Biodiversity and Abundance of Cultured Microfungi from the Permanently Ice-Covered Lake Fryxell, Antarctica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Location
2.2. Sample Collection and Processing
3. Results
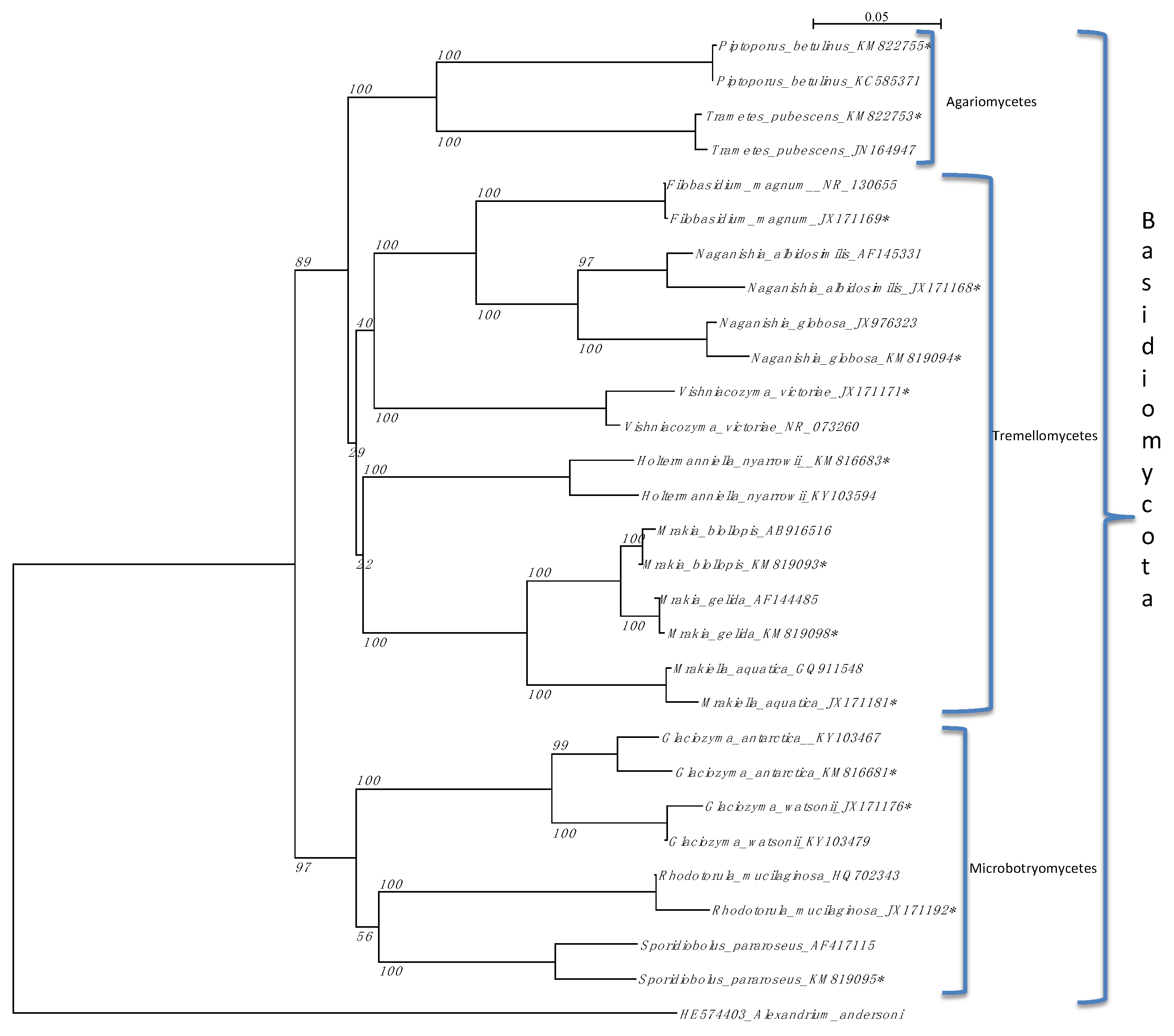
3.1. Fungal Diversity
3.2. Fungal Distribution
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Angino, E.E.; Armitage, K.B.; Tash, J.C. Chemical Stratification in Lake Fryxell, Victoria Land, Antarctica. Science 1962, 138, 34–36. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.S.; Vick-Majors, T.J.; Morgan-Kiss, R.; Takacs-Vesbach, C.; Ducklow, H.W.; Priscu, J.C. Microbial community dynamics in two polar extremes: The lakes of the McMurdo Dry Valleys and the West Antarctic Peninsula marine ecosystem. BioScience 2016, 66, 829–847. [Google Scholar] [CrossRef]
- Brambilla, E.; Hippe, H.; Hagelstein, A.; Tindall, B.J.; Stackebrandt, E. 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 2001, 5, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Karr, E.A.; Ng, J.M.; Belchik, S.M.; Sattley, W.M.; Madigan, M.T.; Achenbach, L.A. Biodiversity of methanogenic and other archaea in the permanently frozen Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 2006, 72, 1663–1666. [Google Scholar] [CrossRef] [PubMed]
- Karr, E.A.; Sattley, W.M.; Jung, D.O.; Madigan, M.T.; Achenbach, L.A. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 2003, 69, 4910–4914. [Google Scholar] [CrossRef] [PubMed]
- Karr, E.A.; Sattley, W.M.; Rice, M.R.; Jung, D.O.; Madigan, M.T.; Achenbach, L.A. Diversity and Distribution of Sulfate-Reducing Bacteria in Permanently Frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2005, 71, 6353–6359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laybourn-Parry, J.; James, M.R.; McKnight, D.M.; Priscu, J.; Spaulding, S.A.; Shiel, R. The microbial plankton of Lake Fryxell, southern Victoria Land, Antarctica during the summers of 1992 and 1994. Polar Biol. 1996, 17, 54–61. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Brambilla, E.; Cousin, S.; Dirks, W.; Pukall, R. Culture-independent analysis of bacterial species from an anaerobic mat from Lake Fryxell, Antarctica: Prokaryotic diversity revisited. Cell Mol. Biol. 2004, 50, 517–524. [Google Scholar] [PubMed]
- Taton, A.; Grubisic, S.; Brambilla, E.; De Wit, R.; Wilmotte, A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): A morphological and molecular approach. Appl. Environ. Microbiol. 2003, 69, 5157–5169. [Google Scholar] [CrossRef] [PubMed]
- Wharton, R.A.; Parker, B.C.; Simmons, G.M.; Seaburg, K.G.; Love, F.G. Biogenic calcite structures forming in Lake Fryxell, Antarctica. Nature 1982, 295, 403–405. [Google Scholar] [CrossRef]
- Ayres, E.; Wall, D.H.; Adams, B.J.; Barrett, J.E.; Virginia, R.A. Unique similarity of faunal communities across aquatic–terrestrial interfaces in a polar desert ecosystem. Ecosystems 2007, 10, 523–535. [Google Scholar] [CrossRef]
- McKnight, D.M.; Howes, B.L.; Taylor, C.D.; Goehringer, D.D. Phytoplankton Dynamics in a Stably Stratified Antarctic Lake during Winter Darkness. J. Phycol. 2000, 36, 852–861. [Google Scholar] [CrossRef]
- Priscu, J.C.; Priscu, L.R.; Howard-Williams, C.H.; Vincent, W.F. Diel patterns of photosynthate biosynthesis by phytoplankton in permanently ice-covered Antarctic lakes under continuous sunlight. J. Plankton Res. 1988, 10, 333–340. [Google Scholar] [CrossRef]
- Priscu, J.C.; Priscu, L.R.; Vincent, W.F.; Howard-Williams, C.H. Photosynthate distribution by microplankton in permanently ice-covered Antarctic desert lakes. Limnol. Oceanogr. 1987, 32, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Roberts, E.C.; Laybourn-Parry, J. Mixotrophic cryptophytes and their predators in the Dry Valley lakes of Antarctica. Freshw. Biol. 1999, 41, 737–746. [Google Scholar] [CrossRef]
- Roberts, E.C.; Laybourn-Parry, J.; McKnight, D.M.; Novarino, G. Stratification and dynamics of microbial loop communities in Lake Fryxell, Antarctica. Freshw. Biol. 2000, 44, 649–661. [Google Scholar] [CrossRef]
- Spaulding, S.A.; McKnight, D.M.; Smith, R.L.; Dufford, R. Phytoplankton population dynamics in perennially ice-covered Lake Fryxell, Antarctica. J. Plankton Res. 1994, 16, 527–541. [Google Scholar] [CrossRef]
- Wharton, R.A., Jr.; Parker, B.C.; Simmons, G.M., Jr. Distribution, species composition and morphology of algal mats in Antarctic dry valley lakes. Phycologia 1983, 22, 355–365. [Google Scholar] [CrossRef]
- Wurzbacher, C.M.; Barlocher, F.; Grossart, H.-P. Fungi in lake ecosystems. Aquat. Microb. Ecol. 2010, 59, 125–149. [Google Scholar] [CrossRef] [Green Version]
- Goto, S.; Sugiyama, J.; Iizuka, H. A Taxonomic Study of Antarctic Yeasts. Mycologia 1969, 61, 748–774. [Google Scholar] [CrossRef] [PubMed]
- Slemmons, C.; Johnson, G.; Connell, L.B. Application of an automated ribosomal intergenic spacer analysis database for identification of cultured Antarctic fungi. Antarct. Sci. 2013, 25, 44–50. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, J., Sninsky, J., White, T.J., Eds.; Academic Press: Orlando, FL, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smit, E.; Leeflang, P.; Glandorf, B.; van Elsas, J.D.; Wernars, K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18s rRNA and temperature gradient electrophoresis. Appl. Environ. Microbiol. 1999, 65, 2614–2621. [Google Scholar] [PubMed]
- Connell, L.B.; Redman, R.S.; Craig, S.D.; Scorzetti, G.; Iszard, M.; Rodriguez, R.J. Diversity of soil yeasts isolated from South Victoria Land, Antarctica. Microb. Ecol. 2008, 56, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 4 June 2018).
- Tsuji, M.; Fujiu, S.; Xiao, N.; Hanada, Y.; Kudoh, S.; Kondo, H.; Tsuda, S.; Hoshino, T. Cold adaptation of fungi obtained from soil and lake sediment in the Skarvsnes ice-free area, Antarctica. FEMS Microbiol. Lett. 2013, 346, 121–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.J.F.; Hendy, C.H. Water column and sediment characteristics of Lake Fryxell, Taylor Valley, Antarctica. N. Z. J. Geol. Geophys. 1985, 28, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Phae, C.-G.; Shoda, M. A new fungus which degrades hydrogen sulfide, methanethiol, dimethyl sulfide and dimethyl disulfide. Biotechnol. Lett. 1991, 13, 375–380. [Google Scholar] [CrossRef]
- Tang, J.; Hu, K.-D.; Hu, L.-Y.; Li, Y.-H.; Liu, Y.-S.; Zhang, H. Hydrogen Sulfide Acts as a Fungicide to Alleviate Senescence and Decay in Fresh-cut Sweetpotato. HortScience 2014, 49, 938–943. [Google Scholar]
- Mosier, A.C.; Murray, A.E.; Fritsen, C.H. Microbiota within the perennial ice cover of Lake Vida, Antarctica. FEMS Microbiol. Ecol. 2007, 59, 274–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priscu, J.C.; Fritsen, C.H.; Adams, E.E.; Giovannoni, S.J.; Paerl, H.W.; McKay, C.P.; Doran, P.T.; Gordon, D.A.; Lanoil, B.D.; Pinckney, J.L. Perennial Antarctic lake ice: An oasis for life in a polar desert. Science 1998, 280, 2095–2098. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.O.; Achenbach, L.A.; Karr, E.A.; Takaichi, S.; Madigan, M. A gas vesiculate planktonic strain of the purple nonsulfur bacterium Rhodoferax antarcticus isolated from Lake Fryxell, Dry Valleys, Antarctica. Arch. Microbiol. 2004, 182, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Bridge, P.D.; Spooner, B.M. Non-lichenized Antarctic fungi: Transient visitors or members of a cryptic ecosystem? Fungal Ecol. 2012, 5, 381–394. [Google Scholar] [CrossRef]





| Species | GenBank Accession # | A1 | B2 | LF above 11 m | LF below 11 m | CG | CWG |
|---|---|---|---|---|---|---|---|
| Filobasidium magnus | JX171169 | X | X | X | |||
| Debaryomyces hansenii | KM816678 | X | X | ||||
| Thelebolus ellipsoideus | JX171195 | X | X | X | X | ||
| Thelebolus globosus | JX171196 | X | X | X | X | ||
| Geomyces sp. 1 | KM816679 | X | X | ||||
| Acremonium sp. | KM816680 | X | X | ||||
| Mrakiella aquatica | JX171181 | X | X | X | |||
| Rhodotorula mucilaginosa | JX171192 | X | X | X | X | ||
| Vishniacozyma victoriae | JX171171 | X | X | ||||
| Glaciozyma antarctica | KM816681 | X | X | ||||
| Geomyces sp. 2 | KM816682 | X | X | X | X | ||
| Glaciozyma watsonii | JX171176 | X | X | ||||
| Naganishia globosa | KM819094 | X | X | X | |||
| Naganishia albidosimilis | JX171168 | X | X | X | |||
| Penicillium dipodomyicola | JX171186 | X | X | ||||
| Holtermanniella nyarrowii | KM816683 | X | X | ||||
| Aureobasidium pullulans | JX171163 | X | X | X | |||
| Toxicocladosporium strelitziae | KM816684 | X | X | ||||
| Cladosporium cladosporoides | KM816685 | X | X | X | |||
| Penicillium commune | JX171184 | X | X | X | |||
| Heydenia alpina | JX171178 | X | X | X | |||
| Clavispora lusitaniae | KM816686 | X | X | ||||
| Mrakia gelida | KM819098 | X | X | ||||
| Mrakia blollopsis | KM819093 | X | X | ||||
| Sporidiobolus pararoseus | KM819095 | X | X | ||||
| Cladosporium sp. | KM819097 | X | X | ||||
| Trichoderma atrovirde | KM822752 | X | X | ||||
| Trametes pubescens | KM822753 | X | X | ||||
| Eutypa lata | KM822754 | X | X | ||||
| Piptoporus betulinus | KM822755 | X | X |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Connell, L.; Segee, B.; Redman, R.; Rodriguez, R.J.; Staudigel, H. Biodiversity and Abundance of Cultured Microfungi from the Permanently Ice-Covered Lake Fryxell, Antarctica. Life 2018, 8, 37. https://doi.org/10.3390/life8030037
Connell L, Segee B, Redman R, Rodriguez RJ, Staudigel H. Biodiversity and Abundance of Cultured Microfungi from the Permanently Ice-Covered Lake Fryxell, Antarctica. Life. 2018; 8(3):37. https://doi.org/10.3390/life8030037
Chicago/Turabian StyleConnell, Laurie, Benjamin Segee, Regina Redman, Russell J. Rodriguez, and Hubert Staudigel. 2018. "Biodiversity and Abundance of Cultured Microfungi from the Permanently Ice-Covered Lake Fryxell, Antarctica" Life 8, no. 3: 37. https://doi.org/10.3390/life8030037
APA StyleConnell, L., Segee, B., Redman, R., Rodriguez, R. J., & Staudigel, H. (2018). Biodiversity and Abundance of Cultured Microfungi from the Permanently Ice-Covered Lake Fryxell, Antarctica. Life, 8(3), 37. https://doi.org/10.3390/life8030037




