Dis. 2016, 62, 1043–1048. [Google Scholar] [CrossRef]
- Bucyibaruta, B.J.; Eyles, J.; Harris, B.; Kabera, G.; Oboirien, K.; Ngyende, B. Patients’ perspectives of acceptability of ART, TB and maternal health services in a subdistrict of Johannesburg, South Africa. BMC Health Serv. Res. 2018, 18, 839. [Google Scholar] [CrossRef]
- Zeleke, E.A.; Stephens, J.H.; Gesesew, H.A.; Gello, B.M.; Ziersch, A. Acceptability and use of HIV self-testing among young people in sub-Saharan Africa: A mixed methods systematic review. BMC Prim. Care 2024, 25, 369. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.J.; Pillai, G.G.; Andrade, C.J.; Ligibel, J.A.; Basu, P.; Cohen, L.; Khan, I.A.; Mustian, K.M.; Puthiyedath, R.; Dhiman, K.S.; et al. Integrative oncology: Addressing the global challenges of cancer prevention and treatment. CA Cancer J. Clin. 2022, 72, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Khorram-Manesh, A.; Burkle, F.M., Jr.; Goniewicz, K. Pandemics: Past, present, and future: Multitasking challenges in need of cross-disciplinary, transdisciplinary, and multidisciplinary collaborative solutions. Osong Public Health Res. Perspect. 2024, 15, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Grossman, C.I.; Ross, A.L.; Auerbach, J.D.; Ananworanich, J.; Dubé, K.; Tucker, J.D.; Noseda, V.; Possas, C.; Rausch, D.M.; International AIDS Society (IAS) ‘Towards an HIV Cure’ Initiative. Towards Multidisciplinary HIV-Cure Research: Integrating Social Science with Biomedical Research. Trends Microbiol. 2016, 24, 5–11. [Google Scholar] [CrossRef]
- Brault, M.A.; Vermund, S.H.; Aliyu, M.H.; Omer, S.B.; Clark, D.; Spiegelman, D. Leveraging HIV Care Infrastructures for Integrated Chronic Disease and Pandemic Management in Sub-Saharan Africa. Int. J. Environ. Res. Public Health 2021, 18, 10751. [Google Scholar] [CrossRef]
- Kennedy, C.E.; Yeh, P.T.; Johnson, C.; Baggaley, R. Should trained lay providers perform HIV testing? A systematic review to inform World Health Organization guidelines. AIDS Care 2017, 29, 1473–1479. [Google Scholar] [CrossRef]
- Adeyemi, O.; Lyons, M.; Njim, T.; Okebe, J.; Birungi, J.; Nana, K.; Claude Mbanya, J.; Mfinanga, S.; Ramaiya, K.; Jaffar, S.; et al. Integration of non-communicable disease and HIV/AIDS management: A review of healthcare policies and plans in East Africa. BMJ Glob. Health 2021, 6, e004669. [Google Scholar] [CrossRef]
- Krishnamoorthy, S.; Dua, A.; Gupta, S. Role of emerging technologies in future IoT-driven Healthcare 4.0 technologies: A survey, current challenges and future directions. J. Ambient Intell. Humaniz. Comput. 2023, 14, 361–407. [Google Scholar] [CrossRef]
- Sebastião, C.S.; Pingarilho, M.; Bathy, J.; Bonfim, E.; Toancha, K.; Miranda, M.N.S.; Martins, M.R.O.; Gomes, P.; Lázaro, L.; Pina-Araujo, I.; et al. MARVEL-minimising the emergence and dissemination of HIV-1 drug resistance in Portuguese-speaking African Countries (PALOP): Low-cost portable NGS platform for HIV-1 surveillance in Africa. BMC Infect. Dis. 2024, 24, 884. [Google Scholar] [CrossRef]
- Del Giovane, S.; Bagheri, N.; Di Pede, A.C.; Chamorro, A.; Ranallo, S.; Migliorelli, D.; Burr, L.; Paoletti, S.; Altug, H.; Porchetta, A. Challenges and perspectives of CRISPR-based technology for diagnostic applications. TrAC Trends Anal. Chem. 2024, 172, 117594. [Google Scholar] [CrossRef]
- Olaboye, J.A.; Maha, C.C.; Kolawole, T.O.; Abdul, S. Integrative analysis of AI-driven optimization in HIV treatment regimens. Comput. Sci. IT Res. J. 2024, 5, 1314–1334. [Google Scholar] [CrossRef]
- Altindiş, M.; Kahraman Kilbaş, E.P. Managing Viral Emerging Infectious Diseases via Current and Future Molecular Diagnostics. Diagnostics 2023, 13, 1421. [Google Scholar] [CrossRef] [PubMed]
- Cuadros, D.F.; Huang, Q.; Mathenjwa, T.; Gareta, D.; Devi, C.; Musuka, G. Unlocking the potential of telehealth in Africa for HIV: Opportunities, challenges, and pathways to equitable healthcare delivery. Front. Digit. Health 2024, 6, 1278223. [Google Scholar] [CrossRef]
- Maggiolo, F.; Bandera, A.; Bonora, S.; Borderi, M.; Calcagno, A.; Cattelan, A.; Cingolani, A.; Gianotti, N.; Lichtner, M.; Lo Caputo, S.; et al. Enhancing care for people living with HIV: Current and future monitoring approaches. Expert Rev. Anti-Infect. Ther. 2021, 19, 443–456. [Google Scholar] [CrossRef] [PubMed]
| Managing Logistical Barriers in HIV Diagnostics | ||||||
|---|---|---|---|---|---|---|
| Identify Barriers | Resource Allocation and Prioritization | Supply Chain Optimization | Training and Capacity Building | Quality Assurance and Monitoring | Data Management and Reporting | Evaluate and Adapt |
| Input | Input | Input | Input | Input | Input | Input |
| List of logistical challenges (e.g., limited transportation, distribution issues, lack of trained personnel, equipment shortages). | Identified barriers and available resources (funding, personnel, and equipment). | Data on local supply chain challenges (supplier delays, regulatory requirements). | List of required skills and training gaps among healthcare personnel. | Established protocols for quality control and diagnostic standards. | Patient and diagnostic data, resource usage, and operational reports. | Ongoing data from monitoring, quality checks, and resource use. |
| Process | Process | Process | Process | Process | Process | Process |
| Conduct needs assessment in target regions, examining barriers specific to transportation, infrastructure, and resources. | Prioritize high-need regions or facilities, allocating resources based on severity of constraints and population needs. | Partner with local suppliers, optimize routes, and work with logistics experts to streamline transport and distribution. | Develop tailored training programs for operating diagnostic equipment, sample handling, and patient data management. | Implement routine quality checks and real-time monitoring using digital tools to track diagnostic accuracy and service delivery. | Use centralized databases to track results, manage patient information, and analyze logistical performance. | Review and assess logistical strategy effectiveness, adjust resource allocation, training, or supply chain processes as needed. |
| Output | Output | Output | Output | Output | Output | Output |
| Detailed report of logistical constraints by region or facility. | Resource allocation plan for phased implementation. | Optimized distribution routes and schedules. | Trained workforce capable of handling diagnostic procedures and equipment efficiently. | Quality-controlled processes and reliable diagnostics, with performance data for continuous improvement. | Comprehensive reporting on diagnostic impact, resource use, and areas for logistical refinement. | Updated logistics model with continuous improvements for scaling and replicating in other regions. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afzal, M.; Agarwal, S.; Elshaikh, R.H.; Babker, A.M.A.; Osman, E.A.I.; Choudhary, R.K.; Jaiswal, S.; Zahir, F.; Prabhakar, P.K.; Abbas, A.M.; et al. Innovative Diagnostic Approaches and Challenges in the Management of HIV: Bridging Basic Science and Clinical Practice. Life 2025, 15, 209. https://doi.org/10.3390/life15020209
Afzal M, Agarwal S, Elshaikh RH, Babker AMA, Osman EAI, Choudhary RK, Jaiswal S, Zahir F, Prabhakar PK, Abbas AM, et al. Innovative Diagnostic Approaches and Challenges in the Management of HIV: Bridging Basic Science and Clinical Practice. Life. 2025; 15(2):209. https://doi.org/10.3390/life15020209
Chicago/Turabian StyleAfzal, Mohd, Shagun Agarwal, Rabab H. Elshaikh, Asaad M. A. Babker, Einas Awad Ibrahim Osman, Ranjay Kumar Choudhary, Suresh Jaiswal, Farhana Zahir, Pranav Kumar Prabhakar, Anass M. Abbas, and et al. 2025. "Innovative Diagnostic Approaches and Challenges in the Management of HIV: Bridging Basic Science and Clinical Practice" Life 15, no. 2: 209. https://doi.org/10.3390/life15020209
APA StyleAfzal, M., Agarwal, S., Elshaikh, R. H., Babker, A. M. A., Osman, E. A. I., Choudhary, R. K., Jaiswal, S., Zahir, F., Prabhakar, P. K., Abbas, A. M., Shalabi, M. G., & Sah, A. K. (2025). Innovative Diagnostic Approaches and Challenges in the Management of HIV: Bridging Basic Science and Clinical Practice. Life, 15(2), 209. https://doi.org/10.3390/life15020209











