Future of the Genetic Code
Abstract
:1. Introduction
2. Synthetic Lifeform Production
3. Anticodons as Identity Elements
4. Misreading by the UNN Anticodon
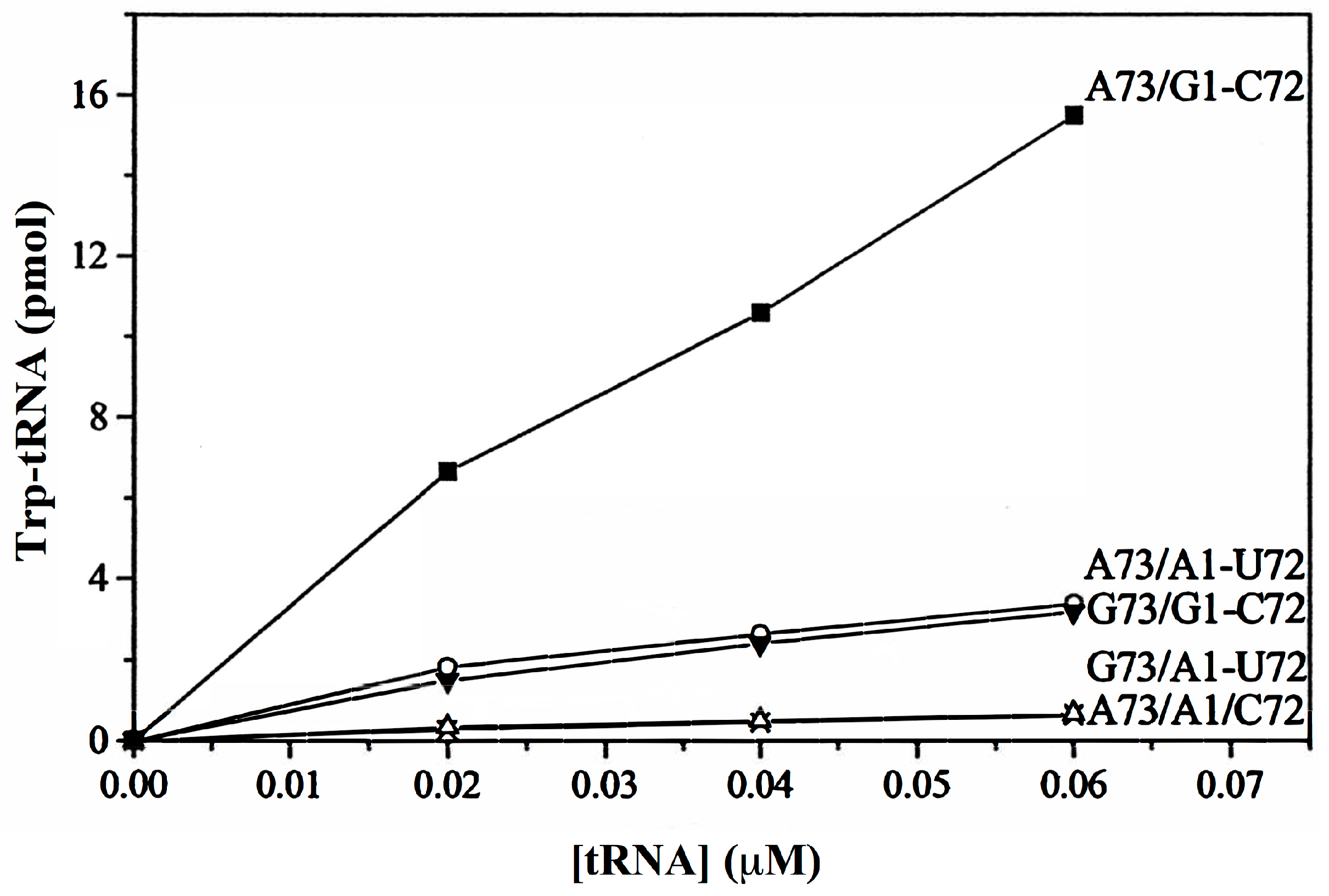
- (a)
- All the cloned tRNA(Trp)s displayed s4U in resemblance to native E. coli tRNA(Trp) even though native B. subtilis and native bovine tRNA(Trp)s were devoid of this modification. Both cloned B. subtilis and native E. coli tRNA(Trp)s contained Cm, although native B. subtilis tRNA(Trp) lacked this modification. Native bovine tRNA(Trp) contained m1A and m2G, whereas cloned bovine and native E. coli tRNA(Trp)s were devoid of these modifications. These findings showed the decisive influence of host enzymes regarding some modifications on exogenous tRNAs.
- (b)
- On the other hand, native and cloned B. subtilus tRNA(Trp)s both contained i6A, but native E. coli tRNA(Trp) did not. Also, native and cloned bovine tRNA(Trp)s both contained Gm, but native E. coli tRNA(Trp) did not. These findings showed that the sequence of an exogenous tRNA could be a more important determinant than host enzymes for other modifications.
5. Discussion
5.1. Protein Structure-Activity Relationships
5.2. Peptide and Protein Drugs
5.3. Enhancement of Biological Fitness
5.4. Metabolic and Biomimetic Engineering
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wong, J.T. Membership mutation of the genetic code: Loss of fitness by tryptophan. Proc. Natl. Acad. Sci. USA 1983, 80, 6303–6306. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. Evolution and mutation of the amino acid code. In Dynamics of Biochemical Systems; Ricard, J., Cornish-Bowden, A., Eds.; Plenum Press: New York, NY, USA, 1984; pp. 247–258. [Google Scholar]
- Bronskill, P.M.; Wong, J.T. Suppression of fluorescence of tryptophan residues in proteins by replacement with 4-fluorotryptophan. Biochem. J. 1988, 249, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Mat, W.K.; Xue, H.; Wong, J.T. Genetic code mutations: The breaking of a three billion year invariance. PLoS ONE 2010, 5, e12206. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.C.; Yim, A.K.; Mat, W.K.; Tong, A.H.; Lok, S.; Xue, H.; Tsui, S.K.; Wong, J.T.; Chan, T.F. Mutations enabling displacement of tryptophan by 4-fluorotryptophan as a canonical amino acid of the genetic code. Genome Biol. Evol. 2014, 6, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Hesman, T. Code breakers: Scientists are altering bacteria in a most fundamental way. Sci. News 2000, 157, 360–362. [Google Scholar] [CrossRef]
- Wong, J.T.; Xue, H. Synthetic genetic codes as the basis of synthetic life. In Chemical Synthetic Biology; Luisi, P.L., Chiarabelli, C., Eds.; Wiley: New York, NY, USA, 2010; pp. 178–199. [Google Scholar]
- Marliere, P.; Patrouix, J.; Doring, V.; Herdewijn, P.; Tricot, S.; Cruveiller, S.; Bouzon, M.; Mutzel, R. Chemical evolution of a bacterium’s genome. Angew Chem. Int. Ed. Engl. 2011, 50, 7109–7114. [Google Scholar] [CrossRef] [PubMed]
- Marliere, P. Charting the xenobiotic continent. In Proceedings of the First Conference on Xenobiology, Genoa, Italy, 6–8 May 2014.
- Acevedo-Rocha, C.G.; Budisa, N. On the road towards chemically modified organisms endowed with a genetic firewall. Angew Chem. Int. Ed. Engl. 2011, 50, 6960–6962. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Chen, F.; Yang, Z. Synthetic biology, tinkering biology and artificial biology: A perspective from chemistry. In Chemical Synthetic Biology; Luisi, P.L., Chiarabelli, C., Eds.; Wiley: New York, NY, USA, 2010; pp. 69–106. [Google Scholar]
- Li, L.; Degardin, M.; Lavergne, T.; Malyshev, D.A.; Kirandeep, D.; Ordoukhanian, P.; Romesberg, F.E. Natural-like replication of an unnatural base pair for the expansion of the genetic alphabet and biotechnology applications. J. Am. Chem. Soc. 2014, 136, 826–829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lamb, B.M.; Feldman, A.W.; Zhou, A.X.; Lavergne, T.; Li, L.; Romesberg, F.E. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc. Nat. Acad. Sci. USA 2017, 114, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Bacher, J.M.; Ellington, A.D. Selection and characterization of Escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 2001, 183, 5414–5425. [Google Scholar] [CrossRef] [PubMed]
- Bacher, J.M.; Bull, J.J.; Ellington, A.D. Evolution of phage with chemically ambiguous proteomes. BMC Evol. Biol. 2003, 3. [Google Scholar] [CrossRef] [PubMed]
- Bacher, J.M.; Hughes, R.A.; Wong, J.T.; Ellington, A.D. Evolving new genetic codes. Trends Ecol. Evol. 2004, 19, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hoesl, M.G.; Oehm, S.; Durkin, P.; Darmon, E.; Peil, L.; Aerni, H.R.; Rappsilber, J.; Rinehart, J.; Leach, D.; Soll, D.; et al. Chemical evolution of a bacterial proteome. Angew Chem. Int. Ed. Engl. 2015, 54, 10030–10034. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Y.; Wong, J.T. Evolutionary relationship between Halobacterium cutirubrum and eukaryotes determined by use of aminoacyl-tRNA synthetases as phylogenetic probes. Can. J. Biochem. 1980, 58, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. Emergence of life: From functional RNA selection to natural selection and beyond. Front Biosci. 2014, 19, 1117–1150. [Google Scholar] [CrossRef]
- Santoro, S.W.; Anderson, J.C.; Lakshman, V.; Schultz, P.G. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 2003, 31, 6700–6709. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, P.; Ling, J.; Wang, Y.S.; Soll, D. Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 2013, 9, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed]
- Hoesl, M.G.; Budisa, N. Recent advances in genetic code engineering in Escherichia coli. Curr. Opin. Biotechnol. 2012, 23, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Greiss, S.; Chin, J.W. Expanding the genetic code of an animal. J. Am. Chem. Soc. 2011, 133, 14196–14199. [Google Scholar] [CrossRef] [PubMed]
- Parrish, A.R.; She, X.; Xiang, Z.; Coin, I.; Shen, Z.; Briggs, S.P.; Dillin, A.; Wang, L. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem. Biol. 2012, 7, 1292–1302. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Riou, M.; Carvalho, S.; Paoletti, P. Expanding the genetic code in Xenopus laevis oocytes. ChemBioChem 2013, 14, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Ellefson, J.W.; Meyer, A.J.; Hughes, R.A.; Cannon, J.R.; Brodbelt, J.S.; Ellington, A.D. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat. Biotechnol. 2014, 32, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Maranhao, A.C.; Ellington, A.D. Evolving orthogonal suppressor tRNAs to incorporate modified amino acids. ACS Synth. Biol. 2016, 27600875. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Wang, K.; Davis, L.; Garcia-Alai, M.; Chin, J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosomes. Nature 2010, 464, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, K.; O’Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Soll, D.; Nureki, O. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature 2009, 457, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Kobayashi, T.; Hino, N.; Yanagishawa, T.; Sakamoto, K.; Yokoyama, S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 2008, 371, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Hancock, S.M.; Uprety, R.; Deiters, A.; Chin, J.W. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 2010, 132, 14819–14824. [Google Scholar] [CrossRef] [PubMed]
- Fekner, T.; Chan, M.K. The pyrrolysine translational machinery as a genetic code-expansion tool. Curr. Opin. Chem. Biol. 2011, 15, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Brocker, M.J.; Prat, L.; Ip, K.; Chirathivat, N.; Felock, A.; Veszpremi, M.; Soll, D. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett. 2015, 589, 2194–2199. [Google Scholar] [CrossRef] [PubMed]
- Thyer, R.; Robotham, S.A.; Brodbelt, J.S.; Ellington, A.D. Evolving tRNASec for efficient incorporation of selenocysteine. J. Am. Chem. Soc. 2015, 137, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, F.J.; Carr, P.A.; Wang, H.H.; Lajoie, M.J.; Sterling, B.; Kraal, L.; Tolonen, A.C.; Gianoulis, T.A.; Goodman, D.B.; Reppas, N.B.; et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 2011, 333, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Yamaguchi, A.; Ohtake, K.; Takahashi, M.; Hayashi, A.; Iraha, F.; Kira, S.; Yanagisawa, T.; Yokoyama, S.; Hoshi, H.; et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acid Res. 2015, 43, 8111–8122. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Prat, L.; Aerni, H.; Ling, J.; Merryman, C.; Glass, J.I.; Rinehart, J.; Söll, D. Transfer RNA misidentification scrambles sense codon recoding. ChemBioChem 2013, 14, 1967–1972. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, W.; Liu, W.R. Toward reassigning the rare AGG codon in Escherichia coli. ChemBioChem 2014, 15, 1750–1754. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, M.J.; Kosuri, S.; Mosberg, J.A.; Gregg, C.J.; Zhang, D.; Church, G.M. Probing the limits of genetic recoding in essential genes. Science 2013, 342, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Bohlke, N.; Budisa, N. Sense codon emancipation for proteome-wide incorporation of noncanonical amino acids; rare isoleucine codon AUA as a target for genetic code expansion. FEMS Mcrobiol. Lett. 2014, 351, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Shen, W.; Giege, R.; Wong, J.T. Identity elements of tRNA(Trp). Identification and evolutionary conservation. J. Biol. Chem. 1993, 268, 9316–9322. [Google Scholar] [PubMed]
- Giege, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acid Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Ling, J. Experimental challenges of sense codon reassignment: An innovative approach to genetic code expansion. FEBS Lett. 2014, 588, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Gong, Q.; Grosjean, H.; Zhu, G.; Wong, J.T.; Xue, H. Recognition by tryptophanyl-tRNA synthetases of discriminator base on the tRNATrp from three biological domains. J. Biol. Chem. 2002, 277, 14343–14349. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. Codon—Anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Wong, J.T.; Ng, S.K.; Mat, W.K.; Hu, T.; Xue, H. Coevolution theory of the genetic code at age forty: Pathway to translation and synthetic life. Life 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Lagerkvist, U. Two-out of three: An alternative method for codon reading. Proc. Natl. Acad. Sci. USA 1978, 75, 1759–1762. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, M.; Karcher, D.; Bock, R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008, 15, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.J.; de Henau, S.; Crothers, D.M. On the physical basis for ambiguity in genetic coding interaction. Proc. Natl. Acad. Sci. USA 1978, 75, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Nishimura, S. Modified nucleosides and codon recognition. In tRNA Structure, Biosynthesis and Function; Soll, D., RajBhandary, U.L., Eds.; American Society for Microbiology Press: Washington, DC, USA, 1995; pp. 207–223. [Google Scholar]
- Van der Gulik, P.T.S.; Hoff, W.D. Unassigned codons, nonsense suppression, and anticodon modifications in the evolution of the genetic code. J. Mol. Evol. 2011, 73, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Van der Gulik, P.T.S.; Hoff, W.D. Anticodon modifications in the tRNA set of LUCA and the fundamental regularity in the standard genetic code. PLoS ONE 2016, 11, e0158342. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Rumer, I.B. On codon systemization in the genetic code. Dokl Akad Nauk SSSR 1966, 167, 1393–1394. [Google Scholar] [PubMed]
- Lehmann, J.; Libchaber, A. Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA 2008, 14, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Curran, J.F. Modified nucleosides in translation. In Modification and Editing of RNA; Grosjean, H., Benne, R., Eds.; American Society for Microbiology Press: Washington, DC, USA, 1998; pp. 493–516. [Google Scholar]
- Bjork, G.R. Biosynthesis and function of modified nucleosides. In tRNA Structure, Biosynthesis and Function; Soll, D., RajBhandary, U.L., Eds.; American Society for Microbiology Press: Washington, DC, USA, 1995; pp. 165–205. [Google Scholar]
- Oba, T.; Andachi, Y.; Muto, A.; Osawa, S. CGG: An unassigned or nonsense codon in Mycoplasma capricolum. Proc. Natl. Acad. Sci. USA 1991, 88, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Kano, A.; Ohama, T.; Abe, R.; Osawa, S. Unassigned or nonsense codons in Micrococcus luteus. J. Mol. Biol. 1993, 230, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L. The formation of organic compounds on the primitive Earth. Ann. N. Y. Acad. Sci. 1957, 69, 260–275. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. Origin of genetically encoded protein synthesis: A model based on selection for RNA peptidation. Ori. Life Evol. Biosph. 1991, 21, 165–176. [Google Scholar] [CrossRef]
- Wong, J.T. A co-evolution theory of the genetic code. Proc. Natl. Acad. Sci. USA 1975, 72, 1909–1912. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. Coevolution theory of the genetic code: A proven theory. Ori. Life Evol. Biosph. 2007, 37, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. Coevolution of the genetic code and amino acid biosynthesis. Trends Biochem. Sci. 1981, 16, 33–35. [Google Scholar] [CrossRef]
- Khoury, G.A.; Baliban, R.C.; Floudas, C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011, 1, 90. [Google Scholar] [CrossRef] [PubMed]
- Naegle, K.M.; Gymrek, M.; Joughin, B.A.; Wagner, J.P.; Welsch, R.E.; Yaffe, M.B.; Lauffenburger, D.A.; White, F.M. PTMScout, a Web resource for analysis of high throughput post-translational proteomics studies. Mol. Cell. Proteom. 2010, 9, 2558–2570. [Google Scholar] [CrossRef] [PubMed]
- Cicardi, M.; Banerji, A.; Bracho, F.; Malbran, A.; Rosenkranz, B.; Riedl, M.; Bork, K.; Lumry, W.; Aberer, W.; Bier, H.; et al. Icatibant, a new bradykinin-receptor antagonist, in hereditary angioedema. N. Engl. J. Med. 2010, 363, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Sim, V. d-amino acid-based peptide inhibitors as early or preventive therapy in Alzheimer disease. Prion 2014, 8. [Google Scholar] [CrossRef]
- Armishaw, C.J.; Daly, N.L.; Nevis, S.T.; Adams, D.J.; Craik, D.J.; Alewood, P.F. Alpha-selenoconotoxins, a new class of potent alpha7 neuronal nicotinic receptor antagonists. J. Biol. Chem. 2006, 281, 14136–14143. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.J.; Love, M.L.; Ma, L.Y.Y.; Blum, M.; Bronskill, P.M.; Bernstein, J.; Grey, A.A.; Hofmann, T.; Camerman, N.; Wong, J.T. Tryptophanyl-tRNA synthetase from Bacillus subtilis: Characterization and role of hydrophobicity in substrate recognition. J. Biol. Chem. 1989, 264, 4304–4311. [Google Scholar] [PubMed]
- Stenflo, J. Contributions of Gla and EGF-like domains to the function of vitamin K-dependent coagulation factors. Crit. Rev. Eukaryot. Gene Expr. 1999, 9, 59–88. [Google Scholar] [PubMed]
- Wang, J.; Zhang, W.; Song, W.; Wang, Y.; Yu, Z.; Li, J.; Wu, M.; Wang, L.; Zang, J.; Lin, Q. A biosynthetic route to photoclick chemistry on proteins. J. Am. Chem. Soc. 2010, 132, 14812–14818. [Google Scholar] [CrossRef] [PubMed]
- Wang, V.; Wu, W. MicroRNA-based therapeutics for cancer. BioDrugs 2009, 23, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.A.; Zamore, P.D. MicroRNA therapeutics. Gene Ther. 2011, 18, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.L.; Bestor, T.H. Cytosine methylation targetted to pre-determined sequences. Nat. Genet. 1997, 17, 376–378. [Google Scholar] [CrossRef] [PubMed]
- Gommans, W.M.; McLaughlin, P.M.; Lindhout, B.I.; Segal, D.J.; Wiegman, D.J.; Haisma, H.J.; van der Zaal, B.J.; Rots, M.G. Engineering zinc finger protein transcription factors to downregulate the epithelial glycoprotein-2 promotor as a novel anti-cancer treatment. Mol. Carcinog. 2007, 46, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Hammerling, M.J.; Ellefson, J.W.; Boutz, D.R.; Marcotte, E.M.; Ellington, A.D.; Barrick, J.E. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 2014, 10, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T.; Bronskill, P.M. Inadequacy of prebiotic synthesis as origin of proteinous amino acids. J. Mol. Evol. 1979, 13, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.L.; Miller, S.L. Reasons for the occurrence of the twenty coded protein amino acids. J. Mol. Evol. 1981, 17, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Chavous, D.A.; Jackson, F.R.; O’Connor, C.M. Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc. Natl. Acad. Sci. USA 2001, 98, 14814–14818. [Google Scholar] [CrossRef] [PubMed]
- Adav, S.S.; Gallart-Palau, X.; Tan, K.H.; Lim, S.K.; Tam, J.P.; Sze, S.K. Dementia-linked amyloidosis is associated with brain protein deamidation as revealed by proteomic profiling of human brain tissues. Mol. Brain 2016, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, E.B.; Buchanan, L.E.; Marek, P.; Cao, P.; Raleigh, D.P.; Zanni, M.T. Deamidation accelerates amyloid formation and alters amylain fiber structure. J. Am. Chem. Soc. 2012, 134, 12658–12667. [Google Scholar] [CrossRef] [PubMed]
- Beld, J.; Woycechowsky, J.; Hilvert, D. Selenoglutathione: Efficient oxidative protein folding by a diselenide. Biochemistry 2007, 46, 5382–5390. [Google Scholar] [CrossRef] [PubMed]
- Metanis, N.; Keinan, E.; Davison, P.E. Synthetic seleno-glutaredoxin 3 analogs are highly reducing oxidoreductases with enhanced catalytic efficiency. J. Am. Chem. Soc. 2006, 128, 16684–16691. [Google Scholar] [CrossRef] [PubMed]
- Commans, S.; Bock, A. Selenocysteine inserting tRNAs: An overview. FEMS Microbiol. Rev. 1999, 23, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Fay, P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 1992, 56, 340–373. [Google Scholar] [PubMed]
- Rowinsky, E.K.; Windle, J.J.; Von Hoff, D.D. Ras protein farnesyltransferase: A strategic target for anticancer therapeutic development. J. Clin. Oncol. 1999, 17, 3631–3652. [Google Scholar] [CrossRef] [PubMed]
- Purchase, R.L.; de Groot, H.J.M. Biosolar cells: Global artificial photosynthesis needs responsive matrices with quantum coherent kinetic control for high yield. Interface Focus 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, A.; Anderlund, M.; Johansson, O.; Lindblad, P.; Lomoth, R.; Polivka, T.; Ott, S.; Stensjö, K.; Styring, S.; Sundström, V.; et al. Biomimetic and microbial approaches to solar fuel generation. Acc. Chem. Res. 2009, 42, 1899–1909. [Google Scholar] [CrossRef] [PubMed]
- Stripp, S.T.; Happe, T. How algae produce hydrogen-news from the photosynthetic hydrogenase. Dalton Trans. 2009. [Google Scholar] [CrossRef] [PubMed]
- English, C.M.; Eckert, C.; Brown, K.; Seibert, M.; King, P.W. Recombinant and in vitro expression systems for hydrogenases: New frontiers in basic and applied studies for biological and synthetic H2 production. Dalton Trans. 2009. [Google Scholar] [CrossRef] [PubMed]
- Tard, C.; Pickett, C.J. Structural and functional analogues of the active sites of the [Fe]-, [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 2009, 109, 2245–2274. [Google Scholar] [CrossRef] [PubMed]
| tRNA(Trp) | s4U | Gm | Cm | m1A | i6A | m2G |
|---|---|---|---|---|---|---|
| Cloned B. subtilis | 0.2 | 0.2 | 1.0 | − | 0.9 | − |
| Cloned bovine | 0.3 | 0.1 | 0.9 | − | − | − |
| Cloned A. fulgidus | 0.2 | 0.1 | − | − | − | − |
| Native E. coli | 0.5 | − | 1.1 | − | − | − |
| Native B. subtilis | − | − | − | − | + | − |
| Native bovine | − | + | + | + | − | + |
| Native H. volcanii | − | − | + | − | − | − |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, H.; Wong, J.T.-F. Future of the Genetic Code. Life 2017, 7, 10. https://doi.org/10.3390/life7010010
Xue H, Wong JT-F. Future of the Genetic Code. Life. 2017; 7(1):10. https://doi.org/10.3390/life7010010
Chicago/Turabian StyleXue, Hong, and J. Tze-Fei Wong. 2017. "Future of the Genetic Code" Life 7, no. 1: 10. https://doi.org/10.3390/life7010010
APA StyleXue, H., & Wong, J. T.-F. (2017). Future of the Genetic Code. Life, 7(1), 10. https://doi.org/10.3390/life7010010





