Freshwater Ammonia-Oxidizing Archaea Retain amoA mRNA and 16S rRNA during Ammonia Starvation
Abstract
:1. Introduction
2. Results
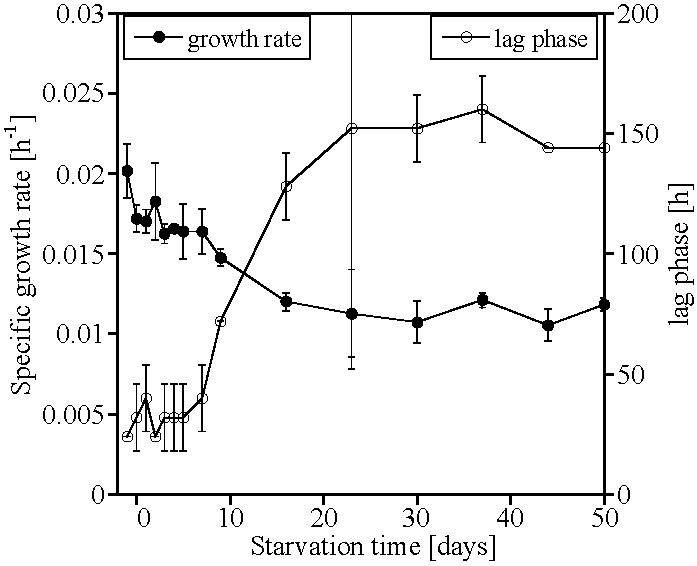



3. Discussion
4. Conclusions
5. Material and Methods
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Kolter, R.; Siegele, D.A.; Tormo, A. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 1993, 47, 855–874. [Google Scholar] [CrossRef] [PubMed]
- Arp, D.; Sayavedra-Soto, L.; Hommes, N. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 2002, 178, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.B.; de la Torre, J.R.; Klotz, M.G.; Urakawa, H.; Pinel, N.; Arp, D.J.; Brochier-Armanet, C.; Chain, P.S.G.; Chan, P.P.; Gollabgir, A.; et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8818–8823. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, A.; Schmidt, I.; Saunders, A.; Nicolaisen, M. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 2005, 71, 1276–1282. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, A.; Bar-Gilissen, M.; Laanbroek, H. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2002, 68, 4751–4757. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, B.; Jones, R. Effects of light and CO on the survival of a marine ammonium-oxidizing bacterium during energy-source deprivation. Appl. Environ. Microbiol. 1988, 54, 2890–2893. [Google Scholar] [PubMed]
- Johnstone, B.; Jones, R. Recovery of a marine chemolithotrophic ammonium-oxidizing bacterium from long-term energy-source deprivation. Can. J. Microbiol. 1988, 34, 1347–1350. [Google Scholar] [CrossRef]
- Jones, R.; Morita, R. Survival of a marine ammonium oxidizer under energy-source deprivation. Mar. Ecol. Prog. Ser. 1985, 26, 175–179. [Google Scholar] [CrossRef]
- Tappe, W.; Laverman, A.; Bohland, M.; Braster, M.; Rittershaus, S.; Groeneweg, J.; van Verseveld, H. Maintenance energy demand and starvation recovery dynamics of Nitrosomonas europaea and Nitrobacter winogradskyi cultivated in a retentostat with complete biomass retention. Appl. Environ. Microbiol. 1999, 65, 2471–2477. [Google Scholar] [PubMed]
- Bollmann, A.; Laanbroek, H. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentrations. FEMS Microbiol. Ecol. 2001, 37, 211–221. [Google Scholar] [CrossRef]
- Koops, H.; Pommerening-Roser, A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 2001, 37, 1–9. [Google Scholar] [CrossRef]
- Pinck, C.; Coeur, C.; Potier, P.; Bock, E. Polyclonal antibodies recognizing the AmoB protein of ammonia oxidizers of the beta-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 2001, 67, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Könneke, M.; Bernhard, A.E.; de la Torré, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Martens-Habbena, W.; Berube, P.M.; Urakawa, H.; de la Torré, J.R.; Stahl, D.A. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 2009, 461, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Stahl, D.A. Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl. Environ. Microbiol. 2013, 79, 6911–6916. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P.; Thrash, J.C.; Nicora, C.D.; Lipton, M.S.; Burnum-Johnson, K.E.; Carini, P.; Smith, R.D.; Giovannoni, S.J. Proteomic and transcriptomic analyses of “Candidatus Pelagibacter ubique” describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium. MBio 2013, 4. [Google Scholar] [CrossRef]
- Bollmann, A.; Sedlacek, C.J.; Norton, J.; Laanbroek, H.J.; Suwa, Y.; Stein, L.Y.; Klotz, M.G.; Arp, D.; Sayavedra-Soto, L.; Lu, M.; et al. Complete genome sequence of Nitrosomonas sp. Is79, an ammonia oxidizing bacterium adapted to low ammonium concentrations. Stand. Genomic Sci. 2013, 7, 469–482. [Google Scholar] [PubMed]
- French, E.; Kozlowski, J.A.; Mukherjee, M.; Bullerjahn, G.; Bollmann, A. Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl. Environ. Microbiol. 2012, 78, 5773–5780. [Google Scholar] [CrossRef] [PubMed]
- French, E.; Kozlowski, J.A.; Bollmann, A. Competition of Ammonia-oxidizing Archaea and Bacteria from freshwater environments. Manuscript under preparation.
- Bollmann, A.; French, E.; Laanbroek, H.J. Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations. Meth. Enzymol. 2011, 486, 55–88. [Google Scholar] [PubMed]
- Belser, L.; Schmidt, E. Growth and oxidation-kinetics of three genera of ammonia-oxidizing nitrifiers. FEMS Microbiol. Lett. 1980, 7, 213–216. [Google Scholar] [CrossRef]
- Keeney, D.R.; Nelson, D.W. Nitrogen-Inorganic forms. In Methods in Soil Analysis—Part 2; Page, A.L., Ed.; American Society of Agronomy: Madison, WI, USA, 1982; pp. 643–698. [Google Scholar]
- Kandeler, E.; Gerber, H. Short-Term Assay of Soil Urease Activity Using Colorimetric Determination of Ammonium. Biol. Fert. Soils 1988, 6, 68–72. [Google Scholar] [CrossRef]
- Shand, C.A.; Williams, B.L.; Coutts, G. Determination of N-species in soil extracts using microplate techniques. Talanta 2008, 74, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Kowalchuk, G.; Stephen, J.; De Boer, W.; Prosser, J.; Embley, T.; Woldendorp, J. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 1997, 63, 1489–1497. [Google Scholar] [PubMed]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
French, E.; Bollmann, A. Freshwater Ammonia-Oxidizing Archaea Retain amoA mRNA and 16S rRNA during Ammonia Starvation. Life 2015, 5, 1396-1404. https://doi.org/10.3390/life5021396
French E, Bollmann A. Freshwater Ammonia-Oxidizing Archaea Retain amoA mRNA and 16S rRNA during Ammonia Starvation. Life. 2015; 5(2):1396-1404. https://doi.org/10.3390/life5021396
Chicago/Turabian StyleFrench, Elizabeth, and Annette Bollmann. 2015. "Freshwater Ammonia-Oxidizing Archaea Retain amoA mRNA and 16S rRNA during Ammonia Starvation" Life 5, no. 2: 1396-1404. https://doi.org/10.3390/life5021396
APA StyleFrench, E., & Bollmann, A. (2015). Freshwater Ammonia-Oxidizing Archaea Retain amoA mRNA and 16S rRNA during Ammonia Starvation. Life, 5(2), 1396-1404. https://doi.org/10.3390/life5021396




