Genome-Wide Identification and Expression of the ERF Gene Family in Populus trichocarpa and Their Responses to Nitrogen and Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the ERF Transcription Factors in P. trichocarpa
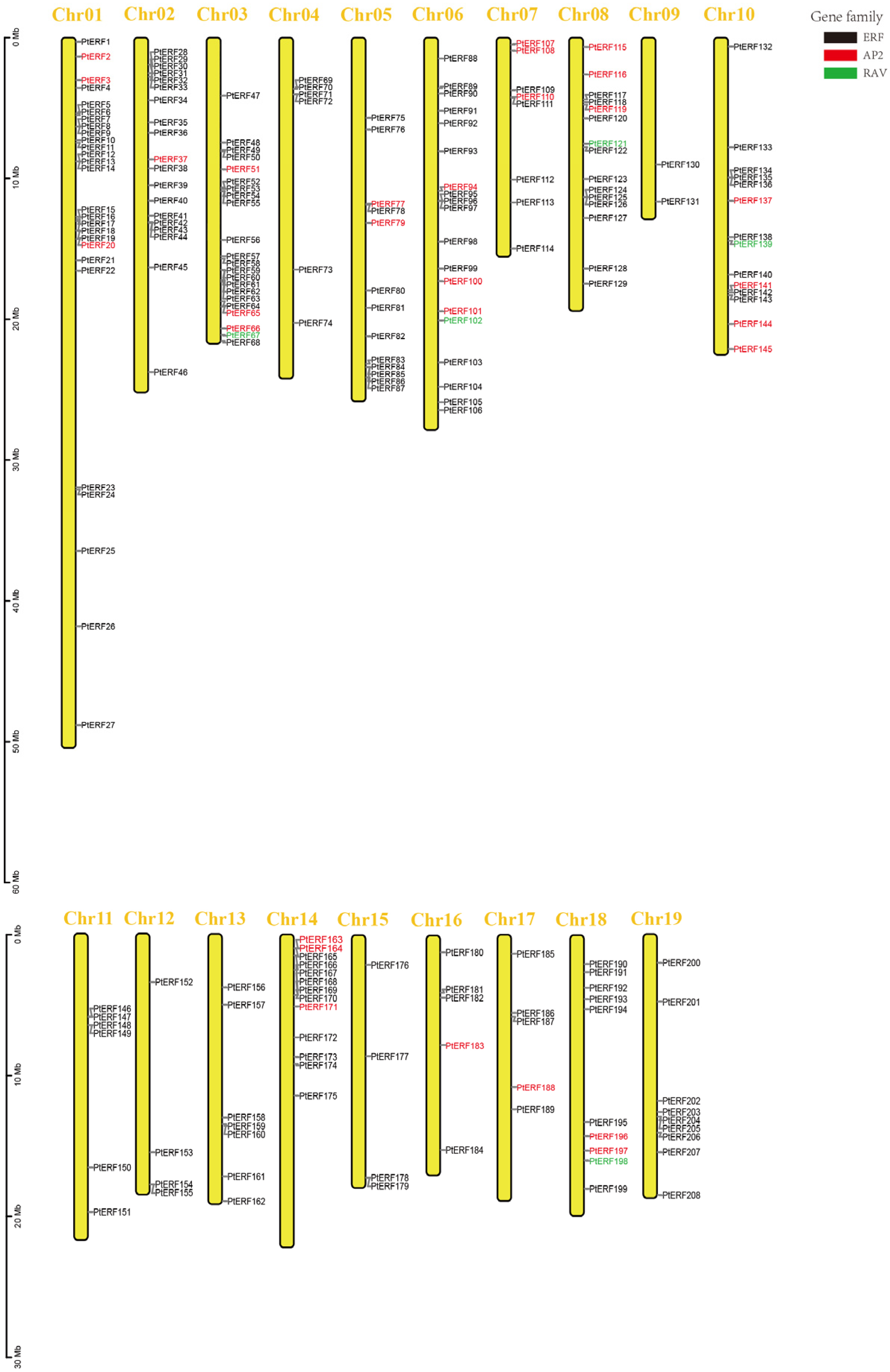
2.2. Chromosomal Distribution and Bioinformatics Analysis
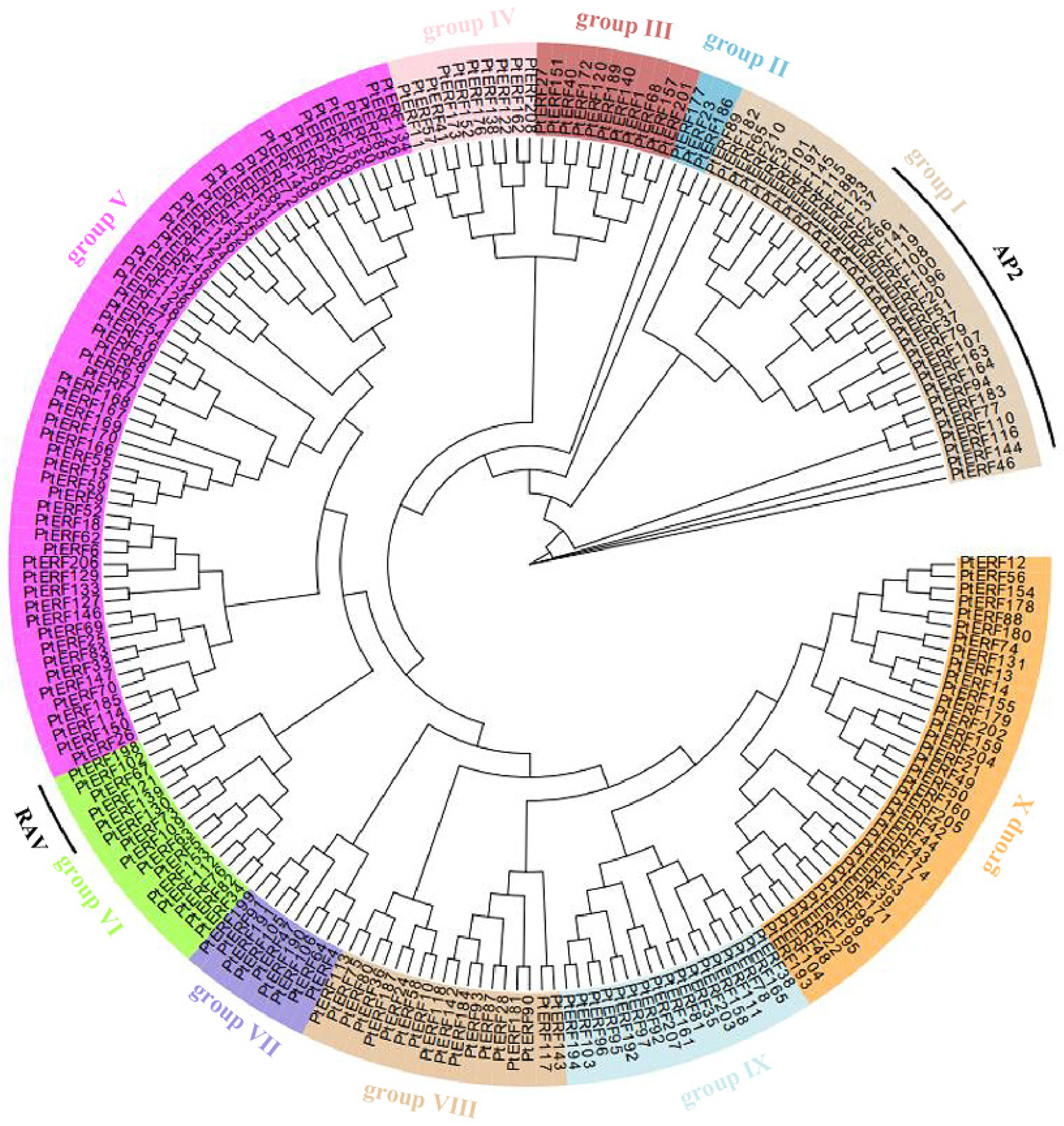
2.3. Phylogenetic and Sequence Analysis
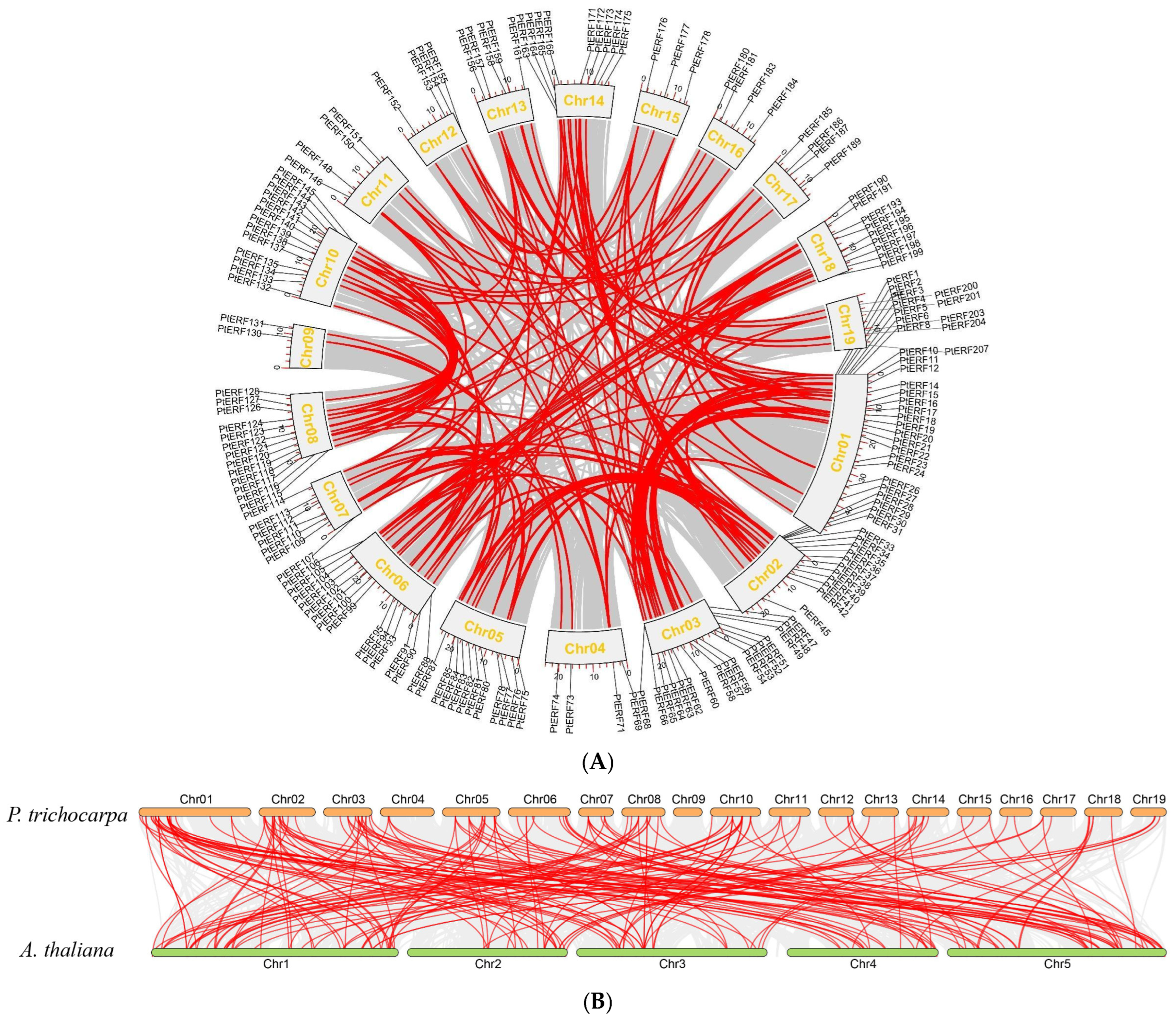
2.4. Analysis of Gene Replication Events
2.5. Analysis of Cis-Acting Elements
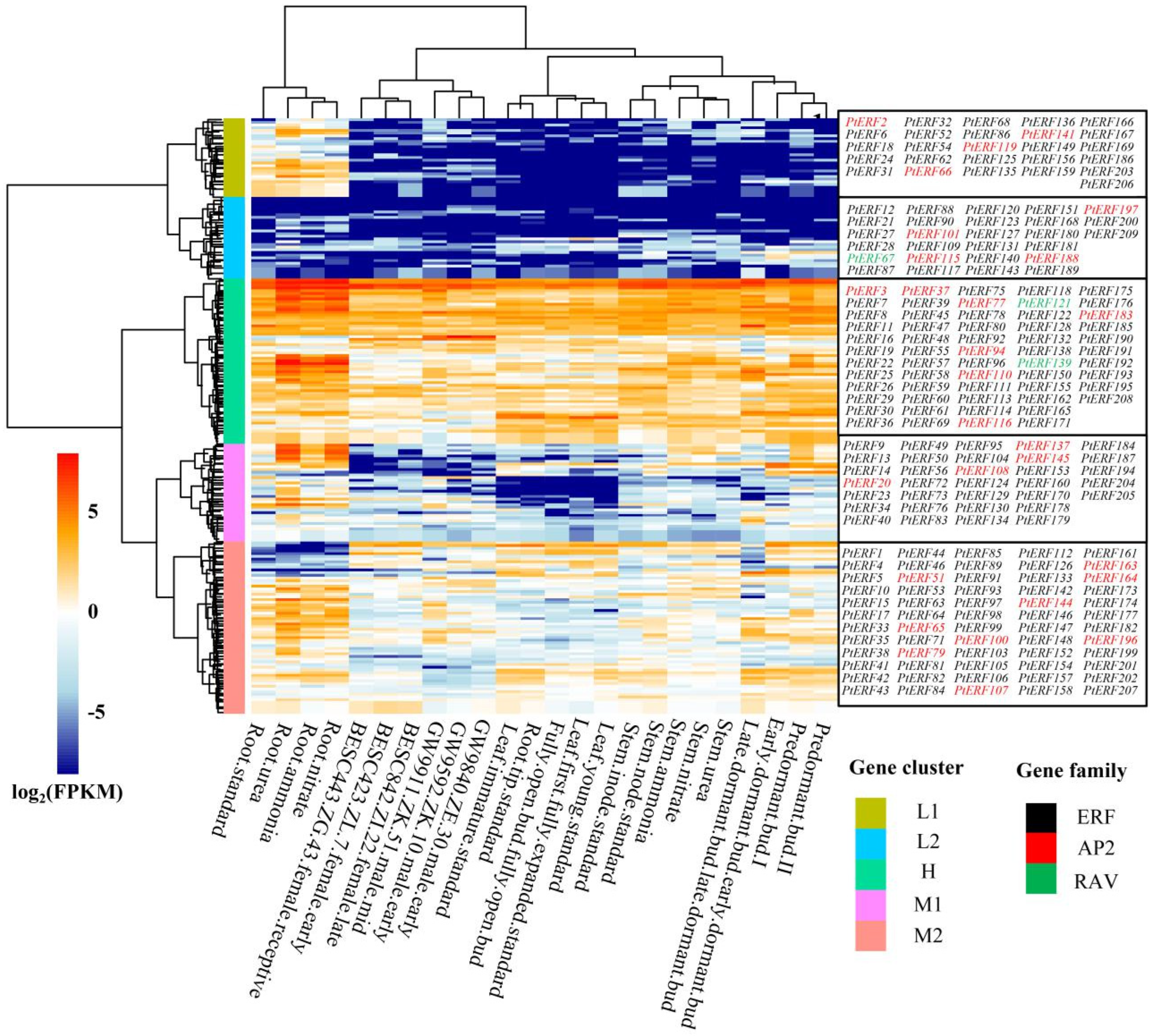
2.6. Gene Expression Analysis
2.7. Co-Expression Network Analysis
3. Results
3.1. Identification and Physicochemical Properties of the PtERF Gene Family Members
3.2. Phylogenetic and Structural Analysis of the PtERF Gene Family Members
3.3. Duplication Events Analysis of PtERF Gene Family
3.4. Cis-Acting Element Analysis of PtERF Gene Family
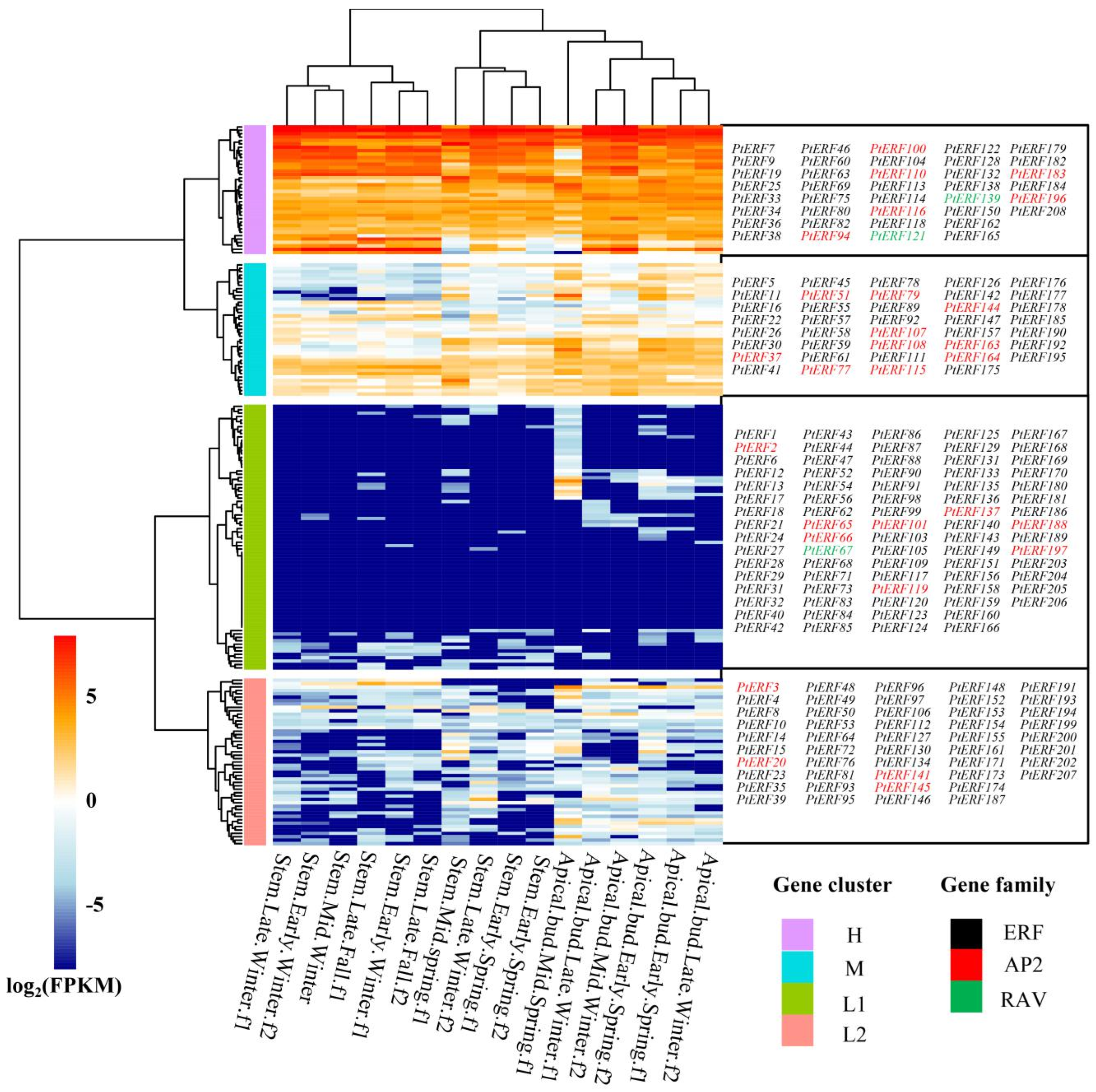
3.5. Tissue-Specific Expression Analysis of PtERF Genes in P. trichocarpa
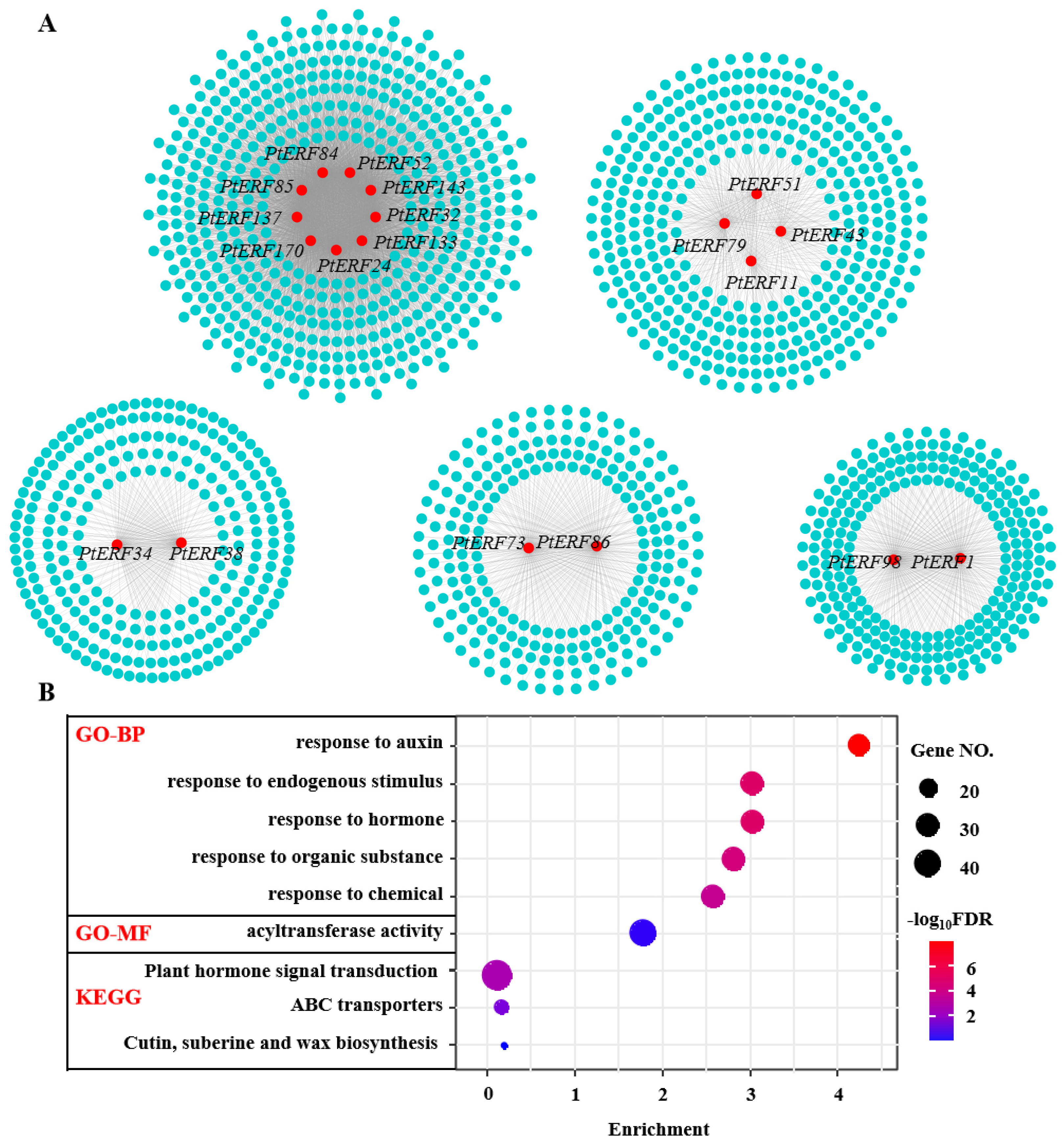
3.6. Co-Expression Analysis of PtERF Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, K.B.; Foley, R.C.; Oate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lim, C.W.; Lee, S.C. Role of pepper MYB transcription factor CaDIM1 in regulation of the drought response. Front. Plant Sci. 2022, 13, 1028392. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Dong, Q.; Wu, J.; Li, H.; Luan, H.; Jia, P.; Zhang, X.; Guo, S.; Yang, M.; Qi, G. Genome-wide analysis of the R2R3-MYB transcription factor gene family expressed in Juglans regia under abiotic and biotic stresses. Ind. Crop. Prod. 2023, 198, 116709. [Google Scholar] [CrossRef]
- Martins Cde, P.; Pedrosa, A.M.; Du, D.; Goncalves, L.P.; Yu, Q.; Gmitter, F.G., Jr.; Costa, M.G. Genome-Wide Characterization and Expression Analysis of Major Intrinsic Proteins during Abiotic and Biotic Stresses in Sweet Orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0138786. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, M.; Zhang, S.; Song, T.; Zhang, M.; Zhou, H.; Wang, Y.; Xiang, J.; Zhang, X. Transcriptomic Identification of Wheat AP2/ERF Transcription Factors and Functional Characterization of TaERF-6-3A in Response to Drought and Salinity Stresses. Int. J. Mol. Sci. 2022, 23, 3272. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Wang, K.; Guo, H.; Yin, Y. AP2/ERF transcription factors and their functions in Arabidopsis responses to abiotic stresses. Environ. Exp. Bot. 2024, 222, 105763. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef]
- Zeng, D.; Dai, L.J.; Li, X.; Li, W.; Qu, G.Z.; Li, S. Genome-Wide Identification of the ERF Transcription Factor Family for Structure Analysis, Expression Pattern, and Response to Drought Stress in Populus alba × Populus glandulosa. Int. J. Mol. Sci. 2023, 24, 3697. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Yang, X.; Wassie, M.; Shi, H. Genome-wide identification and molecular characterization of the AP2/ERF superfamily members in sand pear (Pyrus pyrifolia). BMC Genom. 2023, 24, 32. [Google Scholar] [CrossRef]
- Qiu, X.; Zhao, H.; Abubakar, A.S.; Shao, D.; Chen, J.; Chen, P.; Yu, C.; Wang, X.; Chen, K.; Zhu, A. Genome-Wide Analysis of AP2/ERF Gene Superfamily in Ramie (Boehmeria nivea L.) Revealed Their Synergistic Roles in Regulating Abiotic Stress Resistance and Ramet Development. Int. J. Mol. Sci. 2022, 23, 15117. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, L.; Nie, L.; Zheng, Y.; Zhu, S.; Hou, J.; Li, R.; Chen, G.; Tang, X.; Wang, C.; et al. Genome-wide analysis of the DREB family genes and functional identification of the involvement of BrDREB2B in abiotic stress in wucai (Brassica campestris L.). BMC Genom. 2022, 23, 598. [Google Scholar] [CrossRef]
- Wang, S.; Yao, W.; Zhou, B.; Jiang, T. Structure analysis and expression pattern of the ERF transcription factor family in poplar. Acta Physiol. Plant. 2016, 38, 239. [Google Scholar] [CrossRef]
- Zhao, M.; Haxim, Y.; Liang, Y.; Qiao, S.; Gao, B.; Zhang, D.; Li, X. Genome-wide investigation of AP2/ERF gene family in the desert legume Eremosparton songoricum: Identification, classification, evolution, and expression profiling under drought stress. Front. Plant Sci. 2022, 13, 885694. [Google Scholar] [CrossRef]
- Li, N.; Niu, Y.; Wang, X.; Wang, S.; Chen, Y.; Wang, L. A novel eggplant AP2/ERF transcription factor StPti5 confers resistance to Verticillium wilt across different species. Plant Protect. Sci. 2023, 59, 325–336. [Google Scholar] [CrossRef]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithofer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF Type Transcription Factor OsEREBP1 Confers Biotic and Abiotic Stress Tolerance in Rice. PLoS ONE 2015, 10, e0127831. [Google Scholar] [CrossRef]
- Ma, Z.; Jin, Y.M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef]
- Ma, Z.; Wu, T.; Huang, K.; Jin, Y.M.; Li, Z.; Chen, M.; Yun, S.; Zhang, H.; Yang, X.; Chen, H.; et al. A Novel AP2/ERF Transcription Factor, OsRPH1, Negatively Regulates Plant Height in Rice. Front. Plant Sci. 2020, 11, 709. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The Role of Stress-Responsive Transcription Factors in Modulating Abiotic Stress Tolerance in Plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B. Genomic architecture of promoters and transcriptional regulation of candidate genes in rice involved in tolerance to anaerobic germination. Curr. Plant Biol. 2022, 29, 100236. [Google Scholar] [CrossRef]
- Li, J.; Guo, X.; Zhang, M.; Wang, X.; Zhao, Y.; Yin, Z.; Zhang, Z.; Wang, Y.; Xiong, H.; Zhang, H.; et al. OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 2018, 270, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, Y.; Wang, Y.; Shen, Y.; Yang, J.; Jia, B.; Sun, X.; Sun, M. A comprehensive investigation of the regulatory roles of OsERF096, an AP2/ERF transcription factor, in rice cold stress response. Plant Cell Rep. 2023, 42, 2011–2022. [Google Scholar] [CrossRef]
- Zhuang, J.; Jiang, H.-H.; Wang, F.; Peng, R.-H.; Yao, Q.-H.; Xiong, A.-S. A Rice OsAP23, Functioning as an AP2/ERF Transcription Factor, Reduces Salt Tolerance in Transgenic Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 1336–1345. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Z.; Lei, Y.; Chen, W.; Zhang, Z.; Dai, H. The transcription factor MdERF023 negatively regulates salt tolerance by modulating ABA signaling and Na+/H+ transport in apple. Plant Cell Rep. 2024, 43, 187. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Fu, J.; Zhu, C.; Wang, C.; Liu, L.; Shen, Q.; Xu, D.; Wang, Q. Maize transcription factor ZmEREB20 enhanced salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 159, 257–267. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, J.; Chen, L.; Xiao, W.; Yu, J. ZmEREB46, a maize ortholog of Arabidopsis WAX INDUCER1/SHINE1, is involved in the biosynthesis of leaf epicuticular very-long-chain waxes and drought tolerance. Plant Sci. 2022, 321, 111256. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Zhou, K.; Tian, C.; Aslam, M.; Zhang, B.; Liu, W.; Zou, H. Overexpression of ZmEREBP60 enhances drought tolerance in maize. J. Plant Physiol. 2022, 275, 153763. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zheng, B.; Zhang, C.; Yu, C.; Luo, J. Wood formation in trees responding to nitrogen availability. Ind. Crop. Prod. 2024, 218, 118978. [Google Scholar] [CrossRef]
- Luo, J.; Havé, M.; Clément, G.; Tellier, F.; Balliau, T.; Launay-Avon, A.; Guérard, F.; Zivy, M.; Masclaux-Daubresse, C. Integrating multiple omics to identify common and specific molecular changes occurring in Arabidopsis under chronic nitrate and sulfate limitations. J. Exp. Bot. 2020, 71, 6471–6490. [Google Scholar] [CrossRef]
- Xia, P.; Zhang, Y.; Zhang, X. The Potential Relevance of PnDREBs to Panax notoginseng Nitrogen Sensitiveness. Biochem. Genet. 2024, 62, 2631–2651. [Google Scholar] [CrossRef]
- Joshi, V.; Joshi, M.; Penalosa, A. Comparative analysis of tissue-specific transcriptomic responses to nitrogen stress in spinach (Spinacia oleracea). PLoS ONE 2020, 15, e0232011. [Google Scholar] [CrossRef]
- Li, Z.; Deng, S.; Zhu, D.; Wu, J.; Zhou, J.; Shi, W.; Fayyaz, P.; Luo, Z.-B.; Luo, J. Proteomic reconfigurations underlying physiological alterations in poplar roots in acclimation to changing nitrogen availability. Environ. Exp. Bot. 2023, 211, 105367. [Google Scholar] [CrossRef]
- Zhang, T.T.; Lin, Y.J.; Liu, H.F.; Liu, Y.Q.; Zeng, Z.F.; Lu, X.Y.; Li, X.W.; Zhang, Z.L.; Zhang, S.; You, C.X.; et al. The AP2/ERF transcription factor MdDREB2A regulates nitrogen utilisation and sucrose transport under drought stress. Plant Cell Environ. 2024, 47, 1668–1684. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, J.; Zhao, Y.; Wang, Y.; Zhou, L.; Ning, L.; Shabala, S.; Zhao, H. Transcription factor ZmEREB97 regulates nitrate uptake in maize (Zea mays) roots. Plant Physiol. 2024, 1, 535–550. [Google Scholar] [CrossRef]
- Iniguez, L.P.; Nova-Franco, B.; Hernandez, G. Novel players in the AP2-miR172 regulatory network for common bean nodulation. Plant Signal. Behav. 2015, 10, e1062957. [Google Scholar] [CrossRef]
- Choudhury, S. Computational analysis of the AP2/ERF family in crops genome. BMC Genom. 2024, 25, 102. [Google Scholar] [CrossRef]
- Dou, H.; Wang, T.; Zhou, X.; Feng, X.; Tang, W.; Quan, J.e.; Bi, H. Genome-Wide Identification and Expression of the AP2/ERF Gene Family in Morus notabilis. Forests 2024, 15, 697. [Google Scholar] [CrossRef]
- Chen, G.; Shao, T.; Zhou, Y.; Chen, F.; Zhang, D.; Gu, H.; Yue, Y.; Wang, L.; Yang, X. Analysis of the Aging-Related AP2/ERF Transcription Factor Gene Family in Osmanthus fragrans. Int. J. Mol. Sci. 2024, 25, 8025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhu, C.; Lyu, Y.; Chen, Y.; Zhang, Z.; Lai, Z.; Lin, Y. Genome-wide identification, molecular evolution, and expression analysis provide new insights into the APETALA2/ethylene responsive factor (AP2/ERF) superfamily in Dimocarpus longan Lour. BMC Genom. 2020, 21, 62. [Google Scholar] [CrossRef]
- Zhang, K.; Jiang, L.; Wang, X.; Han, H.; Chen, D.; Qiu, D.; Yang, Y. Transcriptome-wide analysis of AP2/ERF transcription factors involved in regulating Taxol biosynthesis in Taxus × media. Ind. Crop. Prod. 2021, 171, 113972. [Google Scholar] [CrossRef]
- Li, Z.; Guan, L.; Zhang, C.; Zhang, S.; Liu, Y.; Lu, Y.; Luo, J. Nitrogen assimilation genes in poplar: Potential targets for improving tree nitrogen use efficiency. Ind. Crop. Prod. 2024, 216, 118705. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, Y.; Ding, S.; Lv, Y.; Samjhana, W.; Fang, S. Growth, Carbon Storage, and Optimal Rotation in Poplar Plantations: A Case Study on Clone and Planting Spacing Effects. Forests 2020, 11, 842. [Google Scholar] [CrossRef]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef]
- Tuskan, G.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Luo, J.; Liang, Z.; Wu, M.; Mei, L. Genome-wide identification of BOR genes in poplar and their roles in response to various environmental stimuli. Environ. Exp. Bot. 2019, 164, 101–113. [Google Scholar] [CrossRef]
- Li, Z.; Wang, H.; Li, C.; Liu, H.; Luo, J. Genome-Wide Identification of the Remorin Gene Family in Poplar and Their Responses to Abiotic Stresses. Life 2024, 14, 1239. [Google Scholar] [CrossRef]
- Cui, W.; Xu, Z.; Kong, Y.; Yang, L.; Dou, H.; Zhang, D.; Li, M.; Chen, Y.; Ding, S.; Yang, C.; et al. Genome-Wide Identification of the Heat Shock Transcription Factor Gene Family in Rosemary (Salvia rosmarinus). Horticulturae 2024, 10, 1250. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Luo, J.; Nvsvrot, T.; Wang, N. Comparative transcriptomic analysis uncovers conserved pathways involved in adventitious root formation in poplar. Physiol. Mol. Biol. Plants 2021, 27, 1903–1918. [Google Scholar] [CrossRef]
- He, W.; Luo, L.; Xie, R.; Chai, J.; Wang, H.; Wang, Y.; Chen, Q.; Wu, Z.; Yang, S.; Li, M.; et al. Genome-Wide Identification and Functional Analysis of the AP2/ERF Transcription Factor Family in Citrus Rootstock under Waterlogging Stress. Int. J. Mol. Sci. 2023, 24, 8989. [Google Scholar] [CrossRef]
- Li, D.; He, Y.; Li, S.; Shi, S.; Li, L.; Liu, Y.; Chen, H. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef]
- Ren, M.; Yang, W.; Zhang, J.; Zhao, L.; Quan, Y.; He, Z.; Xu, Y.; Zhang, F.; Yin, M.; Wang, Y.; et al. Overexpression of ClRAP2.4 in Chrysanthemum enhances tolerance to cold stress. Funct. Plant Biol. 2023, 50, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Yang, C.; Liu, S.; Li, M.; Gu, L.; Peng, X.; Zhang, Z. Identification of AP2/ERF transcription factors in Tetrastigma hemsleyanum revealed the specific roles of ERF46 under cold stress. Front. Plant Sci. 2022, 13, 936602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, N.; He, C.; Gao, Z.; Chen, G. Comparative transcriptome analysis reveals major genes, transcription factors and biosynthetic pathways associated with leaf senescence in rice under different nitrogen application. BMC Plant Biol. 2024, 24, 419. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, L.; Hu, H.; Tang, N.; Shi, L.; Xu, F.; Wang, S. Arabidopsis ERF012 Is a Versatile Regulator of Plant Growth, Development and Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 6841. [Google Scholar] [CrossRef]
- Liu, J.; Zhong, H.; Cao, C.; Wang, Y.; Zhang, Q.; Wen, Q.; Zhu, H.; Li, Z. Identification of AP2/ERF Transcription Factors and Characterization of AP2/ERF Genes Related to Low-Temperature Stress Response and Fruit Development in Luffa. Agronomy 2024, 14, 2509. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, S.; Zhao, X.; Dong, S.; Chen, J.; Sun, Y.; Sun, Q.; Liu, Q. Genome-wide identification and comprehensive analysis of the AP2/ERF gene family in Prunus sibirica under low-temperature stress. BMC Plant Biol. 2024, 24, 883. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Zhao, G.; Tang, Z.; Bu, T.; Li, C.; et al. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef]
- Luo, J.; Shi, W.; Li, H.; Janz, D.; Luo, Z.-B. The conserved salt-responsive genes in the roots of Populus × canescens and Arabidopsis thaliana. Environ. Exp. Bot. 2016, 129, 48–56. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Xing, L.; Di, Z.; Yang, W.; Liu, J.; Li, M.; Wang, X.; Cui, C.; Wang, X.; Wang, X.; Zhang, R.; et al. Overexpression of ERF1-V from Haynaldia villosa Can Enhance the Resistance of Wheat to Powdery Mildew and Increase the Tolerance to Salt and Drought Stresses. Front. Plant Sci. 2017, 8, 1948. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.; Gao, J.; Wang, B.; Han, H.; Wang, L.; Peng, R.; Yao, Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Park, S.; Kim, Y.; Hyeon, D.Y.; Park, H.; Jeong, J.; Jeong, U.; Yoon, Y.S.; You, D.; Kwak, J.; et al. Ethylene responsive factor34 mediates stress-induced leaf senescence by regulating salt stress-responsive genes. Plant Cell Environ. 2022, 45, 1719–1733. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Zhao, K.; Yao, W.; Li, R.; Zhou, B.; Jiang, T. Over-Expression of ERF38 Gene Enhances Salt and Osmotic Tolerance in Transgenic Poplar. Front. Plant Sci. 2019, 10, 1375. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zou, J.; Cheng, Q.; Fu, R.; Zhang, D.; Lai, Y.; Chen, Y.; Yang, C.; Hu, W.; Ding, S. Genome-Wide Identification and Expression of the ERF Gene Family in Populus trichocarpa and Their Responses to Nitrogen and Abiotic Stresses. Life 2025, 15, 550. https://doi.org/10.3390/life15040550
Li M, Zou J, Cheng Q, Fu R, Zhang D, Lai Y, Chen Y, Yang C, Hu W, Ding S. Genome-Wide Identification and Expression of the ERF Gene Family in Populus trichocarpa and Their Responses to Nitrogen and Abiotic Stresses. Life. 2025; 15(4):550. https://doi.org/10.3390/life15040550
Chicago/Turabian StyleLi, Mingwan, Jun Zou, Qian Cheng, Ran Fu, Dangquan Zhang, Yong Lai, Yuanyuan Chen, Chaochen Yang, Wentao Hu, and Shen Ding. 2025. "Genome-Wide Identification and Expression of the ERF Gene Family in Populus trichocarpa and Their Responses to Nitrogen and Abiotic Stresses" Life 15, no. 4: 550. https://doi.org/10.3390/life15040550
APA StyleLi, M., Zou, J., Cheng, Q., Fu, R., Zhang, D., Lai, Y., Chen, Y., Yang, C., Hu, W., & Ding, S. (2025). Genome-Wide Identification and Expression of the ERF Gene Family in Populus trichocarpa and Their Responses to Nitrogen and Abiotic Stresses. Life, 15(4), 550. https://doi.org/10.3390/life15040550




