Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution
Abstract
:1. Introduction
2. Intron-Containing tRNAs on the Domains of Life Tree

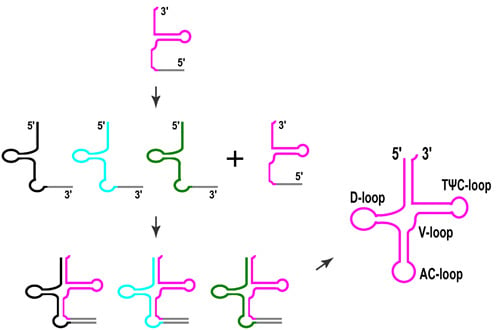
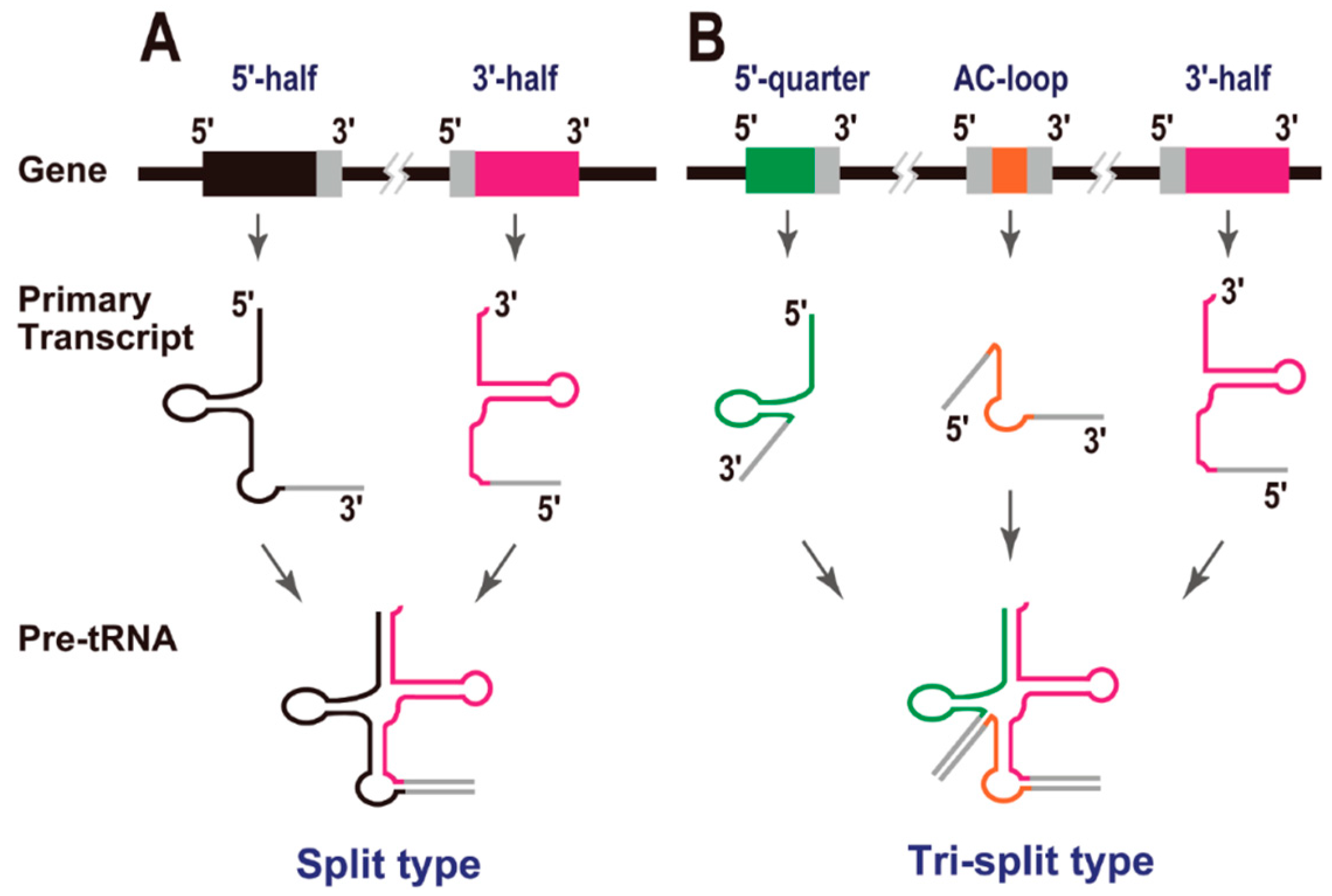
3. Split tRNAs and Tri-Split tRNAs
4. tRNA-Derived Fragments (tRFs)

5. Possible Evolutionary Scenarios for tRNA Molecule

6. Conclusions
- (a)
- Asymmetric combination of 5' and 3' tRNA halves may have generated the diversity of tRNA molecules.
- (b)
- Even recently, tRNA genes have divided into 2–3 segments, and tRNA molecules themselves can fragment post-transcriptionally.
Acknowledgments
Conflicts of Interest
References
- Sugahara, J.; Kikuta, K.; Fujishima, K.; Yachie, N.; Tomita, M.; Kanai, A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008, 25, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Munch, R.; Hohn, M.J.; Jahn, D.; Soll, D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5'- and 3'-halves. Nature 2005, 433, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, K.; Sugahara, J.; Kikuta, K.; Hirano, R.; Sato, A.; Tomita, M.; Kanai, A. Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl. Acad. Sci. USA 2009, 106, 2683–2687. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Cozen, A.E.; Lowe, T.M. Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Pearson, M.; Soll, D. The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 2005, 579, 2945–2947. [Google Scholar] [CrossRef] [PubMed]
- Soma, A.; Onodera, A.; Sugahara, J.; Kanai, A.; Yachie, N.; Tomita, M.; Kawamura, F.; Sekine, Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 2007, 318, 450–453. [Google Scholar] [CrossRef]
- Soma, A.; Sugahara, J.; Onodera, A.; Yachie, N.; Kanai, A.; Watanabe, S.; Yoshikawa, H.; Ohnuma, M.; Kuroiwa, H.; Kuroiwa, T.; et al. Identification of highly-disrupted tRNA genes in nuclear genome of the red alga, Cyanidioschyzon merolae 10D. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, S.; Sugahara, J.; Kanai, A.; Nozaki, H. Permuted tRNA genes in the nuclear and nucleomorph genomes of photosynthetic eukaryotes. Mol. Biol. Evol. 2010, 27, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Trotta, C.R.; Miao, F.; Arn, E.A.; Stevens, S.W.; Ho, C.K.; Rauhut, R.; Abelson, J.N. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 1997, 89, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, K.; Sugahara, J.; Miller, C.S.; Baker, B.J.; Di Giulio, M.; Takesue, K.; Sato, A.; Tomita, M.; Banfield, J.F.; Kanai, A. A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity. Nucleic Acids Res. 2011, 39, 9695–9704. [Google Scholar] [CrossRef] [PubMed]
- Marck, C.; Grosjean, H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA 2003, 9, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Tocchini-Valentini, G.D.; Fruscoloni, P.; Tocchini-Valentini, G.P. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. USA 2005, 102, 15418–15422. [Google Scholar] [CrossRef] [PubMed]
- Belfort, M.; Weiner, A. Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell 1997, 89, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, R.; Mohr, G.; Belfort, M.; Lambowitz, A.M. Group I and group II introns. FASEB J. 1993, 7, 15–24. [Google Scholar] [PubMed]
- Tanner, M.; Cech, T. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA 1996, 2, 74–83. [Google Scholar] [PubMed]
- Kanai, A. Molecular evolution of disrupted transfer RNA genes and their introns in archea. In Evolutionary Biology: Exobiology and Evolutionary Mechanisms; Pontarotti, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 181–193. [Google Scholar]
- Fujishima, K.; Sugahara, J.; Tomita, M.; Kanai, A. Large-scale tRNA intron transposition in the archaeal order thermoproteales represents a novel mechanism of intron gain. Mol. Biol. Evol. 2010, 27, 2233–2243. [Google Scholar] [CrossRef] [PubMed]
- Randau, L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- Waters, E.; Hohn, M.J.; Ahel, I.; Graham, D.E.; Adams, M.D.; Barnstead, M.; Beeson, K.Y.; Bibbs, L.; Bolanos, R.; Keller, M.; et al. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 2003, 100, 12984–12988. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, K.; Kanai, A. tRNA gene diversity in the three domains of life. Front. Genet. 2014, 5. [Google Scholar] [CrossRef]
- Raina, M.; Ibba, M. tRNAs as regulators of biological processes. Front. Genet. 2014, 5. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12. [Google Scholar] [CrossRef]
- Jochl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Huttenhofer, A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- Haiser, H.J.; Karginov, F.V.; Hannon, G.J.; Elliot, M.A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008, 36, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Nakamura, M.; Takahashi, Y.; Sandelin, A.; Katayama, S.; Fukuda, S.; Daub, C.O.; Kai, C.; Kawai, J.; Yasuda, J.; et al. Hidden layers of human small RNAs. BMC Genomics 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Chen, X.; Inukai, S.; Zhao, H.; Slack, F.J. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 2011, 17, 1804–1820. [Google Scholar] [CrossRef] [PubMed]
- Sobala, A.; Hutvagner, G. Transfer RNA-derived fragments: Origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2011, 2, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Fujishima, K.; Tomita, M.; Kanai, A. Metatranscriptomic analysis of microbes in an Oceanfront deep-subsurface hot spring reveals novel small RNAs and type-specific tRNA degradation. Appl. Environ. Microbiol. 2012, 78, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The origin of the tRNA molecule: Independent data favor a specific model of its evolution. Biochimie 2012, 94, 1464–1466. [Google Scholar] [CrossRef] [PubMed]
- Larkin, D.C.; Williams, A.M.; Martinis, S.A.; Fox, G.E. Identification of essential domains for Escherichia coli tRNAleu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 2002, 30, 2103–2113. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.Y.; Weiner, A.M.; Maizels, N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA 1998, 4, 276–284. [Google Scholar] [PubMed]
- Turk, R.M.; Chumachenko, N.V.; Yarus, M. Multiple translational products from a five-nucleotide ribozyme. Proc. Natl. Acad. Sci. USA 2010, 107, 4585–4589. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kikuchi, Y. Origin of the cloverleaf shape of transfer RNA—The double-hairpin model: Implication for the role of tRNA intron and long extra loop. Viva Orig. 2001, 29, 134–142. [Google Scholar]
- Fujishima, K.; Sugahara, J.; Tomita, M.; Kanai, A. Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5' and 3' tRNA halves. PLoS One 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anolles, G.; Sun, F.J. The natural history of transfer RNA and its interactions with the ribosome. Front. Genet. 2014, 5. [Google Scholar] [CrossRef]
- Randau, L.; Soll, D. Transfer RNA genes in pieces. EMBO Rep. 2008, 9, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Nunoura, T.; Takaki, Y.; Kakuta, J.; Nishi, S.; Sugahara, J.; Kazama, H.; Chee, G.J.; Hattori, M.; Kanai, A.; Atomi, H.; et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011, 39, 3204–3223. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, J.; Fujishima, K.; Nunoura, T.; Takaki, Y.; Takami, H.; Takai, K.; Tomita, M.; Kanai, A. Genomic heterogeneity in a natural archaeal population suggests a model of tRNA gene disruption. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanai, A. Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution. Life 2015, 5, 321-331. https://doi.org/10.3390/life5010321
Kanai A. Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution. Life. 2015; 5(1):321-331. https://doi.org/10.3390/life5010321
Chicago/Turabian StyleKanai, Akio. 2015. "Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution" Life 5, no. 1: 321-331. https://doi.org/10.3390/life5010321
APA StyleKanai, A. (2015). Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution. Life, 5(1), 321-331. https://doi.org/10.3390/life5010321