Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study
Abstract
1. Introduction
1.1. Stroke Statistics, Causes of Stroke, and Need for Screening
1.2. Importance of Imaging Modalities and Plaque Quantification
1.3. Brief Background of AI Literature
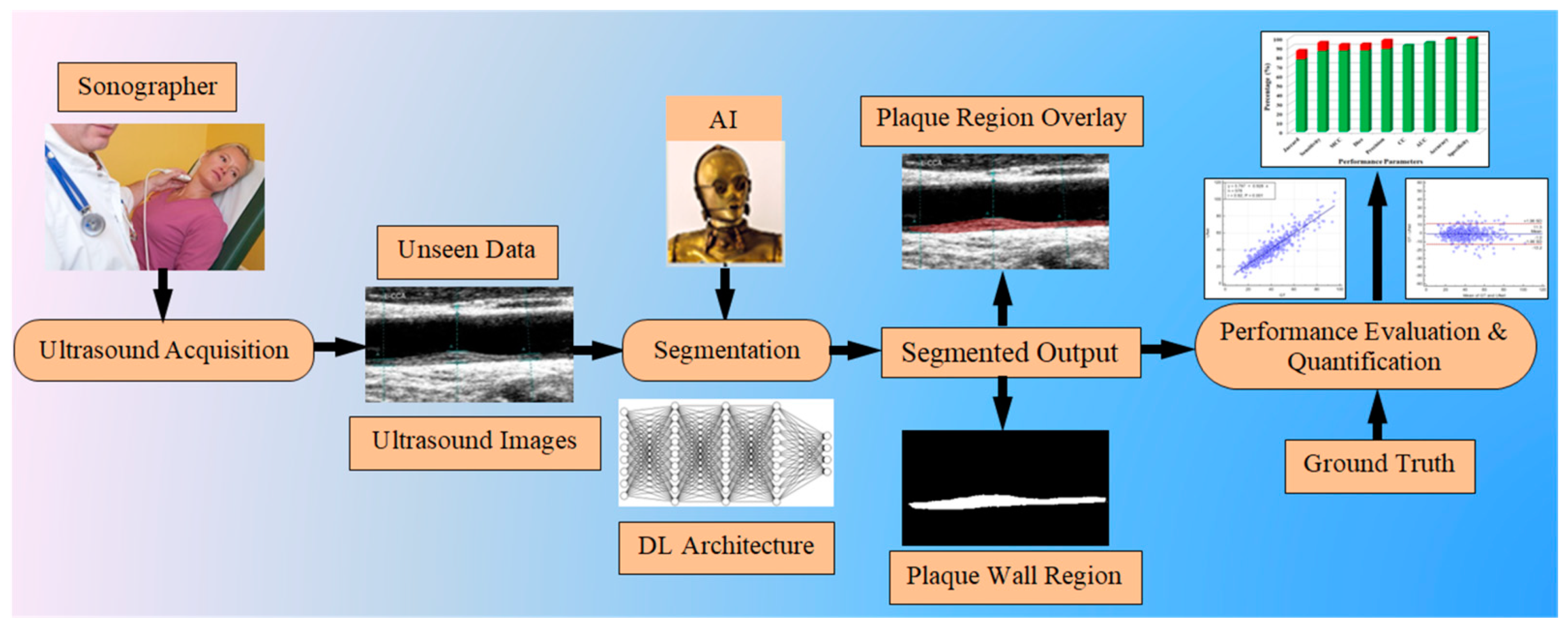
1.4. Motivation, Hypothesis of Unseen AI, and Concept of Global Segmentation System
1.5. Layout of This Study
2. Methodology
2.1. Patient Demographics, Data Collection, and Data Preparation
2.1.1. Patient Demographics for the First Group: Japanese Cohort
2.1.2. Patient Demographics for the Second Group: Hong Kong Cohort
2.1.3. Data Acquisition and Ultrasound Imaging for the Two Ethnic Groups
2.1.4. Ground-Truth Data Preparation
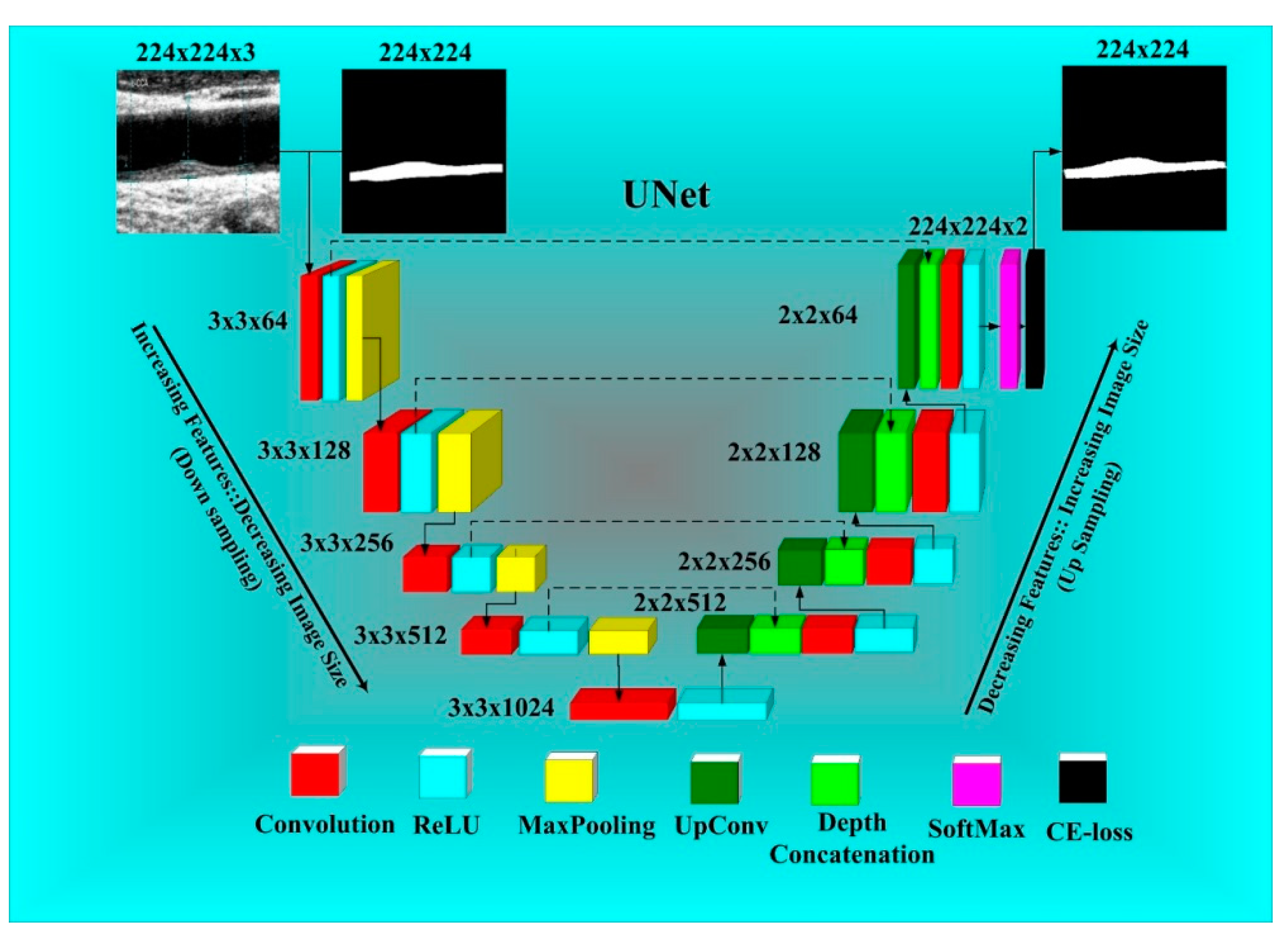
2.2. UNet-Based Deep Learning Architecture
2.3. Experimental Protocol
2.3.1. Unseen AI Data Experiments
2.3.2. Seen AI Data Experiments
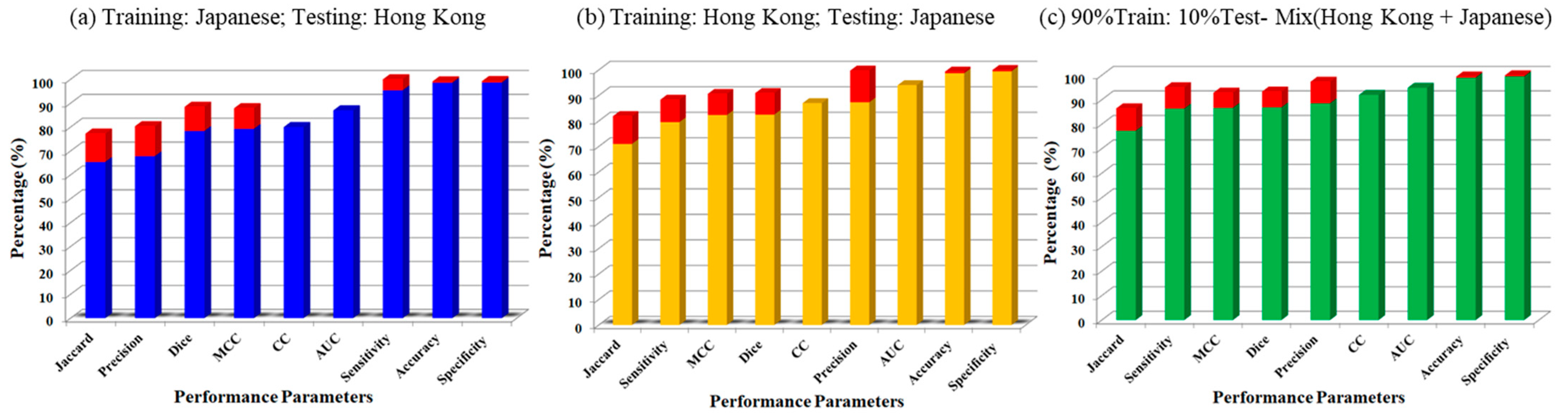
3. Results
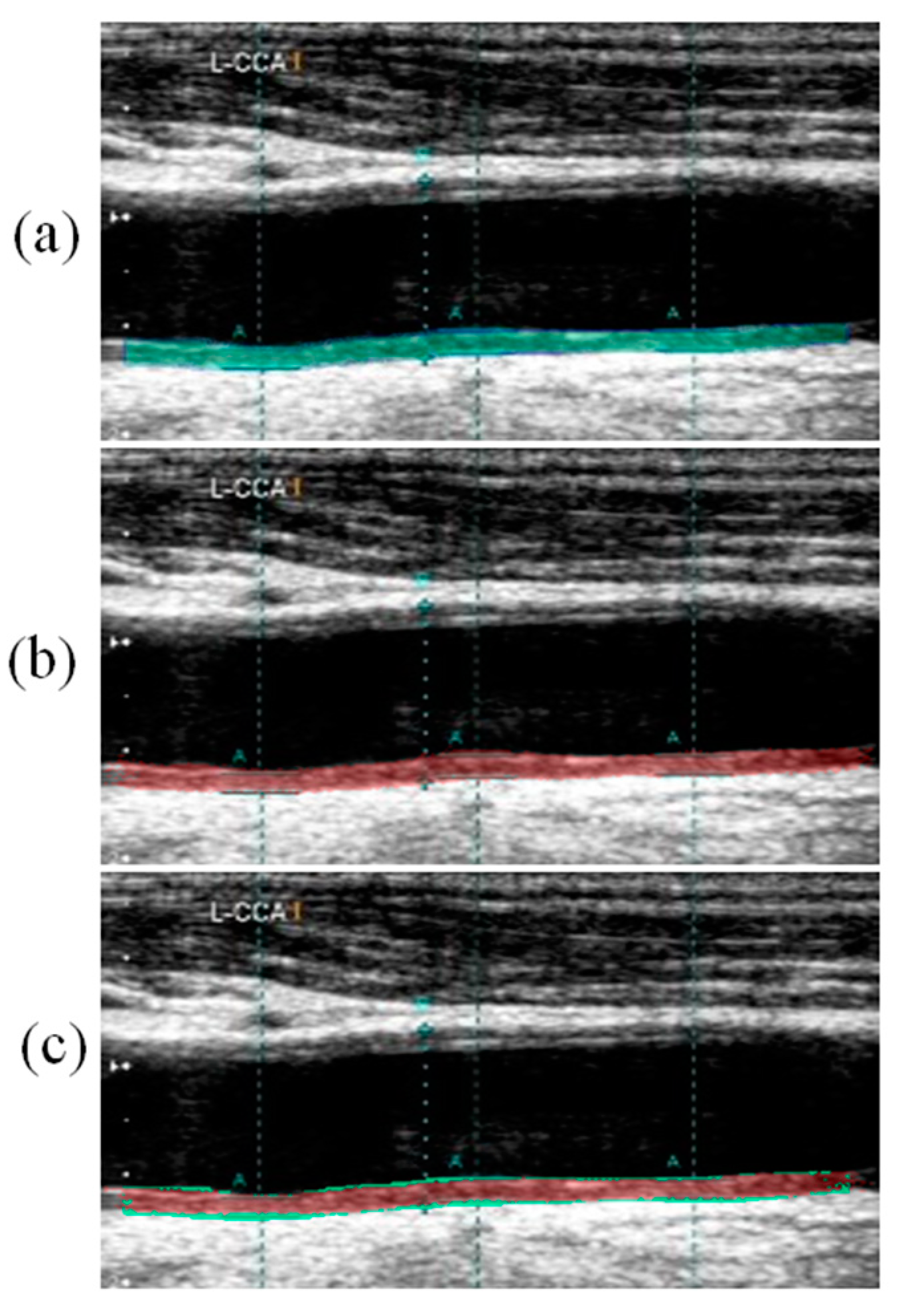
Visual Segmentation Results
4. Performance Evaluation
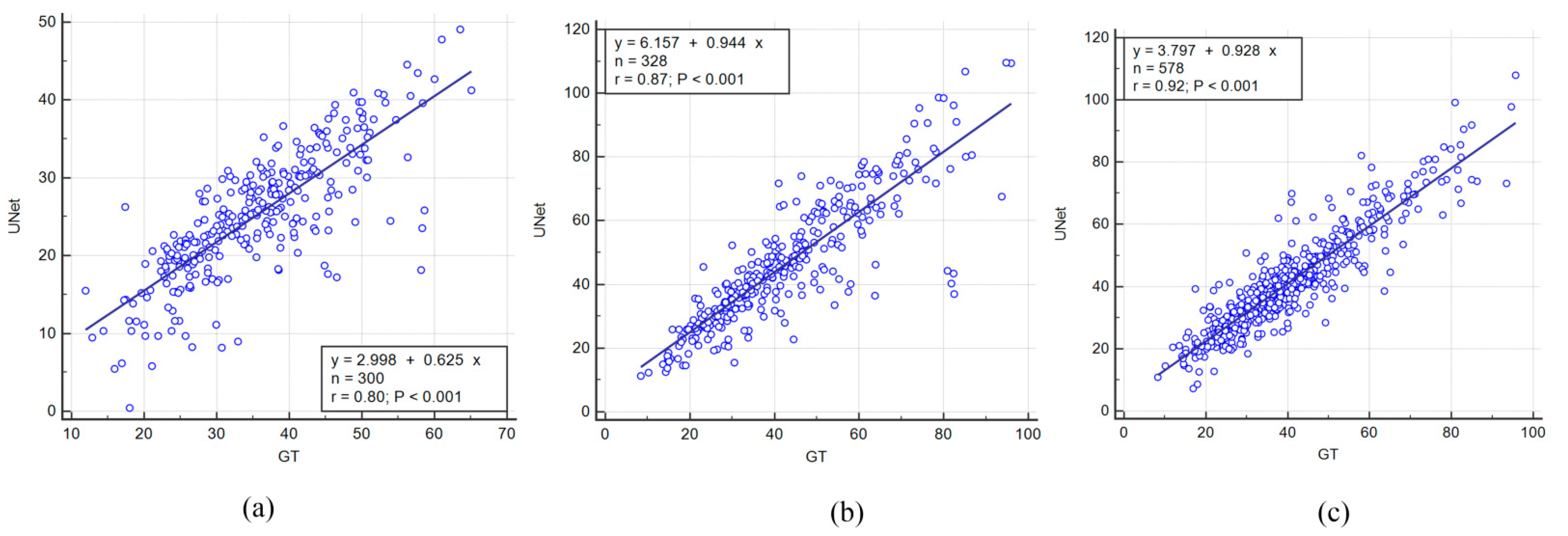
4.1. Correlation between AI Models and Ground Truth
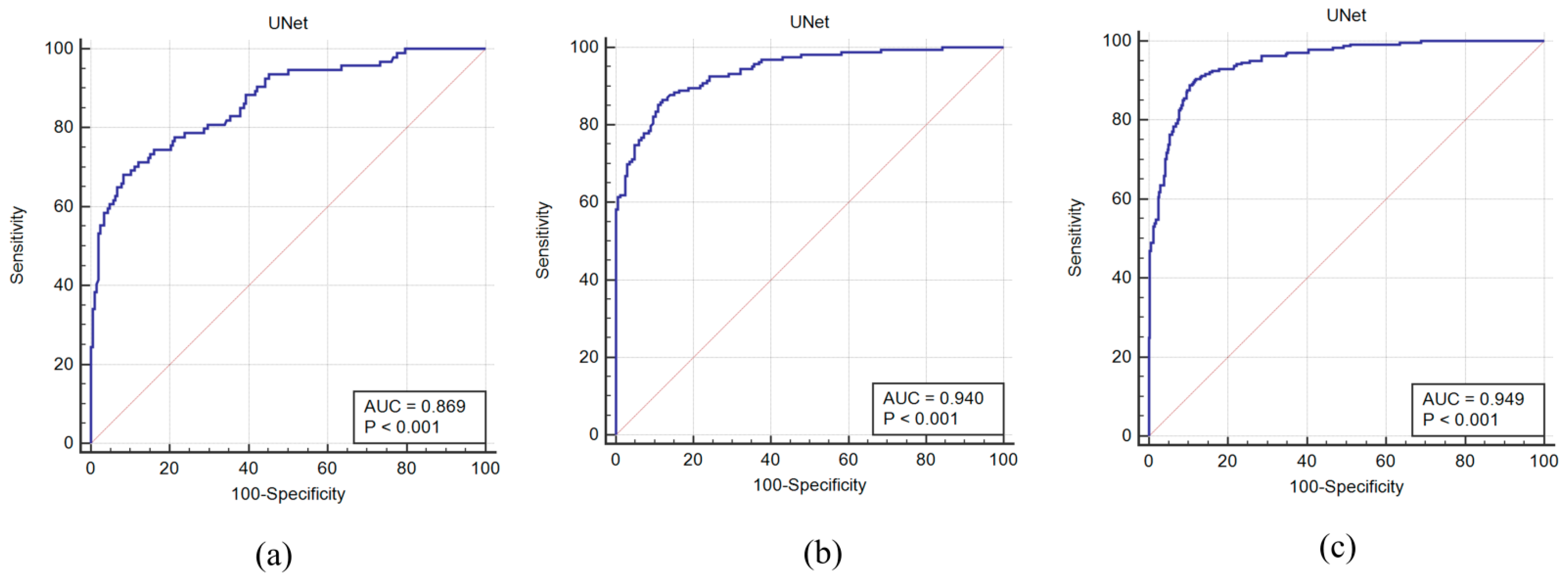
4.2. Receiver Operating Characteristics and AUC
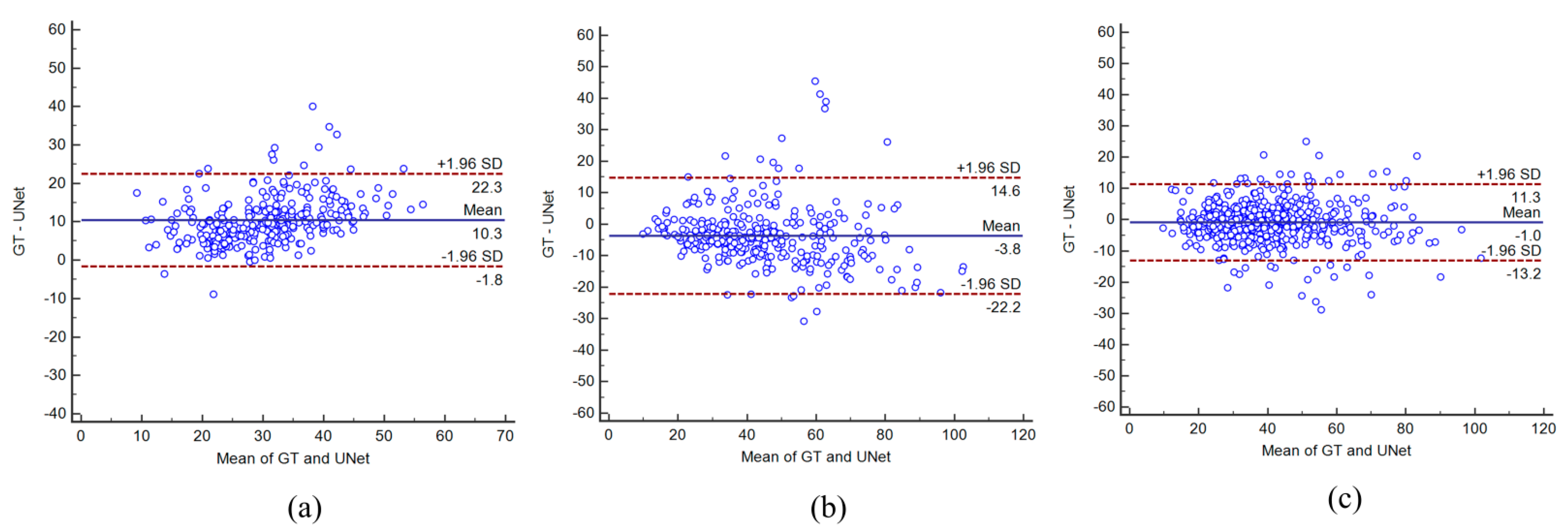
4.3. Bland–Altman Plots
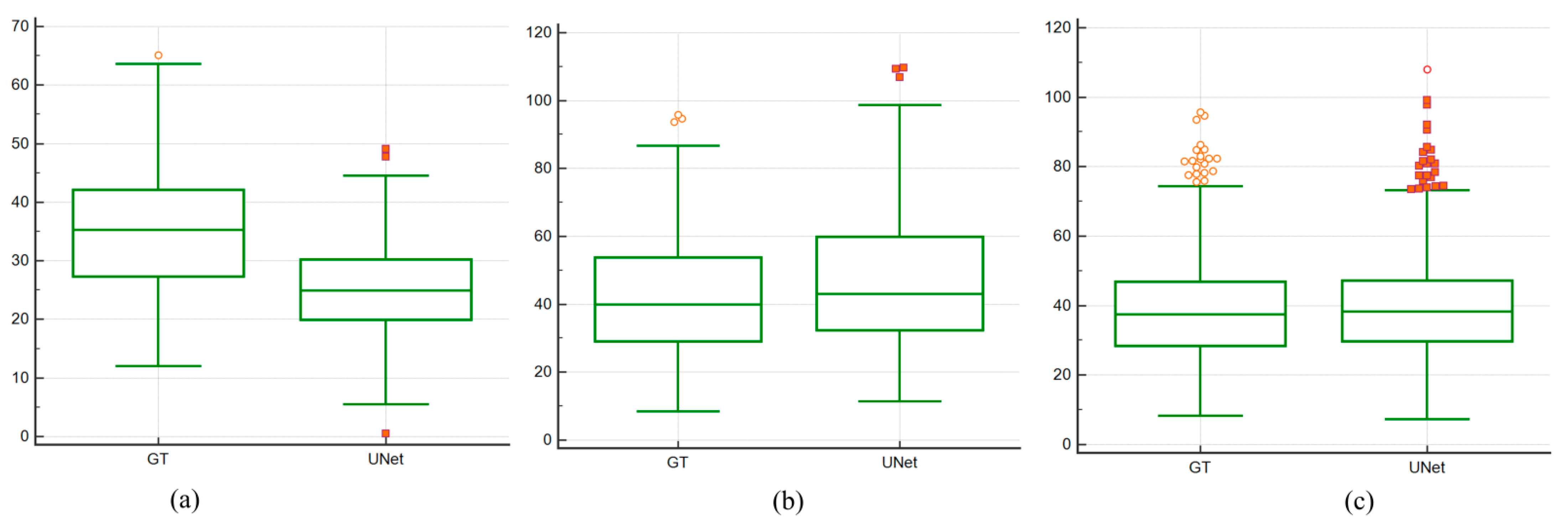
4.4. Paired Sample t-Test and ANOVA Test
4.5. Figure of Merit
5. Discussion
5.1. Benchmarking
5.2. Short Note on Image Quality
5.3. Strength, Limitations and Future Extensions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Centers of Disease Control and Prevention. U.S. Department of Health & Human Services; Centers of Disease Control and Prevention: Atlanta, GA, USA, 2018.
- Suri, J.S.; Kathuria, C.; Molinari, F. Atherosclerosis Disease Management; Springer Inc.: New York, NY, USA, 2010; p. 944. [Google Scholar]
- Libby, P.; Buring, J.; Badimon, L.; Hansson, G.; Deanfield, J.; Bittencourt, M.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56. [Google Scholar] [CrossRef]
- Patel, A.; Suri, H.S.; Singh, J.; Kumar, D.; Shafique, S.; Nicolaides, A.; Jain, S.K.; Saba, L.; Gupta, A.; Laird, J.R.; et al. A Review on Atherosclerotic Biology, Wall Stiffness, Physics of Elasticity, and Its Ultrasound-Based Measurement. Curr. Atheroscler. Rep. 2016, 18, 83. [Google Scholar] [CrossRef]
- Viswanathan, V.; Jamthikar, A.D.; Gupta, D.; Shanu, N.; Puvvula, A.; Khanna, N.N.; Saba, L.; Omerzum, T.; Viskovic, K.; Mavrogeni, S.; et al. Low-Cost Preventive Screening Using Carotid Ultrasound in Patients with Diabetes. Front. Biosci. 2020, 25, 1132–1171. [Google Scholar]
- Porcu, M.; Mannelli, L.; Melis, M.; Suri, J.S.; Gerosa, C.; Cerrone, G.; DeFazio, G.; Faa, G.; Saba, L. Carotid Plaque Imaging Profiling in Subjects with Risk Factors (Diabetes and Hypertension). Cardiovasc. Diagn. Ther. 2020, 10, 1005–1018. [Google Scholar] [CrossRef]
- Jamthikar, A.D.; Puvvula, A.; Gupta, D.; Johri, A.M.; Nambi, V.; Khanna, N.N.; Saba, L.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; et al. Cardiovascular Disease and Stroke Risk Assessment in Patients with Chronic Kidney Disease Using Integration of Estimated Glomerular Filtration Rate, Ultrasonic Image Phenotypes, and Artificial Intelligence: A Narrative Review. Int. Angiol. 2020, 40, 150–164. [Google Scholar] [CrossRef]
- Corrias, G.; Cocco, D.; Suri, J.S.; Meloni, L.; Cademartiri, F.; Saba, L. Heart Applications of 4d Flow. Cardiovasc. Diagn. Ther. 2020, 10, 1140–1149. [Google Scholar] [CrossRef]
- Suri, J.S.; Laxminarayan, S. Angiography and Plaque Imaging: Advanced Segmentation Techniques; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Biswas, M.; Saba, L.; Chakrabartty, S.; Khanna, N.N.; Song, H.; Suri, H.S.; Sfikakis, P.P.; Mavrogeni, S.; Viskovic, K.; Laird, J.R.; et al. Two-Stage Artificial Intelligence Model for Jointly Measurement of Atherosclerotic Wall Thickness and Plaque Burden in Carotid Ultrasound: A Screening Tool for Cardiovascular/Stroke Risk Assessment. Comput. Biol. Med. 2020, 123, 103847. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Suri, J.S. Automatic Vessel Indentification for Angiographic Screening. U.S. Patent 6845260B2, 18 January 2005. [Google Scholar]
- Saba, L.; Suri, J.S. Multi-Detector Ct Imaging: Principles, Head, Neck, and Vascular Systems; CRC Press: Boca Raton, FL, USA, 2013; Volume 1. [Google Scholar]
- Seabra, J.; Sanches, J. Ultrasound Imaging: Advances and Applications; Springer: New York, NY, USA, 2012. [Google Scholar]
- Suri, J.S. Advances in Diagnostic and Therapeutic Ultrasound Imaging; Artech House: London, UK, 2008. [Google Scholar]
- Molinari, F.; Meiburger, K.M.; Saba, L.; Acharya, U.R.; Ledda, G.; Zeng, G.; Ho, S.Y.S.; Ahuja, A.T.; Ho, S.C.; Nicolaides, A.; et al. Ultrasound IMT Measurement on a Multi-Ethnic and Multi-Institutional Database: Our Review and Experience Using Four Fully Automated and One Semi-Automated Methods. Comput. Methods Programs Biomed. 2012, 108, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Meiburger, K.M.; Molinari, F.; Ledda, G.; Anzidei, M.; Acharya, U.R.; Zeng, G.; Shafique, S.; Nicolaides, A.; Suri, J.S. Carotid Imt Variability (Imtv) and Its Validation in Symptomatic Versus Asymptomatic Italian Population: Can This Be a Useful Index for Studying Symptomaticity? Echocardiography 2012, 29, 1111–1119. [Google Scholar] [CrossRef]
- Lucatelli, P.; Raz, E.; Saba, L.; Argiolas, G.M.; Montisci, R.; Wintermark, M.; King, K.S.; Molinari, F.; Ikeda, N.; Siotto, P.; et al. Relationship between Leukoaraiosis, Carotid Intima-Media Thickness and Intima-Media Thickness Variability: Preliminary Results. Eur. Radiol. 2016, 26, 4423–4431. [Google Scholar] [CrossRef] [PubMed]
- Johri, A.M.; Lajkosz, K.A.; Grubic, N.; Islam, S.; Li, T.Y.; Simpson, C.S.; Ewart, P.; Suri, J.S.; Hétu, M.-F. Maximum Plaque Height in Carotid Ultrasound Predicts Cardiovascular Disease Outcomes: A Population-Based Validation Study of the American Society of Echocardiography’s Grade II–III Plaque Characterization and Protocol. Int. J. Cardiovasc. Imaging 2021, 37, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Puvvula, A.; Jamthikar, A.D.; Gupta, D.; Khanna, N.N.; Porcu, M.; Saba, L.; Viskovic, K.; Ajuluchukwu, J.N.A.; Gupta, A.; Mavrogeni, S.; et al. Morphological Carotid Plaque Area Is Associated with Glomerular Filtration Rate: A Study of South Asian Indian Patients With Diabetes and Chronic Kidney Disease. Angiology 2020, 71, 520–535. [Google Scholar] [CrossRef]
- Kotsis, V.; Jamthikar, A.D.; Araki, T.; Gupta, D.; Laird, J.R.; Giannopoulos, A.A.; Saba, L.; Suri, H.S.; Mavrogeni, S.; Kitas, G.D.; et al. Echolucency-Based Phenotype in Carotid Atherosclerosis Disease for Risk Stratification of Diabetes Patients. Diabetes Res. Clin. Pract. 2018, 143, 322–331. [Google Scholar] [CrossRef]
- Cuadrado-Godia, E.; Maniruzzaman; Araki, T.; Puvvula, A.; Rahman, J.; Saba, L.; Suri, H.S.; Gupta, A.; Banchhor, S.K.; Teji, J.S.; et al. Morphologic TPA (mTPA) and Composite Risk Score for Moderate Carotid Atherosclerotic Plaque Is Strongly Associated with Hba1c in Diabetes Cohort. Comput. Biol. Med. 2018, 101, 128–145. [Google Scholar] [CrossRef]
- Kumar, P.K.; Araki, T.; Rajan, J.; Laird, J.R.; Nicolaides, A.; Suri, J.S. State-of-the-Art Review on Automated Lumen and Adventitial Border Delineation and Its Measurements in Carotid Ultrasound. Comput. Methods Programs Biomed. 2018, 163, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.K.; Araki, T.; Rajan, J.; Saba, L.; Lavra, F.; Ikeda, N.; Sharma, A.M.; Shafique, S.; Nicolaides, A.; Laird, J.R.; et al. Accurate Lumen Diameter Measurement in Curved Vessels in Carotid Ultrasound: An Iterative Scale-Space and Spatial Transformation Approach. Med. Biol. Eng. Comput. 2017, 55, 1415–1434. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Banchhor, S.K.; Londhe, N.D.; Araki, T.; Laird, J.R.; Gupta, A.; Nicolaides, A.; Suri, J.S. Web-Based Accurate Measurements of Carotid Lumen Diameter and Stenosis Severity: An Ultrasound-Based Clinical Tool for Stroke Risk Assessment During Multicenter Clinical Trials. Comput. Biol. Med. 2017, 91, 306–317. [Google Scholar] [CrossRef]
- Saba, L.; Jamthikar, A.; Gupta, D.; Khanna, N.N.; Viskovic, K.; Suri, H.S.; Gupta, A.; Mavrogeni, S.; Turk, M.; Laird, J.R.; et al. Global Perspective on Carotid Intima-Media Thickness and Plaque: Should the Current Measurement Guidelines Be Revisited? Int. Angiol. 2020, 38, 451–465. [Google Scholar] [CrossRef]
- Saba, L.; Agarwal, N.; Cau, R.; Gerosa, C.; Sanfilippo, R.; Porcu, M.; Montisci, R.; Cerrone, G.; Qi, Y.; Balestrieri, A.; et al. Review of Imaging Biomarkers for the Vulnerable Carotid Plaque. JVS Vasc. Sci. 2021, 2, 149–158. [Google Scholar] [CrossRef]
- Saba, L.; Banchhor, S.K.; Araki, T.; Viskovic, K.; Londhe, N.D.; Laird, J.R.; Suri, H.S.; Suri, J.S. Intra- and Inter-Operator Reproducibility of Automated Cloud-Based Carotid Lumen Diameter Ultrasound Measurement. Indian Heart J. 2018, 70, 649–664. [Google Scholar] [CrossRef]
- Saba, L.; Banchhor, S.K.; Araki, T.; Suri, H.S.; Londhe, N.D.; Laird, J.R.; Višković, K.; Suri, J.S. Intra- and Inter-Operator Reproducibility Analysis of Automated Cloud-based Carotid Intima Media Thickness Ultrasound Measurement. J. Clin. Diagn. Res. 2018, 12, 1–7. [Google Scholar] [CrossRef]
- Molinari, F.; Zeng, G.; Suri, J.S. Intima-Media Thickness: Setting a Standard for a Completely Automated Method of Ultrasound Measurement. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2010, 57, 1112–1124. [Google Scholar] [CrossRef]
- Molinari, F.; Pattichis, C.S.; Zeng, G.; Saba, L.; Acharya, U.R.; Sanfilippo, R.; Nicolaides, A.; Suri, J.S. Completely Automated Multiresolution Edge Snapper—A New Technique for an Accurate Carotid Ultrasound Imt Measurement: Clinical Validation and Benchmarking on a Multi-Institutional Database. IEEE Trans. Image Process. 2012, 21, 1211–1222. [Google Scholar] [CrossRef]
- Ayman El-Baz, J.S.S. Artificial Intelligence in Cancer Diagnosis and Prognosis. In Ipem-Iop Series in Physics and Engineering in Medicine and Biology; IOP Press: Bristol, UK, 2022; Volume 1. [Google Scholar]
- Rangayyan, R.M.; Suri, J.S. Recent Advances in Breast Imaging, Mammography, and Computer-Aided Diagnosis of Breast Cancer; SPIE Publications: Bellingham, WA, USA, 2006. [Google Scholar]
- Saba, L.; Jain, P.; Suri, H.S.; Ikeda, N.; Araki, T.; Singh, B.K.; Nicolaides, A.; Shafique, S.; Gupta, A.; Laird, J.R.; et al. Plaque Tissue Morphology-Based Stroke Risk Stratification Using Carotid Ultrasound: A Polling-Based Pca Learning Paradigm. J. Med. Syst. 2017, 41, 98. [Google Scholar] [CrossRef]
- Araki, T.; Jain, P.; Suri, H.S.; Londhe, N.D.; Ikeda, N.; El-Baz, A.; Shrivastava, V.; Saba, L.; Nicolaides, A.; Shafique, S.; et al. Stroke Risk Stratification and Its Validation Using Ultrasonic Echolucent Carotid Wall Plaque Morphology: A Machine Learning Paradigm. Comput. Biol. Med. 2017, 80, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Maniruzzaman, M.; Rahman, J.; Hasan, A.-M.; Suri, H.S.; Abedin, M.; El-Baz, A.; Suri, J.S. Accurate Diabetes Risk Stratification Using Machine Learning: Role of Missing Value and Outliers. J. Med. Syst. 2018, 42, 92. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Krishnan, M.M.R.; Krishnananda, N.; Ranjan, S.; Umesh, P.; Suri, J.S. Automated Classification of Patients with Coronary Artery Disease Using Grayscale Features from Left Ventricle Echocardiographic Images. Comput. Methods Programs Biomed. 2013, 112, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Sharma, N.; Giannopoulos, A.A.; Saba, L.; Nicolaides, A.; Suri, J.S. Hybrid Deep Learning Segmentation Models for Atherosclerotic Plaque in Internal Carotid Artery B-Mode Ultrasound. Comput. Biol. Med. 2021, 136, 104721. [Google Scholar] [CrossRef]
- Biswas, M.; Kuppili, V.; Edla, D.R.; Suri, H.S.; Saba, L.; Marinhoe, R.T.; Sanches, J.; Suri, J.S. Symtosis: A Liver Ultrasound Tissue Characterization and Risk Stratification in Optimized Deep Learning Paradigm. Comput. Methods Programs Biomed. 2018, 155, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Gupta, S.; Bhavsar, A.; Nigam, A.; Sharma, N. Localization of Common Carotid Artery Transverse Section in B-Mode Ultrasound Images Using Faster Rcnn: A Deep Learning Approach. Med. Biol. Eng. Comput. 2020, 58, 471–482. [Google Scholar] [CrossRef]
- Saba, L.; Biswas, M.; Suri, H.S.; Viskovic, K.; Laird, J.R.; Cuadrado-Godia, E.; Nicolaides, A.; Khanna, N.N.; Viswanathan, V.; Suri, J.S. Ultrasound-Based Carotid Stenosis Measurement and Risk Stratification in Diabetic Cohort: A Deep Learning Paradigm. Cardiovasc. Diagn. Ther. 2019, 9, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Johri, A.M.; Sharma, A.M.; Kolluri, R.; Bhatt, D.L.; Nicolaides, A.; Suri, J.S. Ultrasound-Based Internal Carotid Artery Plaque Characterization Using Deep Learning Paradigm on a Supercomputer: A Cardiovascular Disease/Stroke Risk Assessment System. Int. J. Cardiovasc. Imaging 2021, 37, 1511–1528. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. Multimodality Carotid Plaque Tissue Characterization and Classification in the Artificial Intelligence Paradigm: A Narrative Review for Stroke Application. Ann. Transl. Med. 2021, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Saba, L.; Gupta, S.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; Sfikakis, P.P.; et al. Wilson Disease Tissue Classification and Characterization Using Seven Artificial Intelligence Models Embedded with 3D Optimization Paradigm on a Weak Training Brain Magnetic Resonance Imaging Datasets: A Supercomputer Application. Med. Biol. Eng. Comput. 2021, 59, 511–533. [Google Scholar] [CrossRef]
- Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; Nicolaides, A.; et al. State-of-the-Art Review on Deep Learning in Medical Imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar]
- Saba, L.; Biswas, M.; Kuppili, V.; Godia, E.C.; Suri, H.S.; Edla, D.R.; Omerzu, T.; Laird, J.R.; Khanna, N.N.; Mavrogeni, S.; et al. The Present and Future of Deep Learning in Radiology. Eur. J. Radiol. 2019, 114, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Saxena, S.; Nayak, G.K.; Saba, L.; Sharma, N.; Suri, J.S. Artificial Intelligence-Based Hybrid Deep Learning Models for Image Classification: The First Narrative Review. Comput. Biol. Med. 2021, 137, 104803. [Google Scholar] [CrossRef]
- Zhou, R.; Guo, F.; Azarpazhooh, M.R.; Hashemi, S.; Cheng, X.; Spence, J.D.; Ding, M.; Fenster, A. Deep Learning-Based Measurement of Total Plaque Area in B-Mode Ultrasound Images. IEEE J. Biomed. Health Inform. 2021, 25, 2967–2977. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M.; et al. Systematic Review of Artificial Intelligence in Acute Respiratory Distress Syndrome for COVID-19 Lung Patients: A Biomedical Imaging Perspective. IEEE J. Biomed. Health Inform. 2021, 25, 4128–4139. [Google Scholar] [CrossRef]
- Molinari, F.; Meiburger, K.M.; Saba, L.; Acharya, U.R.; Famiglietti, L.; Georgiou, N.; Nicolaides, A.; Mamidi, R.S.; Kuper, H.; Suri, J.S. Automated Carotid Imt Measurement and Its Validation in Low Contrast Ultrasound Database of 885 Patient Indian Population Epidemiological Study: Results of Atheroedge® Software. In Multi-Modality Atherosclerosis Imaging and Diagnosis; Springer: Berlin/Heidelberg, Germany, 2014; pp. 209–219. [Google Scholar]
- Saba, L.; Banchhor, S.K.; Suri, H.S.; Londhe, N.D.; Araki, T.; Ikeda, N.; Viskovic, K.; Shafique, S.; Laird, J.R.; Gupta, A.; et al. Accurate Cloud-Based Smart IMT Measurement, Its Validation and Stroke Risk Stratification in Carotid Ultrasound: A Web-Based Point-of-Care Tool for Multicenter Clinical Trial. Comput. Biol. Med. 2016, 75, 217–234. [Google Scholar] [CrossRef]
- Araki, T.; Aimbe, F.; Kumar, P.K.; Suri, H.S.; Ikeda, N.; Gupta, A.; Saba, L.; Rajan, J.; Lavra, F.; Sharma, A.M.; et al. Two Automated Techniques for Carotid Lumen Diameter Measurement: Regional Versus Boundary Approaches. J. Med. Syst. 2016, 40, 1–19. [Google Scholar] [CrossRef]
- Molinari, F.; Liboni, W.; Giustetto, P.; Badalamenti, S.; Suri, J.S. Automatic Computer-Based Tracings (Act) in Longitudinal 2-D Ultrasound Images Using Different Scanners. J. Mech. Med. Biol. 2009, 9, 481–505. [Google Scholar] [CrossRef]
- Molinari, F.; Zeng, G.; Suri, J.S. An Integrated Approach to Computer-Based Automated Tracing and Its Validation for 200 Common Carotid Arterial Wall Ultrasound Images: A New Technique. J. Ultrasound Med. 2010, 29, 399–418. [Google Scholar] [CrossRef]
- Molinari, F.; Liboni, W.; Pantziaris, M.; Suri, J.S. CALSFOAM-Completed Automated Local Statistics Based First Order Absolute Moment for Carotid Wall Recognition, Segmentation and IMT Measurement: Validation and Benchmarking on a 300 Patient Database. Int. Angiol. 2011, 30, 227–241. [Google Scholar] [PubMed]
- Meiburger, K.M.; Molinari, F.; Acharya, U.R.; Saba, L.; Rodrigues, P.; Liboni, W.; Nicolaides, A.; Suri, J.S. Automated Carotid Artery Intima Layer Regional Segmentation. Phys. Med. Biol. 2011, 56, 4073–4090. [Google Scholar] [CrossRef] [PubMed]
- Molinari, F.; Meiburger, K.M.; Acharya, U.R.; Zeng, G.; Rodrigues, P.S.; Saba, L.; Nicolaides, A.; Suri, J.S. Cares 3.0: A Two Stage System Combining Feature-Based Recognition and Edge-Based Segmentation for CIMT Measurement on a Multi-Institutional Ultrasound Database of 300 Images. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011. [Google Scholar]
- Molinari, F.; Meiburger, K.M.; Zeng, G.; Acharya, U.R.; Liboni, W.; Nicolaides, A.; Suri, J.S. Carotid Artery Recognition System: A Comparison of Three Automated Paradigms for Ultrasound Images. Med. Phys. 2011, 39, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Molinari, F.; Acharya, U.R.; Zeng, G.; Meiburger, K.M.; Suri, J.S. Completely Automated Robust Edge Snapper for Carotid Ultrasound IMT Measurement on a Multi-Institutional Database of 300 Images. Med. Biol. Eng. Comput. 2011, 49, 935–945. [Google Scholar] [CrossRef]
- Molinari, F.; Zeng, G.; Suri, J.S. A State of the Art Review on Intima–Media Thickness (IMT) Measurement and Wall Segmentation Techniques for Carotid Ultrasound. Comput. Methods Programs Biomed. 2010, 100, 201–221. [Google Scholar] [CrossRef]
- Saba, L.; Gao, H.; Raz, E.; Sree, S.V.; Mannelli, L.; Tallapally, N.; Molinari, F.; Bassareo, P.P.; Acharya, U.R.; Poppert, H.; et al. Semiautomated Analysis of Carotid Artery Wall Thickness in MRI. J. Magn. Reson. Imaging 2014, 39, 1457–1467. [Google Scholar] [CrossRef]
- Ikeda, N.; Gupta, A.; Dey, N.; Bose, S.; Shafique, S.; Arak, T.; Godia, E.C.; Saba, L.; Laird, J.R.; Nicolaides, A.; et al. Improved Correlation between Carotid and Coronary Atherosclerosis Syntax Score Using Automated Ultrasound Carotid Bulb Plaque IMT Measurement. Ultrasound Med. Biol. 2015, 41, 1247–1262. [Google Scholar] [CrossRef]
- Ikeda, N.; Dey, N.; Sharma, A.; Gupta, A.; Bose, S.; Acharjee, S.; Shafique, S.; Cuadrado-Godia, E.; Araki, T.; Saba, L.; et al. Automated Segmental-IMT Measurement in Thin/Thick Plaque with Bulb Presence in Carotid Ultrasound from Multiple Scanners: Stroke Risk Assessment. Comput. Methods Programs Biomed. 2017, 141, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.; Korcarz, C.E.; Gepner, A.D.; Kaufman, J.D.; Post, W.; Tracy, R.; Gassett, A.J.; Ma, N.; McClelland, R.L.; Stein, J.H. Ultrasound Carotid Plaque Features, Cardiovascular Disease Risk Factors and Events: The Multi-Ethnic Study of Atherosclerosis. Atherosclerosis 2018, 276, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Jamthikar, A.; Gupta, D.; Cuadrado-Godia, E.; Puvvula, A.; Khanna, N.N.; Saba, L.; Viskovic, K.; Mavrogeni, S.; Turk, M.; Laird, J.R. Ultrasound-Based Stroke/Cardiovascular Risk Stratification Using Framingham Risk Score and Ascvd Risk Score Based on “Integrated Vascular Age” Instead of “Chronological Age”: A Multi-Ethnic Study of Asian Indian, Caucasian, and Japanese Cohorts. Cardiovasc. Diagn. Ther. 2020, 10, 939. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.K.; Darshan, P.; Kumar, S.; Ravindra, R.; Rajan, J.; Saba, L.; Suri, J.S. Magnetic Resonance Image Denoising Using Nonlocal Maximum Likelihood Paradigm in DCT-Framework. Int. J. Imaging Syst. Technol. 2015, 25, 256–264. [Google Scholar] [CrossRef]
- Saba, L.; Di Martino, M.; Siotto, P.; Anzidei, M.; Argiolas, G.M.; Porcu, M.; Suri, J.S.; Wintermark, M. Radiation Dose and Image Quality of Computed Tomography of the Supra-Aortic Arteries: A Comparison between Single-Source and Dual-Source CT Scanners. J. Neuroradiol. 2018, 45, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Wei, L.; Turlapati, R.; Suri, J.S. Improvement in C-Arm Acquired DSA Image Quality via Combined Effect of Inverse Consistent Motion Correction and Nonlinear Normalization. In Proceedings of the 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007. [Google Scholar]
- Suri, J.; Wu, D.; Gao, J.; Singh, S.; Laxminarayan, S. A Comparison of State-of-the-Art Diffusion Imaging Techniques for Smoothing Medical/Non-Medical Image Data. In Proceedings of the Object Recognition Supported by User Interaction for Service Robots, Quebec City, QC, Canada, 11–15 August 2002. [Google Scholar]
- Sudeep, P.; Palanisamy, P.; Rajan, J.; Baradaran, H.; Saba, L.; Gupta, A.; Suri, J.S. Speckle Reduction in Medical Ultrasound Images Using an Unbiased Non-Local Means Method. Biomed. Signal Process. Control. 2016, 28, 1–8. [Google Scholar] [CrossRef]
- Suri, J.; Liu, K.; Reden, L.; Laxminarayan, S. White and Black Blood Volumetric Angiographic Filtering: Ellipsoidal Scale-Space Approach. IEEE Trans. Inf. Technol. Biomed. 2002, 6, 142–158. [Google Scholar] [CrossRef]
- Banchhor, S.K.; Araki, T.; Londhe, N.D.; Ikeda, N.; Radeva, P.; Elbaz, A.; Saba, L.; Nicolaides, A.; Shafique, S.; Laird, J.R.; et al. Five Multiresolution-Based Calcium Volume Measurement Techniques from Coronary IVUS Videos: A Comparative Approach. Comput. Methods Programs Biomed. 2016, 134, 237–258. [Google Scholar] [CrossRef]
- Suri, J.; Liu, K.; Singh, S.; Laxminarayan, S.; Zeng, X.; Reden, L. Shape Recovery Algorithms Using Level Sets in 2-D/3-D Medical Imagery: A State-of-the-Art Review. IEEE Trans. Inf. Technol. Biomed. 2002, 6, 8–28. [Google Scholar] [CrossRef]
- Suri, J.S.; Laxminarayan, S. Pde and Level Sets; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Molinari, F.; Saba, L.; Nicolaides, A.; Suri, J.S. An Accurate and Generalized Approach to Plaque Characterization in 346 Carotid Ultrasound Scans. IEEE Trans. Instrum. Meas. 2012, 61, 1045–1053. [Google Scholar] [CrossRef]
- Saba, L.; Tallapally, N.; Gao, H.; Molinari, F.; Anzidei, M.; Piga, M.; Sanfilippo, R.; Suri, J.S. Semiautomated and Automated Algorithms for Analysis of the Carotid Artery Wall on Computed Tomography and Sonography: A Correlation Study. J. Ultrasound Med. 2013, 32, 665–674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Sharma, A.; Cuadrado-Godia, E.; Laird, J.R.; Nicolaides, A.; Suri, J.S. Deep Learning Fully Convolution Network for Lumen Characterization in Diabetic Patients Using Carotid Ultrasound: A Tool for Stroke Risk. Med. Biol. Eng. Comput. 2019, 57, 543–564. [Google Scholar] [CrossRef] [PubMed]



| Exp # | Name of Exp | Training DB | Testing DB | Training Protocol |
|---|---|---|---|---|
| Exp #1 | Unseen AI-1 (Tr: JAP, Te: HK) | Japanese 330 | Hong Kong 300 | All Japanese DB for training |
| Exp #2 | Unseen AI-2 (Tr: HK, Te: JAP) | Hong Kong 300 | Japanese 330 | All Hong Kong DB for training |
| Exp #3 | Seen AI-1; CV w/ Mixed | Japanese (330) + Hong Kong (250) | Japanese (330) + Hong Kong (250) | 10-fold cross-validation |
| Exp #4 | Seen AI-2; CV w/ JAP | Japanese (330) | Japanese (330) | 10-fold cross-validation |
| Exp #5 | Seen AI-3; CV w/ HK | Hong Kong (300) | Hong Kong (300) | 10-fold cross-validation |
| Experiment # | UNet Experiments | ACC | Sens | Spec | Prec | MCC | DSC | JI |
|---|---|---|---|---|---|---|---|---|
| Exp #1 | Unseen AI-1 (Tr: JAP, Te: HK) | 98.55 ± 0.57 | 95.41 ± 5.29 | 98.64 ± 0.62 | 67.82 ± 12.55 | 79.29 ± 8.64 | 78.38 ± 10.11 | 65.42 ± 11.84 |
| Exp #2 | Unseen AI-2 (Tr: HK, Te: JAP) | 98.67 ± 0.67 | 79.52 ± 8.84 | 99.47 ± 0.67 | 87.29 ± 12.45 | 82.29 ± 8.34 | 82.49 ± 8.44 | 70.98 ± 10.90 |
| Exp #3 | Seen AI-1 CV w/ Mixed | 99.01 ± 0.44 | 86.37 ± 8.69 | 99.52 ± 0.41 | 88.55 ± 8.82 | 86.68 ± 6.19 | 86.89 ± 6.43 | 77.34 ± 9.15 |
| Exp #4 | Seen AI-2 CV w/ JAP | 98.99 ± 0.58 | 91.25 ± 8.13 | 99.26 ± 0.64 | 81.01 ± 14.80 | 84.88 ± 9.49 | 84.65 ± 10.68 | 74.62 ± 13.54 |
| Exp #5 | Seen AI-3 CV w/ HK | 98.96 ± 0.39 | 87.27 ± 7.70 | 99.43 ± 0.42 | 86.50 ± 10.45 | 86.04 ± 7.72 | 86.29 ± 8.31 | 76.59 ± 9.96 |
| Experiment # | UNet Experiment | CC | AUC | FoM |
|---|---|---|---|---|
| Exp #1 | Unseen AI-1 (Tr: JAP, Te: HK) | 0.8 | 0.87 | 70.96 |
| Exp #2 | Unseen AI-2 (Tr: HK, Te: JAP) | 0.87 | 0.94 | 91.14 |
| Exp #3 | Seen AI-1, CV w/ Mixed | 0.92 | 0.95 | 97.57 |
| Exp #4 | Seen AI-2, CV w/ JAP | 0.87 | 0.93 | 88.89 |
| Exp #5 | Seen AI-3, CV w/ HK | 0.89 | 0.95 | 99.14 |
| #Exp | Comparison of Experiments | CC | AUC | ACC | Sens | Spec | Prec | MCC | DSC | JI |
|---|---|---|---|---|---|---|---|---|---|---|
| 3-1 | Seen AI-1 CV w/ Mixed-Unseen AI-1 (Tr: JAP, Te: HK) | 13.04 | 8.42 | 0.46 | −10.47 | 0.88 | 23.41 | 8.53 | 9.79 | 15.41 |
| ~ | √ | √ | √ | √ | ~ | √ | √ | ~ | ||
| 3-2 | Seen AI-1 CV w/ Mixed-Unseen AI-2 (Tr: HK, Te: JAP) | 5.43 | 1.05 | 0.34 | 7.93 | 0.05 | 1.42 | 5.06 | 5.06 | 8.22 |
| √ | √ | √ | √ | √ | √ | √ | √ | √ | ||
| 4-1 | Seen AI-2, CV w/ JAP-Unseen AI-1 (Tr: JAP, Te: HK) | 8.05 | 6.45 | 0.44 | −4.56 | 0.62 | 16.28 | 6.59 | 7.41 | 12.33 |
| √ | √ | √ | √ | √ | ~ | √ | √ | ~ | ||
| 4-2 | Seen AI-2, CV w/ JAP-Unseen AI-2 (Tr: HK, Te: JAP) | 0.00 | −1.08 | 0.32 | 12.85 | −0.21 | −7.75 | 3.05 | 2.55 | 4.88 |
| √ | √ | √ | ~ | √ | √ | √ | √ | √ | ||
| 5-1 | Seen AI-3, CV w/ H -Unseen AI-1 (Tr: JAP, Te: HK) | 10.11 | 8.42 | 0.41 | −9.33 | 0.79 | 21.60 | 7.85 | 9.17 | 14.58 |
| √ | √ | √ | √ | √ | ~ | √ | √ | ~ | ||
| 5-2 | Seen AI-3, CV w/ H -Unseen AI-2 (Tr: HK, Te: JAP) | 2.25 | 1.05 | 0.29 | 8.88 | −0.04 | −0.91 | 4.36 | 4.40 | 7.32 |
| √ | √ | √ | √ | √ | √ | √ | √ | √ | ||
| 3-4 | Seen AI-1 CV w/ Mixed-Seen AI-2, CV w/ JAP | 5.43 | 2.11 | 0.02 | −5.65 | 0.26 | 8.51 | 2.08 | 2.58 | 3.52 |
| √ | √ | √ | √ | √ | √ | √ | √ | √ | ||
| 3-5 | Seen AI-1 CV w/ Mixed-Seen AI-3, CV w/ HK | 3.26 | 0.00 | 0.05 | −1.04 | 0.09 | 2.32 | 0.74 | 0.69 | 0.97 |
| √ | √ | √ | √ | √ | √ | √ | √ | √ |
| Sr# | Authors and Year | Cohorts | Images | Purpose | Model |
|---|---|---|---|---|---|
| 1 | Molinari et al., 2012 [15] | Torino (n1) Nicosia (n2) Cagliari (n3) Porto (n4) Hong Kong (n5) | n1 = 200 n2 = 100 n3 = 42 n4 = 23 n5 = 300 | IMT measurement using auto and semi-auto methods | ML |
| 2 | Ikeda et al., 2013 [62] | Japanese (n1) Italy (n2) Hong Kong (n3) | n1 = 259 n2 = 98 n3 = 300 | IMT measurement in Bulb area | ML |
| 3 | Zhou et al., 2020 [47] | SPARC (n1) Chinese * (n2) | n1 = 510 n2 = 638 | Plaque area measurement in ICA and CCA images | DL |
| 4 | Carol et al., 2018 [63] | White (n1) Chinese (n2) Black (n3) Hispanic (n4) | n1 = 946 n2 = 185 n3 = 595 n4 = 479 | Carotid plaque analysis using manual method | Statistical method |
| 5 | Jamathikar et al., 2020 [64] | Japanese (n1) Asian-Indian (n2) Spanish (n3) | n1 = 404 n2 = 628 n3 = 264 | Framingham risk score-based stroke risk stratification | ML |
| 6 | Proposed method | Japanese (n1) Hong Kong (n2) | n1 = 330 n2 = 300 | Plaque area measurement in CCA images | DL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Laird, J.R.; Nicolaides, A.N.; Suri, J.S. Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study. Diagnostics 2021, 11, 2257. https://doi.org/10.3390/diagnostics11122257
Jain PK, Sharma N, Saba L, Paraskevas KI, Kalra MK, Johri A, Laird JR, Nicolaides AN, Suri JS. Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study. Diagnostics. 2021; 11(12):2257. https://doi.org/10.3390/diagnostics11122257
Chicago/Turabian StyleJain, Pankaj K., Neeraj Sharma, Luca Saba, Kosmas I. Paraskevas, Mandeep K. Kalra, Amer Johri, John R. Laird, Andrew N. Nicolaides, and Jasjit S. Suri. 2021. "Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study" Diagnostics 11, no. 12: 2257. https://doi.org/10.3390/diagnostics11122257
APA StyleJain, P. K., Sharma, N., Saba, L., Paraskevas, K. I., Kalra, M. K., Johri, A., Laird, J. R., Nicolaides, A. N., & Suri, J. S. (2021). Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study. Diagnostics, 11(12), 2257. https://doi.org/10.3390/diagnostics11122257








