Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers
Abstract
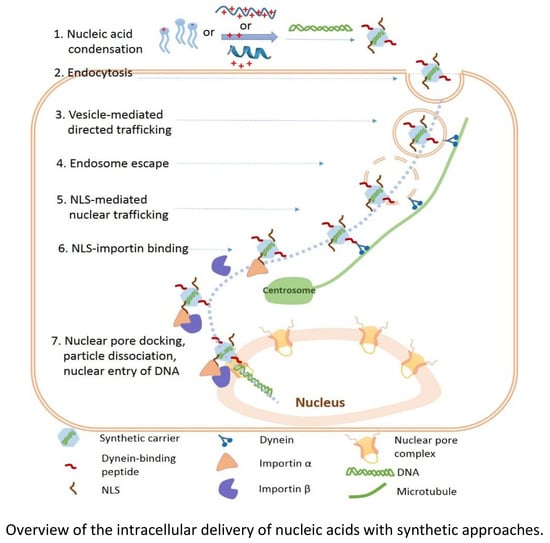
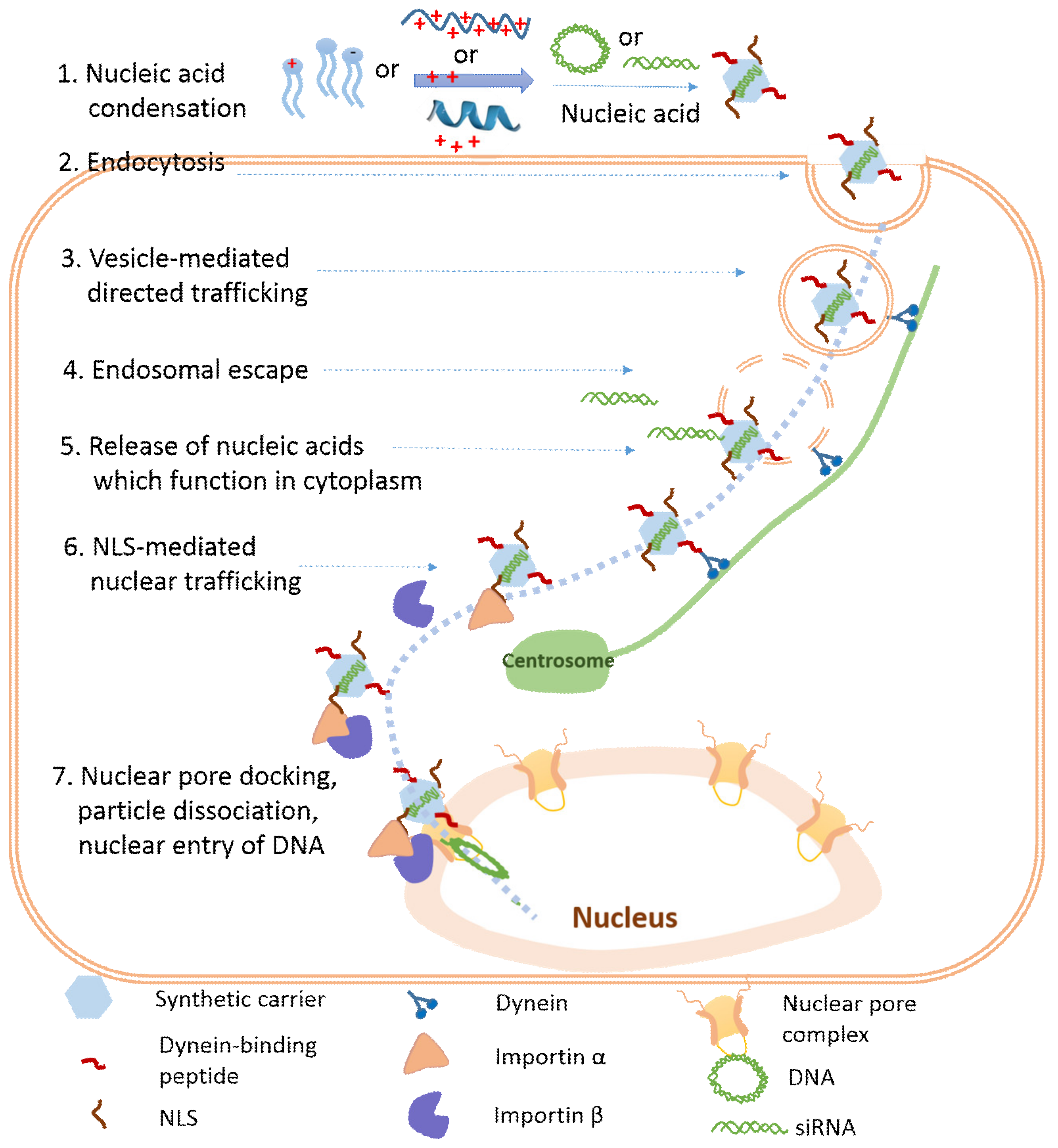
:1. Introduction
2. Natural and Synthetic Formulation Materials-Induced Nucleic Acid Condensation and Protection
2.1. Lipid-Based Lipoplexes
2.2. Polymer-Based Polyplexes
2.3. Peptide-Based Polyplexes
2.4. Nature-Inspired Artificial Viruses
3. Cellular Internalization of Nucleic Acids
3.1. Cellular Uptake of Lipoplexes
3.2. Cellular Uptake of Polymer-Based Polyplexes
3.3. Cellular Uptake of Peptide-Based Polyplexes and Artificial Viruses
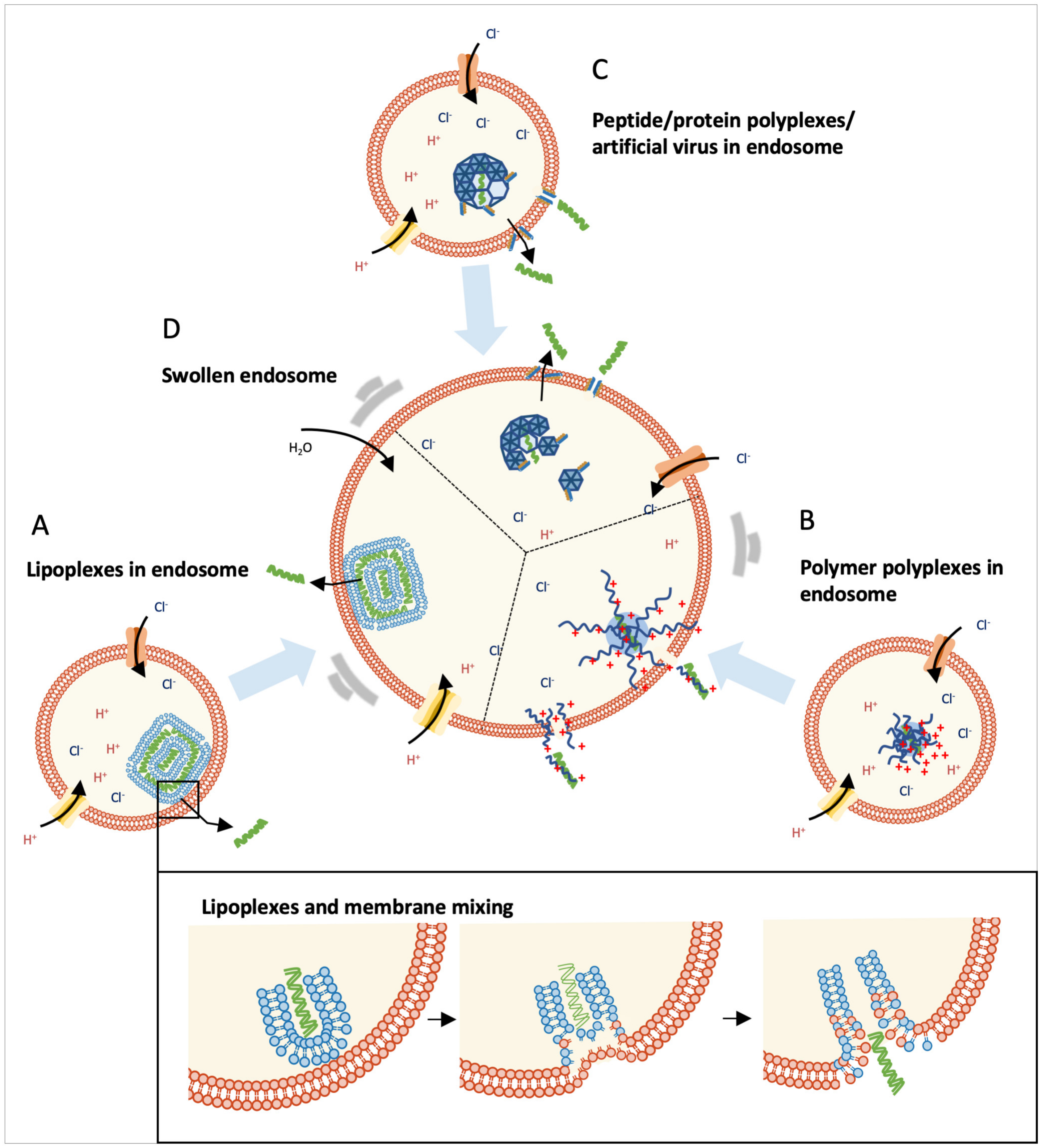
4. Endosomal Escape of Nucleic Acids
4.1. Environmental Changes during the Endosome Maturation Process
4.2. Lipid-Mediated Endosomal Escape
4.3. Polymer-Mediated Endosomal Escape
4.4. Peptide-Mediated Endosomal Escape
5. Nucleic Acid Release from the Carriers
6. Nuclear Trafficking of DNA
6.1. Challenges of Free DNA in Cytoplasm
6.2. Vesicle and Ligand-Guided Active Nuclear Trafficking of DNA
7. Nuclear Entry of DNA
7.1. Passive and Active Nuclear Accumulation of DNA
7.2. NLS-Mediated Active Nuclear Entry of DNA
7.3. Other Strategy-Facilitated Nuclear Entry of DNA
8. Summary and Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mansoori, B.; Shotorbani, S.S.; Baradaran, B. RNA Interference and its Role in Cancer Therapy. Adv. Pharm. Bull. 2014, 4, 313–321. [Google Scholar] [PubMed] [Green Version]
- Yang, B.; Jeang, J.; Yang, A.; Wu, T.C.; Hung, C.-F. DNA vaccine for cancer immunotherapy. Hum. Vaccines Immunother. 2014, 10, 3153–3164. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-Y.; Cao, C.-Y. The application of CRISPR-Cas9 genome editing tool in cancer immunotherapy. Briefings Funct. Genom. 2019, 18, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Filley, A.C.; Henriquez, M.; Dey, M. CART Immunotherapy: Development, Success, and Translation to Malignant Gliomas and Other Solid Tumors. Front. Oncol. 2018, 8, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.A.; Shalaev, E.; Karami, T.K.; Cunningham, J.; Slater, N.K.H.; Rivers, H.M. Pharmaceutical Development of AAV-Based Gene Therapy Products for the Eye. Pharm. Res. 2019, 36, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Mahato, R.I. Lipid and polymeric carrier-mediated nucleic acid delivery. Expert Opin. Drug Deliv. 2010, 7, 1209–1226. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Torchilin, V.P. Dendrimers for siRNA delivery. Pharmaceuticals 2013, 6, 161–183. [Google Scholar] [CrossRef]
- Rezaee, M.; Oskuee, R.K.; Nassirli, H.; Malaekeh-Nikouei, B. Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. J. Control. Release 2016, 236, 1–14. [Google Scholar] [CrossRef]
- Ozpolat, B.; Sood, A.K.; Lopez-Berestein, G. Liposomal siRNA nanocarriers for cancer therapy. Adv. Drug Deliv. Rev. 2014, 66, 110–116. [Google Scholar] [CrossRef]
- Démoulins, T.; Milona, P.; Englezou, P.C.; Ebensen, T.; Schulze, K.; Suter, R.; Pichon, C.; Midoux, P.; Guzmán, C.A.; Ruggli, N.; et al. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 711–722. [Google Scholar] [CrossRef]
- Crombez, L.; Aldrian-Herrada, G.; Konate, K.; Nguyen, Q.N.; McMaster, G.K.; Brasseur, R.; Heitz, F.; Divita, G. A New Potent Secondary Amphipathic Cell–penetrating Peptide for siRNA Delivery Into Mammalian Cells. Mol. Ther. 2009, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Elouahabi, A.; Ruysschaert, J.-M. Formation and Intracellular Trafficking of Lipoplexes and Polyplexes. Mol. Ther. 2005, 11, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Estrada, L.C.; Digman, M.A.; Pozzi, D.; Cardarelli, F.; Gratton, E.; Caracciolo, G. Intracellular trafficking of cationic liposome–DNA complexes in living cells. Soft Matter 2012, 8, 7919–7927. [Google Scholar] [CrossRef] [PubMed]
- Lechardeur, D.; Lukacs, G. Intracellular Barriers to Non-Viral Gene Transfer. Curr. Gene Ther. 2002, 2, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Dominska, M.; Dykxhoorn, D.M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010, 123, 1183–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughan, E.E.; Dean, D.A. Intracellular Trafficking of Plasmids during Transfection Is Mediated by Microtubules. Mol. Ther. 2006, 13, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Panté, N.; Kann, M. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of about 39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhi, D.; Huang, L. Lipid-based vectors for siRNA delivery. J. Drug Target. 2012, 20, 724–735. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.; Lächelt, U.; Bartek, J.; Wagner, E.; Moghimi, S.M. Polyplex Evolution: Understanding Biology, Optimizing Performance. Mol. Ther. 2017, 25, 1476–1490. [Google Scholar] [CrossRef] [Green Version]
- Scholz, C.; Wagner, E. Therapeutic plasmid DNA versus siRNA delivery: Common and different tasks for synthetic carriers. J. Control. Release 2012, 161, 554–565. [Google Scholar] [CrossRef]
- Guan, S.; Rosenecker, J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017, 24, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Siepmann, J.; Faham, A.; Clas, S.-D.; Boyd, B.J.; Jannin, V.; Bernkop-Schnürch, A.; Zhao, H.; Lecommandoux, S.; Evans, J.C.; Allen, C.; et al. Lipids and polymers in pharmaceutical technology: Lifelong companions. Int. J. Pharm. 2019, 558, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Zheng, M.; Tao, W.; Chung, R.; Jin, D.; Ghaffari, D.; Farokhzad, O.C. Challenges in DNA Delivery and Recent Advances in Multifunctional Polymeric DNA Delivery Systems. Biomacromolecules 2017, 18, 2231–2246. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, A.; Miyata, K.; Kataoka, K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv. Rev. 2016, 104, 61–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lächelt, U.; Wagner, E. Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef] [PubMed]
- Palamà, I.E.; Cortese, B.; D’Amone, S.; Gigli, G. mRNA delivery using non-viral PCL nanoparticles. Biomater. Sci. 2015, 3, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Neuberg, P.; Kichler, A. Recent Developments in Nucleic Acid Delivery with Polyethylenimines; Elsevier: Amsterdam, The Netherlands, 2014; Volume 88. [Google Scholar]
- Wagner, E. Polymers for Nucleic Acid Transfer—An Overview; Elsevier: Amsterdam, The Netherlands, 2014; Volume 88. [Google Scholar]
- Li, J.; Liang, H.; Liu, J.; Wang, Z. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int. J. Pharm. 2018, 546, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Zylberberg, C.; Gaskill, K.; Pasley, S.; Matosevic, S. Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther. 2017, 24, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Rietwyk, S.; Peer, D. Next-Generation Lipids in RNA Interference Therapeutics. ACS Nano 2017, 11, 7572–7586. [Google Scholar] [CrossRef]
- Meisel, J.W.; Gokel, G.W. A Simplified Direct Lipid Mixing Lipoplex Preparation: Comparison of Liposomal-, Dimethylsulfoxide-, and Ethanol-Based Methods. Sci. Rep. 2016, 6, 27662. [Google Scholar] [CrossRef]
- Koltover, I.; Salditt, T.; Rädler, J.O.; Safinya, C.R. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 1998, 281, 78–81. [Google Scholar] [CrossRef]
- Rädler, J.O.; Ra, J.O.; Koltover, I.; Salditt, T.; Safinya, C.R. DNA Intercalation in Multilamellar Membranes in Structure of DNA—Cationic Liposome Complexes: DNA Intercalation in Multilamellar Membranes in Distinct Interhelical Packing Regimes. Science 2007, 810, 1–5. [Google Scholar]
- Ewert, K.K.; Evans, H.M.; Zidovska, A.; Bouxsein, N.F.; Ahmad, A.; Safinya, C.R. A Columnar Phase of Dendritic Lipid-Based Cationic Liposome−DNA Complexes for Gene Delivery: Hexagonally Ordered Cylindrical Micelles Embedded in a DNA Honeycomb Lattice. J. Am. Chem. Soc. 2006, 128, 3998–4006. [Google Scholar] [CrossRef] [PubMed]
- Smisterová, J.; Wagenaar, A.; Stuart, M.C.A.; Polushkin, E.; Brinke, G.T.; Hulst, R.; Engberts, J.B.F.N.; Hoekstra, D. Molecular Shape of the Cationic Lipid Controls the Structure of Cationic Lipid/Dioleylphosphatidylethanolamine-DNA Complexes and the Efficiency of Gene Delivery. J. Biol. Chem. 2001, 276, 47615–47622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, N. Lipid-Nucleic Acid Supramolecular Complexes: Lipoplex Structure and the Kinetics of Formation. AIMS Biophys. 2015, 2, 163–183. [Google Scholar] [CrossRef]
- Mouritsen, O.G. Lipids, curvature, and nano-medicine*. Eur. J. Lipid Sci. Technol. 2011, 113, 1174–1187. [Google Scholar] [CrossRef] [PubMed]
- Dan, N.; Danino, D. Structure and kinetics of lipid–nucleic acid complexes. Adv. Colloid Interface Sci. 2014, 205, 230–239. [Google Scholar] [CrossRef]
- Wang, X.; Yu, B.; Ren, W.; Mo, X.; Zhou, C.; He, H.; Jia, H.; Wang, L.; Jacob, S.T.; Lee, R.J.; et al. Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J. Control. Release 2015, 172, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Zhi, D.; Zhang, S.; Wang, B.; Zhao, Y.; Yang, B.; Yu, S. Transfection Efficiency of Cationic Lipids with Different Hydrophobic Domains in Gene Delivery. Bioconjugate Chem. 2010, 21, 563–577. [Google Scholar] [CrossRef]
- Movahedi, F.; Hu, R.G.; Becker, D.L.; Xu, C. Stimuli-responsive liposomes for the delivery of nucleic acid therapeutics. Nanomed. Nanotechnol.Biol. Med. 2015, 11, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Puras, G.; Mashal, M.; Zarate, J.; Agirre, M.; Ojeda, E.; Grijalvo, S.; Eritja, R.; Díaz-Tahoces, A.; Navarrete, G.M.; Avilés-Trigueros, M.; et al. A novel cationic niosome formulation for gene delivery to the retina. J. Control. Release 2014, 174, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Kolate, A.; Baradia, D.; Patil, S.; Vhora, I.; Kore, G.; Misra, A. PEG—A versatile conjugating ligand for drugs and drug delivery systems. J. Control. Release 2014, 192, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Lee, R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016, 99, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Colosimo, A.; Serafino, A.; Sangiuolo, F.; Di Sario, S.; Bruscia, E.; Amicucci, P.; Novelli, G.; Dallapiccola, B.; Mossa, G. Gene transfection efficiency of tracheal epithelial cells by DC-Chol–DOPE/DNA complexes. Biochim. Biophys. Acta 1999, 1419, 186–194. [Google Scholar] [CrossRef]
- Yue, Y.; Jin, F.; Deng, R.; Cai, J.; Dai, Z.; Lin, M.C.; Kung, H.-F.; Mattebjerg, M.A.; Andresen, T.L.; Wu, C. Revisit complexation between DNA and polyethylenimine—Effect of length of free polycationic chains on gene transfection. J. Control. Release 2011, 152, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. Alnylam launches era of RNAi drugs. Nat. Biotechnol. 2018, 36, 777–778. [Google Scholar] [CrossRef] [PubMed]
- Molecules, U.; Index, S.; Gelbart, W.M.; Bruinsma, R.F.; Pincus, P.A.; Parsegian, V.A. DNA condensation. Curr. Opin. Struct. Biol. 1996, 6, 334–341. [Google Scholar]
- Rinkenauer, A.C.; Schubert, S.; Traeger, A. The influence of polymer architecture on in vitro pDNA transfection. J. Mater. Chem. B 2015, 3, 7477–7493. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, X.; Xu, L.; Wang, X.; Hennink, W.E.; Zhuo, R. Novel Reduction-Responsive Cross-Linked Polyethylenimine Derivatives by Click Chemistry for Nonviral Gene Delivery. Bioconjugate Chem. 2010, 21, 1827–1835. [Google Scholar] [CrossRef]
- Taranejoo, S.; Liu, J.; Verma, P.; Hourigan, K. A review of the developments of characteristics of PEI derivatives for gene delivery applications. J. Appl. Polym. Sci. 2015, 132, 2–9. [Google Scholar] [CrossRef]
- Wang, M.; Guo, Y.; Yu, M.; Ma, P.X.; Mao, C.; Lei, B. Photoluminescent and biodegradable polycitrate-polyethylene glycol-polyethyleneimine polymers as highly biocompatible and efficient vectors for bioimaging-guided siRNA and miRNA delivery. Acta Biomater. 2017, 54, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.; Hu, Q.; Tang, G.; Li, J. FGFR-targeted gene delivery mediated by supramolecular assembly between β-cyclodextrin-crosslinked PEI and redox-sensitive PEG. Biomaterials 2013, 34, 6482–6494. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, C.; Pezzoli, D.; Malloggi, C.; Candeo, A.; Capelli, G.; Bassi, A.; Volonterio, A.; Taroni, P.; Candiani, G. The study of polyplex formation and stability by time-resolved fluorescence spectroscopy of SYBR Green I-stained DNA. Photochem. Photobiol. Sci. 2014, 13, 1680–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mady, M.M.; Mohammed, W.A.; El-Guendy, N.M.; Elsayed, A.A. Effect of Polymer Molecular Weight on the Dna/Pei Polyplexes Properties. J. Biophys. 2011, 21, 151–165. [Google Scholar]
- Chiper, M.; Tounsi, N.; Kole, R.; Kichler, A.; Zuber, G. Self-aggregating 1.8 kDa polyethylenimines with dissolution switch at endosomal acidic pH are delivery carriers for plasmid DNA, mRNA, siRNA and exon-skipping oligonucleotides. J. Control. Release 2017, 246, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, M.; Zhang, Z.; Gong, T.; Sun, X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2016, 23, 2596–2607. [Google Scholar] [CrossRef]
- Dai, Z.; Wu, C. How Does DNA Complex with Polyethylenimine with Different Chain Lengths and Topologies in Their Aqueous Solution Mixtures? Macromolecules 2012, 45, 4346–4353. [Google Scholar] [CrossRef]
- Alameh, M.; Lavertu, M.; Tran-Khanh, N.; Chang, C.Y.; Lesage, F.; Bail, M.; Darras, V.; Chevrier, A.; Buschmann, M.D. SiRNA Delivery with Chitosan: Influence of Chitosan Molecular Weight, Degree of Deacetylation, and Amine to Phosphate Ratio on in Vitro Silencing Efficiency, Hemocompatibility, Biodistribution, and in Vivo Efficacy. Biomacromolecules 2018, 19, 112–131. [Google Scholar] [CrossRef] [PubMed]
- Supaprutsakul, S.; Chotigeat, W.; Wanichpakorn, S.; Kedjarune-Leggat, U. Transfection efficiency of depolymerized chitosan and epidermal growth factor conjugated to chitosan–DNA polyplexes. J. Mater. Sci. Mater. Electron. 2010, 21, 1553–1561. [Google Scholar] [CrossRef]
- Sah, E.; Sah, H. Recent Trends in Preparation of Poly(lactide- co -glycolide) Nanoparticles by Mixing Polymeric Organic Solution with Antisolvent. J. Nanomater. 2015, 2015, 1–22. [Google Scholar] [CrossRef]
- Tahara, K.; Sakai, T.; Yamamoto, H.; Takeuchi, H.; Kawashima, Y. Establishing chitosan coated PLGA nanosphere platform loaded with wide variety of nucleic acid by complexation with cationic compound for gene delivery. Int. J. Pharm. 2008, 354, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Udhayakumar, V.K.; De Beuckelaer, A.; McCaffrey, J.; McCrudden, C.M.; Kirschman, J.L.; Vanover, D.; van Hoecke, L.; Roose, K.; Deswarte, K.; De Geest, B.G.; et al. Arginine-Rich Peptide-Based mRNA Nanocomplexes Efficiently Instigate Cytotoxic T Cell Immunity Dependent on the Amphipathic Organization of the Peptide. Adv. Heal. Mater. 2017, 6, 1601412. [Google Scholar] [CrossRef] [PubMed]
- Tönges, L.; Lingor, P.; Egle, R.; Dietz, G.P.; Fahr, A.; Bähr, M. Stearylated octaarginine and artificial virus-like particles for transfection of siRNA into primary rat neurons. RNA 2006, 12, 1431–1438. [Google Scholar] [CrossRef] [Green Version]
- Deshayes, S.; Gerbal-Chaloin, S.; Morris, M.C.; Aldrian-Herrada, G.; Charnet, P.; Divita, G.; Heitz, F. On the mechanism of non-endosomial peptide-mediated cellular delivery of nucleic acids. Biochim. Biophys. Acta. 2004, 1667, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crombez, L.; Morris, M.C.; Dufort, S.; Aldrian-Herrada, G.; Nguyen, Q.; Mc Master, G.; Coll, J.-L.; Heitz, F.; Divita, G. Targeting cyclin B1 through peptide-based delivery of siRNA prevents tumour growth. Nucleic Acids Res. 2009, 37, 4559–4569. [Google Scholar] [CrossRef]
- Chen, S.; Lei, Q.; Li, S.-Y.; Qin, S.-Y.; Jia, H.-Z.; Cheng, Y.-J.; Zhang, X.-Z. Fabrication of dual responsive co-delivery system based on three-armed peptides for tumor therapy. Biomaterials 2016, 92, 25–35. [Google Scholar] [CrossRef]
- Chen, Q.; Qi, R.; Chen, X.; Yang, X.; Wu, S.; Xiao, H.; Dong, W. A Targeted and Stable Polymeric Nanoformulation Enhances Systemic Delivery of mRNA to Tumors. Mol. Ther. 2017, 25, 92–101. [Google Scholar] [CrossRef] [Green Version]
- Crowley, S.T.; Poliskey, J.A.; Baumhover, N.J.; Rice, K.G. Efficient Expression of Stabilized mRNAPEG-Peptide Polyplexes in Liver. Gene Ther. 2016, 22, 993–999. [Google Scholar] [CrossRef]
- Dileo, J.; Banerjee, R.; Whitmore, M.; Nayak, J.V.; Falo, L.D.; Huang, L. Lipid–protamine–DNA-mediated antigen delivery to antigen-presenting cells results in enhanced anti-tumor immune responses. Mol. Ther. 2003, 7, 640–648. [Google Scholar] [CrossRef]
- Singh, P.; Gonzalez, M.J.; Manchester, M. Viruses and their uses in nanotechnology. Drug Dev. Res. 2006, 67, 23–41. [Google Scholar] [CrossRef]
- Jeevanandam, J.; Pal, K.; Danquah, M.K. Virus-like nanoparticles as a novel delivery tool in gene therapy. Biochimie 2019, 157, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Jekhmane, S.; De Haas, R.; Filho, O.P.D.S.; Van Asbeck, A.H.; Favretto, M.E.; Garcia, A.H.; Brock, R.; de Vries, R. Virus-Like Particles of mRNA with Artificial Minimal Coat Proteins: Particle Formation, Stability, and Transfection Efficiency. Nucleic Acid Ther. 2017, 27, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Zhitnyuk, Y.; Gee, P.; Lung, M.S.; Sasakawa, N.; Xu, H.; Saito, H.; Hotta, A. Efficient mRNA delivery system utilizing chimeric VSVG-L7Ae virus-like particles. Biochem. Biophys. Res. Commun. 2018, 505, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.-N.; Yang, Y.-H.; Wu, M.-S.; Chou, M.-C.; Fang, C.-Y.; Lin, M.-C.; Tai, C.-K.; Shen, C.-H.; Chen, P.-L.; Chang, D.; et al. Gene therapy for human glioblastoma using neurotropic JC virus-like particles as a gene delivery vector. Sci. Rep. 2018, 8, 2213. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.-N.; Huang, Y.-L.; Lin, M.-C.; Fang, C.-Y.; Shen, C.-H.; Chen, P.-L.; Wang, M.; Chang, D.; Tseng, C.-E. Inhibition of human diffuse large B-cell lymphoma growth by JC polyomavirus-like particles delivering a suicide gene. J. Transl. Med. 2015, 13, 3909. [Google Scholar] [CrossRef]
- Chao, C.-N.; Lin, M.-C.; Fang, C.-Y.; Chen, P.-L.; Chang, D.; Shen, C.-H.; Wang, M. Gene Therapy for Human Lung Adenocarcinoma Using a Suicide Gene Driven by a Lung-Specific Promoter Delivered by JC Virus-Like Particles. PLoS ONE 2016, 11, 0157865. [Google Scholar] [CrossRef]
- Chen, L.-S.; Wang, M.; Ou, W.-C.; Fung, C.-Y.; Chen, P.-L.; Chang, C.-F.; Huang, W.-S.; Wang, J.-Y.; Lin, P.Y.; Chang, D. Efficient gene transfer using the human JC virus-like particle that inhibits human colon adenocarcinoma growth in a nude mouse model. Gene Ther. 2010, 17, 1033–1041. [Google Scholar] [CrossRef] [Green Version]
- Ni, R.; Chau, Y. Structural Mimics of Viruses Through Peptide/DNA Co-Assembly. J. Am. Chem. Soc. 2014, 136, 17902–17905. [Google Scholar] [CrossRef]
- Ni, R.; Chau, Y. Tuning the Inter-nanofibril Interaction To Regulate the Morphology and Function of Peptide/DNA Co-assembled Viral Mimics. Angew. Chem. Int. Ed. 2017, 56, 9356–9360. [Google Scholar] [CrossRef]
- Hernandez-Garcia, A.; Kraft, D.J.; Janssen, A.F.J.; Bomans, P.H.H.; Sommerdijk, N.A.J.M.; Thies-Weesie, D.M.E.; Favretto, M.E.; Brock, R.; De Wolf, F.A.; Werten, M.W.T.; et al. Design and self-assembly of simple coat proteins for artificial viruses. Nat. Nanotechnol. 2014, 9, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Ruff, Y.; Moyer, T.; Newcomb, C.J.; Demeler, B.; Stupp, S.I. Precision Templating with DNA of a Virus-like Particle with Peptide Nanostructures. J. Am. Chem. Soc. 2013, 135, 6211–6219. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.-B.; Lee, E.; Yoon, Y.-R.; Lee, M.S.; Lee, M. Filamentous Artificial Virus from a Self-Assembled Discrete Nanoribbon. Angew. Chem. 2008, 120, 4601–4604. [Google Scholar] [CrossRef]
- El-Sayed, A.; Harashima, H. Endocytosis of Gene Delivery Vectors: From Clathrin-dependent to Lipid Raft-mediated Endocytosis. Mol. Ther. 2013, 21, 1118–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, N.N.; Vandenbroucke, R.; Wagner, E.; Ogris, M.; Vongersdorff, K.; Desmedt, S.; Von Gersdorff, K.; De Smedt, S.C. The Internalization Route Resulting in Successful Gene Expression Depends on both Cell Line and Polyethylenimine Polyplex Type. Mol. Ther. 2006, 14, 745–753. [Google Scholar]
- Suen, W.L.L.; Chau, Y. Size-dependent internalisation of folate-decorated nanoparticles via the pathways of clathrin and caveolae-mediated endocytosis in ARPE-19 cells. J. Pharm. Pharmacol. 2014, 66, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, Y.S.; Pang, B.; Hyun, D.C.; Yang, M.; Xia, Y. Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem. Int. Ed. 2014, 53, 12320–12364. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Tang, P.S.; Chan, W.C. The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.J.; Langer, R.; Chen, J. A Novel Mechanism Is Involved in Cationic Lipid-Mediated Functional siRNA Delivery Accessed articles. Mol. Pharm. 2009, 6, 763–771. [Google Scholar] [CrossRef]
- Lazebnik, M.; Keswani, R.K.; Pack, D.W. Endocytic Transport of Polyplex and Lipoplex siRNA Vectors in HeLa Cells. Pharm. Res. 2016, 33, 2999–3011. [Google Scholar] [CrossRef]
- Pozzi, D.; Marchini, C.; Cardarelli, F.; Amenitsch, H.; Garulli, C.; Bifone, A.; Caracciolo, G. Transfection efficiency boost of cholesterol-containing lipoplexes. Biochim. Biophys. Acta 2012, 1818, 2335–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hama, S.; Akita, H.; Ito, R.; Mizuguchi, H.; Hayakawa, T.; Harashima, H. Quantitative Comparison of Intracellular Trafficking and Nuclear Transcription between Adenoviral and Lipoplex Systems. Mol. Ther. 2006, 13, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Marchini, C.; Pozzi, D.; Montani, M.; Alfonsi, C.; Amici, A.; Amenitsch, H.; De Sanctis, S.C.; Caracciolo, G. Tailoring Lipoplex Composition to the Lipid Composition of Plasma Membrane: A Trojan Horse for Cell Entry? Langmuir 2010, 26, 13867–13873. [Google Scholar] [CrossRef] [PubMed]
- Gabrielson, N.P.; Pack, D.W. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J. Control. Release 2009, 136, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Chau, Y. Different oligoarginine modifications alter endocytic pathways and subcellular trafficking of polymeric nanoparticles. Biomater. Sci. 2016, 4, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, K.; Ruthardt, N.; von Gersdorff, K.; Bausinger, R.; Wagner, E.; Ogris, M.; Bräuchle, C. Cellular dynamics of EGF receptor-targeted synthetic viruses. Mol. Ther. 2007, 15, 1297–1305. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Du, J.; Ren, K.; Wang, Y. Cell penetrating peptide-based polyplexes shelled with polysaccharide to improve stability and gene transfection. Nanoscale 2015, 7, 8476–8484. [Google Scholar] [CrossRef]
- Simeoni, F. Insight into the mechanism of the peptide-based gene delivery system MPG: Implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003, 31, 2717–2724. [Google Scholar] [CrossRef]
- Deshayes, S.; Konate, K.; Aldrian, G.; Crombez, L.; Heitz, F.; Divita, G. Structural polymorphism of non-covalent peptide-based delivery systems: Highway to cellular uptake. Biochim. Biophys. Acta 2010, 1798, 2304–2314. [Google Scholar] [CrossRef]
- Deshayes, S.; Heitz, A.; Morris, M.C.; Charnet, P.; Divita, G.; Heitz, F. Insight into the Mechanism of Internalization of the Cell-Penetrating Carrier Peptide Pep-1 through Conformational Analysis. Biochemistry 2004, 43, 1449–1457. [Google Scholar] [CrossRef]
- Bus, T.; Traeger, A.; Schubert, U.S. The great escape: How cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B 2018, 6, 6904–6918. [Google Scholar] [CrossRef]
- Durymanov, M.; Reineke, J. Non-viral Delivery of Nucleic Acids: Insight Into Mechanisms of Overcoming Intracellular Barriers. Front. Pharmacol. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohdanowicz, M.; Grinstein, S. Role of Phospholipids in Endocytosis, Phagocytosis, and Macropinocytosis. Physiol. Rev. 2013, 93, 69–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelphati, O.; Szoka, F.C. Mechanism of oligonucleotide release from cationic liposomes. Proc. Natl. Acad. Sci. USA 1996, 93, 11493–11498. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Munye, M.M.; Tagalakis, A.D.; Manunta, M.D.I.; Hart, S.L. The Role of the Helper Lipid on the DNA Transfection Efficiency of Lipopolyplex Formulations. Sci. Rep. 2014, 4, 7107. [Google Scholar] [CrossRef] [PubMed]
- Essex, S.; Navarro, G.; Sabhachandani, P.; Chordia, A.; Trivedi, M.; Movassaghian, S.; Torchilin, V.P. Phospholipid-modified PEI-based nanocarriers for in vivo siRNA therapeutics against multidrug-resistant tumors. Gene Ther. 2015, 22, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, F.; Pozzi, D.; Bifone, A.; Marchini, C.; Caracciolo, G. Cholesterol-dependent macropinocytosis and endosomal escape control the transfection efficiency of lipoplexes in CHN living cells. Mol. Pharm. 2012, 9, 334–340. [Google Scholar] [CrossRef]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. 2012, 124, 8657–8661. [Google Scholar] [CrossRef]
- Benjaminsen, R.V.; Mattebjerg, M.A.; Henriksen, J.R.; Moghimi, S.M.; Andresen, T.L. The possible “proton sponge” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol. Ther. 2013, 21, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Behr, J.P. The proton sponge. A trick to enter cells the viruses did not exploit. Chimia (Aarau) 1997, 51, 34–36. [Google Scholar]
- Nguyen, J.; Szoka, F.C. Nucleic acid delivery: The missing pieces of the puzzle? Accounts Chem. Res. 2012, 45, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Brans, T.; Samal, S.K.; Dubruel, P.; Demeester, J.; De Smedt, S.C.; Remaut, K.; Braeckmans, K.; Vermeulen, L.M. Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano 2018, 12, 2332–2345. [Google Scholar] [Green Version]
- Aoki, Y.; Hosaka, S.; Kawa, S.; Kiyosawa, K. Potential tumor-targeting peptide vector of histidylated oligolysine conjugated to a tumor-homing RGD motif. Cancer Gene Ther. 2001, 8, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.; Mixson, A.J. Modified branched peptides with a histidine-rich tail enhance in vitro gene transfection. Nucleic Acids Res. 2005, 33, e40. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-L.; Higuchi, Y.; Kawakami, S.; Yamashita, F.; Hashida, M. Efficient Gene Transfection by Histidine-Modified Chitosan through Enhancement of Endosomal Escape. Bioconjugate Chem. 2010, 21, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; Gao, X. Functional peptides for siRNA delivery. Adv. Drug Deliv. Rev. 2017, 110–111, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Van Rossenberg, S.M.W.; Hickabottom, M.; Parker, G.A.; Freemont, P.; Crook, T.; Allday, M.J. Targeted Lysosome Disruptive Elements for Improvement of Parenchymal Liver Cell-specific Gene Delivery. J. Biol. Chem. 2002, 277, 45803–45810. [Google Scholar] [CrossRef] [Green Version]
- Kusumoto, K.; Akita, H.; Santiwarangkool, S.; Harashima, H. Advantages of ethanol dilution method for preparing GALA-modified liposomal siRNA carriers on the in vivo gene knockdown efficiency in pulmonary endothelium. Int. J. Pharm. 2014, 473, 144–147. [Google Scholar] [CrossRef] [Green Version]
- Freulon, I.; Roche, A.-C.; Monsigny, M.; Mayer, R. Delivery of oligonucleotides into mammalian cells by anionic peptides: Comparison between monomeric and dimeric peptides. Biochem. J. 2001, 354, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Nicol, F.; Szoka, F.C. GALA: A designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv. Drug Deliv. Rev. 2004, 56, 967–985. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.C.; Malhotra, M.; Sweeney, K.; Darcy, R.; Nelson, C.C.; Hollier, B.G.; O’Driscoll, C.M. Folate-targeted amphiphilic cyclodextrin nanoparticles incorporating a fusogenic peptide deliver therapeutic siRNA and inhibit the invasive capacity of 3D prostate cancer tumours. Int. J. Pharm. 2017, 532, 511–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, H.M.; Maksimenko, A.; Urbinati, G.; Chapuis, H.; Raouane, M.; Desmaële, D.; Yasuhiro, H.; Harashima, H.; Couvreur, P.; Massaad-Massade, L. Effects of Silencing the RET/PTC1 Oncogene in Papillary Thyroid Carcinoma by siRNA-Squalene Nanoparticles With and Without Fusogenic Companion GALA-Cholesterol. Thyroid 2013, 24, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Leong, K.W.; Grigsby, C.L. Balancing protection and release of DNA: Tools to address a bottleneck of non-viral gene delivery. J. R. Soc. Interface 2010, 7, S67–S82. [Google Scholar]
- Hirsch, M.; Helm, M. Live cell imaging of duplex siRNA intracellular trafficking. Nucleic Acids Res. 2015, 43, 4650–4660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittrup, A.; Ai, A.; Liu, X.; Hamar, P.; Trifonova, R.; Charisse, K.; Manoharan, M.; Kirchhausen, T.; Lieberman, J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015, 33, 870–876. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, D.V.; Fidelman, N.A.; Dan, N.; Lauffenburger, D.A. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 2000, 67, 598–606. [Google Scholar] [CrossRef]
- Huth, S.; Hoffmann, F.; Von Gersdorff, K.; Laner, A.; Reinhardt, D.; Rosenecker, J.; Rudolph, C. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J. Gene Med. 2006, 8, 1416–1424. [Google Scholar] [CrossRef]
- Forrest, M.L.; Meister, G.E.; Koerber, J.T.; Pack, D.W. Partial Acetylation of Polyethylenimine Enhances In Vitro Gene Delivery. Pharm. Res. 2004, 21, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Gabrielson, N.P.; Pack, D.W. Acetylation of Polyethylenimine Enhances Gene Delivery via Weakened Polymer/DNA Interactions. Biomacromolecules 2006, 7, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, J.W.; Lynn, D.M. Addition of ‘Charge-Shifting’ Side Chains to Linear Poly(ethyleneimine) Enhances Cell Transfection Efficiency. Biomacromolecules 2008, 9, 2063–2071. [Google Scholar] [CrossRef] [PubMed]
- Köping-Höggård, M.; Vårum, K.M.; Issa, M.; Danielsen, S.; Christensen, B.E.; Stokke, B.T.; Artursson, P. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004, 11, 1441–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiang, T.; Wen, J.; Lim, H.W.; Leong, K.W. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials 2004, 25, 5293–5301. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-J.; Wang, Z.-Y.; Wang, Y.-C. Redox-sensitive dendrimersomes assembled from amphiphilic Janus dendrimers for siRNA delivery. Biomater. Sci. 2018, 6, 2122–2129. [Google Scholar] [CrossRef]
- Xu, X.; Wu, J.; Liu, S.; Saw, P.E.; Tao, W.; Li, Y.; Krygsman, L.; Yegnasubramanian, S.; De Marzo, A.M.; Shi, J.; et al. Redox-Responsive Nanoparticle-Mediated Systemic RNAi for Effective Cancer Therapy. Small 2008, 14, 1802565. [Google Scholar] [CrossRef]
- Lukacs, G.L.; Haggie, P.; Lechardeur, D.; Freedman, N.; Verkman, S.; Verkman, A.S. Cell Biology and Metabolism: Size-dependent DNA Mobility in Cytoplasm and Nucleus Size-dependent DNA Mobility in Cytoplasm and Nucleus. J. Biol. Chem. 2000, 275, 1625–1629. [Google Scholar] [CrossRef]
- Bai, H.; Lester, G.M.S.; Petishnok, L.C.; Dean, D.A. Cytoplasmic transport and nuclear import of plasmid DNA. Biosci. Rep. 2017, 37, 20160616. [Google Scholar] [CrossRef]
- Lechardeur, D.; Sohn, K.J.; Haardt, M.; Joshi, P.B.; Monck, M.; Graham, R.W.; Beatty, B.; Squire, J.; O’Brodovich, H.; Lukacs, G.L. Metabolic instability of plasmid DNA in the cytosol: A potential barrier to gene transfer. Gene Ther. 2002, 6, 482–497. [Google Scholar] [CrossRef]
- Cohen, R.N.; Van Der Aa, M.A.; Macaraeg, N.; Lee, A.P.; Szoka, F.C., Jr. Quantification of Plasmid DNA Copies in the Nucleus after Lipoplex and Polyplex Transfection. J. Control. Release 2009, 135, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Ondrej, V.; Lukásová, E.; Falk, M.; Kozubek, S. The role of actin and microtubule networks in plasmid DNA intracellular trafficking. Acta Biochim. Pol. 2007, 54, 657–663. [Google Scholar] [PubMed]
- Sauer, A.; De Bruin, K.; Ruthardt, N.; Mykhaylyk, O.; Plank, C.; Bräuchle, C. Dynamics of magnetic lipoplexes studied by single particle tracking in living cells. J. Control. Release 2009, 137, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Mieruszynski, S.; Digman, M.A.; Gratton, E.; Jones, M.R. Characterization of exogenous DNA mobility in live cells through fluctuation correlation spectroscopy. Sci. Rep. 2015, 5, 13848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loubéry, S.; Wilhelm, C.; Hurbain, I.; Neveu, S.; Louvard, D.; Coudrier, E. Different Microtubule Motors Move Early and Late Endocytic Compartments. Traffic 2008, 9, 492–509. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, F.; Digiacomo, L.; Marchini, C.; Amici, A.; Salomone, F.; Fiume, G.; Rossetta, A.; Gratton, E.; Pozzi, D.; Caracciolo, G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016, 6, 25879. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.; Wirtz, D.; Hanes, J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl. Acad. Sci. USA 2003, 100, 3878–3882. [Google Scholar] [CrossRef] [Green Version]
- Bausinger, R.; Von Gersdorff, K.; Braeckmans, K.; Ogris, M.; Wagner, E.; Bräuchle, C.; Zumbusch, A. The Transport of Nanosized Gene Carriers Unraveled by Live-Cell Imaging. Angew. Chem. 2006, 118, 1598–1602. [Google Scholar] [CrossRef]
- Akita, H.; Enoto, K.; Masuda, T.; Mizuguchi, H.; Tani, T.; Harashima, H. Particle Tracking of Intracellular Trafficking of Octaarginine-modified Liposomes: A Comparative Study With Adenovirus. Mol. Ther. 2010, 18, 955–964. [Google Scholar] [CrossRef]
- King, S.J.; Schroer, T.A. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2000, 2, 20–24. [Google Scholar] [CrossRef]
- Kim, A.J.; Boylan, N.J.; Suk, J.S.; Lai, S.K.; Hanes, J. Non-degradative intracellular trafficking of highly compacted polymeric DNA nanoparticles. J. Control. Release 2012, 158, 102–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, J.; Vallee, R.B. Adenovirus Recruits Dynein by an Evolutionary Novel Mechanism Involving Direct Binding to pH-Primed Hexon. Viruses 2011, 3, 1417–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mabit, H.; Nakano, M.Y.; Prank, U.; Saam, B.; Döhner, K.; Sodeik, B.; Greber, U.F. Intact Microtubules Support Adenovirus and Herpes Simplex Virus Infections. J. Virol. 2002, 76, 9962–9971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, M.W.; Diefenbach, R.; Homa, F.L.; Miranda-Saksena, M.; Rixon, F.J.; Vittone, V.; Byth, K.; Cunningham, A. Herpes Simplex Virus Type 1 Capsid Protein VP26 Interacts with Dynein Light Chains RP3 and Tctex1 and Plays a Role in Retrograde Cellular Transport. J. Biol. Chem. 2004, 279, 28522–28530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akita, H.; Enoto, K.; Tanaka, H.; Harashima, H. Particle Tracking Analysis for the Intracellular Trafficking of Nanoparticles Modified with African Swine Fever Virus Protein p54-derived Peptide. Mol. Ther. 2013, 21, 309–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludtke, J.J.; Zhang, G.; Sebestyén, M.G.; A Wolff, J. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J. Cell Sci. 1999, 112, 2033–2041. [Google Scholar] [PubMed]
- Kirchenbuechler, I.; Kirchenbuechler, D.; Elbaum, M. Correlation between cationic lipid-based transfection and cell division. Exp. Cell Res. 2016, 345, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Durymanov, M.O.; Yarutkin, A.V.; Khramtsov, Y.V.; Rosenkranz, A.A.; Sobolev, A.S. Live imaging of transgene expression in Cloudman S91 melanoma cells after polyplex-mediated gene delivery. J. Control. Release 2015, 215, 73–81. [Google Scholar] [CrossRef]
- Kamiya, H.; Fujimura, Y.; Matsuoka, I.; Harashima, H. Live cell imaging of duplex siRNA intracellular trafficking. Biochem. Biophys. Res. Commun. 2002, 298, 591–597. [Google Scholar] [CrossRef]
- Grandinetti, G.; Smith, A.E.; Reineke, T.M. Membrane and Nuclear Permeabilization by Polymeric pDNA Vehicles: Efficient Method for Gene Delivery or Mechanism of Cytotoxicity? Mol. Pharm. 2012, 9, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Breuzard, G.; Tertil, M.; Gonçalves, C.; Cheradame, H.; Géguan, P.; Pichon, C.; Midoux, P. Nuclear delivery of NFκB-assisted DNA/polymer complexes: Plasmid DNA quantitation by confocal laser scanning microscopy and evidence of nuclear polyplexes by FRET imaging. Nucleic Acids Res. 2008, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl. Acad. Sci. USA 2002, 96, 5177–5181. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.P.; Dean, D.A. Progress and prospects: Nuclear import of nonviral vectors. Gene Ther. 2010, 17, 439–447. [Google Scholar] [CrossRef] [PubMed]
- McLane, L.M.; Corbett, A.H. Nuclear localization signals and human disease. IUBMB Life 2009, 61, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Reaa, J.C.; Giblyb, R.F.; Barronc, A.E.; Shea, L.D. Self-assembling peptide–lipoplexes for substrate-mediated gene delivery. Acta Biomater. 2009, 5, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mitin, A.; Vinogradov, S.V. Efficient Nuclear Targeting by NLS-PEG-Acridine Conjugates in the Transfection of Blood-Brain Barrier Endothelial Cells with Lipoplexes and Polyplexes. Bioconjugate Chem. 2009, 20, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wang, J.; Shen, J.; Liu, M.; Jin, X.; Tang, G.; Chu, P.K. Intracellular pathways and nuclear localization signal peptide-mediated gene transfection by cationic polymeric nanovectors. Biomaterials 2012, 33, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.M.; Dean, D.A. Tissue-specific and transcription factor-mediated nuclear entry of DNA. Adv. Drug Deliv. Rev. 2009, 61, 603–613. [Google Scholar] [CrossRef]
- Munkonge, F.M.; Amin, V.; Hyde, S.C.; Green, A.-M.; Pringle, I.A.; Gill, D.R.; Smith, J.W.S.; Hooley, R.P.; Xenariou, S.; Ward, M.A.; et al. Identification and Functional Characterization of Cytoplasmic Determinants of Plasmid DNA Nuclear Import. J. Biol. Chem. 2009, 284, 26978–26987. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Kube, D.M.; Cooper, M.J.; Davis, P.B. Cell Surface Nucleolin Serves as Receptor for DNA Nanoparticles Composed of Pegylated Polylysine and DNA. Mol. Ther. 2008, 16, 333–342. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Jans, D.A. Nucleocytoplasmic transport of DNA: Enhancing non-viral gene transfer. Biochem. J. 2007, 406, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yao, J.; Du, R.; Hou, L.; Zhou, J.; Lu, Y.; Meng, Q.; Zhang, Q. Ternary complexes with core-shell bilayer for double level targeted gene delivery: In vitro and in vivo evaluation. Pharm. Res. 2013, 30, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Fan, Y.; Li, Y.; Huang, L. Strategies on the nuclear-targeted delivery of genes. J. Drug Target. 2013, 21, 926–939. [Google Scholar] [CrossRef] [PubMed]






| Formulation Material | Experimental Nucleic Acids | Experimental Cell Type | Size (nm) | Surface Charge (mV) | Ref. |
|---|---|---|---|---|---|
| Lipid | |||||
| DOTAP/DOPE/cholesterol | siRNA | BSC-40; 293FT; HeLa cells | 100–200 | N/A | [92] |
| DOTAP/DOPE/cholesterol; PEI | siRNA | HeLa cells | 468 ± 19; 209 ± 17 | N/A | [93] |
| DC-Cholesterol/DOPE | pDNA: luciferase | CHO cells | 60–70 | 58.4–64.7 | [94] |
| DC-cholesterol/DOPE;DOPC/DOTAP; DC-cholesterol/DOPE/DOTAP/DOPC | pDNA: luciferase | NIH 3T3 cells | 180 ± 2; 234 ± 4; 205 ± 2 | 48.3 ± 1.2; 43.3 ± 1.5; 46.7 ± 1.2 | [96] |
| Lipofectamine Plus | pDNA: luciferase | A549 cells | N/A | N/A | [95] |
| Polymer | |||||
| PEI (disulfide cross-linked) | pDNA: luciferase | HEK293 T; HeLa cells | ~200 | ~20 | [52] |
| POCG-PEG-PEI polymer | DNA; siRNA; miRNA | MCF-7; C2C12 | 151; 172; 245 | ~30 | [54] |
| MPC/Ad-SS-PEG | pDNA: GFP | Hep G2 cells; HeLa cells; SKOV-3 cells; PC-3 cells | 100–200 | 0.5–5 | [55] |
| PEG-PEI (1.8 kDa) (PEI amines are modified with aromatic rings) | pDNA: luciferase mRNA: luciferase siRNA: anti-luciferase | HeLa cells; U87 cells | 180–240 | N/A | [58] |
| PEI-stearic acid copolymer | mRNA: ACRA mRNA: HIV-1 gag antigen | DC 2.4 cells | 117.77 ± 3.894 | N/A | [59] |
| Folic acid-PEI; transferrin-PEI | pDNA: luciferase | HeLa cells | N/A | N/A | [97] |
| mPEG-PCL; R-PEG-PCL; RRRR-PEG-PCL; RRRRRRRR-PEG-PCL | N/A | HeLa cells | 80–110 | 20–40 | [98] |
| EGF-PEG-PEI | DNA | HuH7 cells | 266 ± 26 | N/A | [99] |
| Peptide/protein | |||||
| RALA (WEARLARALARALARHLARALARALRACEA) | mRNA: GFP mRNA: ovalbumin | DC2.4 cells | 89(N/P 5); 91(N/P 10) | 14.6(N/P 5); 26.3(N/P 10) | [65] |
| MPG-8-cholesterol (MPG-8: β-AFLGWLGAWGTMGWSPKKKRK-Cya) | siRNA: Cyc-B1 | HS68; HeLa; PC-3; MCF-7; SCK3-Her2 cells | 120 ± 50 | 16 ± 3 | [68] |
| (CRR)2KRRC and (CHH)2KHHC cross-linked peptide | pDNA: p53 | NIH3T3 cells; HeLa cells | ~164; ~172 | ~30; ~20 | [69] |
| Virus-like particle (VLP) (Vesicular stomatitis virus and Archeoglobus fulgidus-based) | mRNA: GFP | HEK293T cells; THP-1 cells; Human iPS cells | N/A | N/A | [76] |
| VLP (neurotropic JC polyomavirus: JCPyV)) | pDNA: suicide gene (HSV-TK) | U87-MG cells | N/A | N/A | [77] |
| VLP (JCPyV)) | pDNA: suicide gene (HSV-TK) | Toledo and HT cells; SU-DHL-2 cells | N/A | N/A | [78] |
| VLP (JCPyV)) | pDNA: suicide gene (pSPB-tk) | A549 cell; H460 cell | N/A | N/A | [79] |
| VLP (JCPyV)) | pDNA: suicide gene, (HSV-TK) | COLO-320 HSR cell | N/A | N/A | [80] |
| Self-assembled peptide: K3C6SPD (KKKC6-WLVFFAQQ-GSPD) | pDNA: GFP | Hek293 cells | ~70 | 25 | [81,82] |
| K12-(GAGAGAGQ)10-407-amino-acid hydrophilic random coil | mRNA: GFP and luciferase; pDNA: YFP | Hek293 cells; HeLa cells | ~150 (average length) | -5 | [75,83] |
| Spermine-Coiled-coil peptide-PEG (Coiled coil peptide: REGVAKALRAVANALHYNASALEEVADALQKVKM) | N/A | N/A | N/A | N/A | [84] |
| Glucose-GSGSGSKKKKKKKKGGSGGSWKWEWKWEWKWEWG | siRNA: GFP | HeLa cells | ~70 | ~0 | [85] |
| CPP-based polyplexes shelled with polysaccharide (CPP: RRRRRRRR) | pDNA: luciferase | HEK293 T; Cos7 cells | Able to be modified | Able to be modified | [100] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, R.; Feng, R.; Chau, Y. Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers. Life 2019, 9, 59. https://doi.org/10.3390/life9030059
Ni R, Feng R, Chau Y. Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers. Life. 2019; 9(3):59. https://doi.org/10.3390/life9030059
Chicago/Turabian StyleNi, Rong, Ruilu Feng, and Ying Chau. 2019. "Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers" Life 9, no. 3: 59. https://doi.org/10.3390/life9030059