Radiation Biomarkers in Large Scale Human Health Effects Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection and Blood Sampling
2.2. Ex Vivo Irradiation of Separated Lymphocytes and Whole Blood
2.3. Gamma H2AX Assay for Initial and Residual Radiation-Induced Foci
2.4. Dicentric and Micronucleus Assays
2.5. DCA Calibration Curve Construction
2.6. Statistical Methods
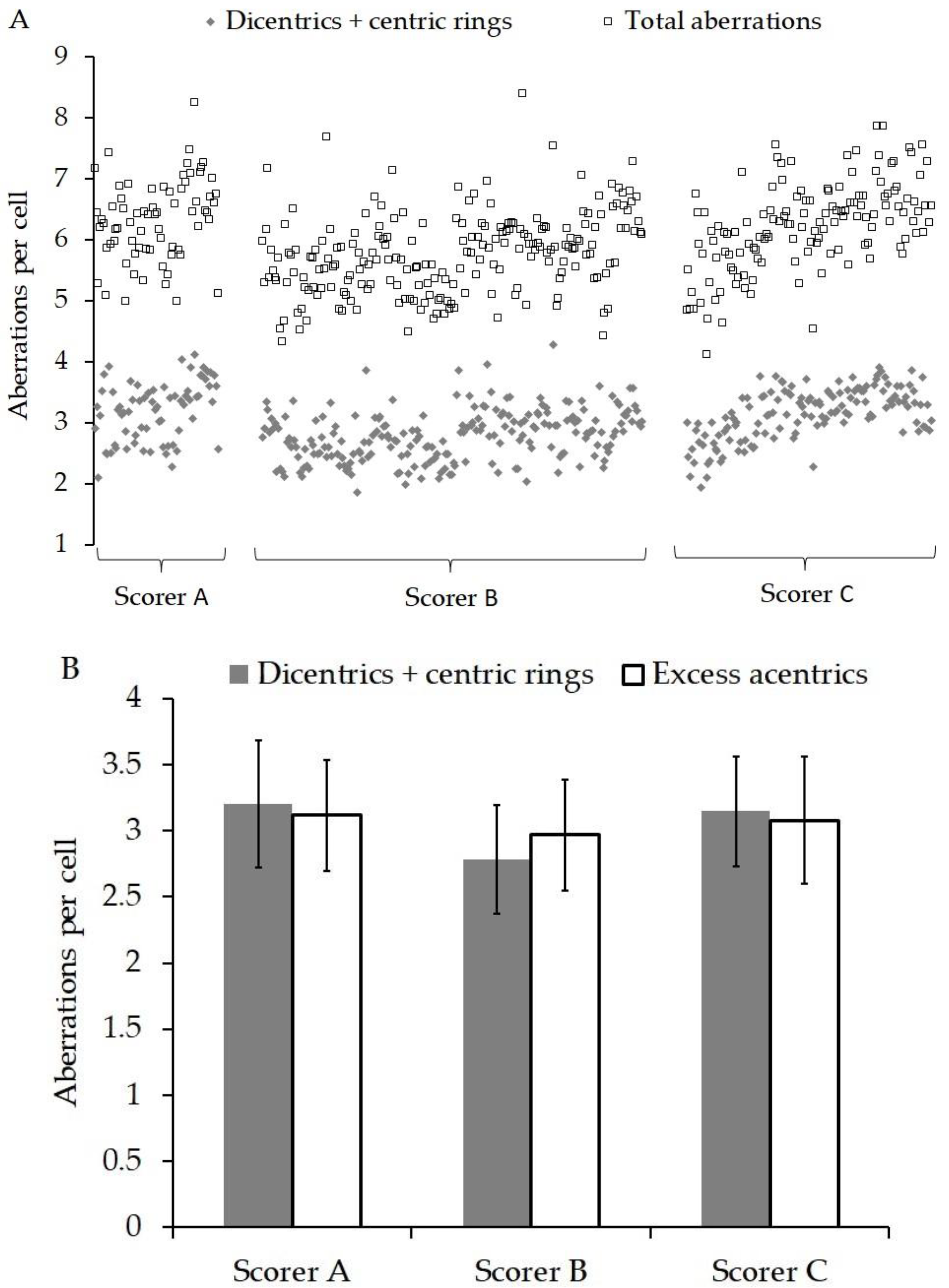
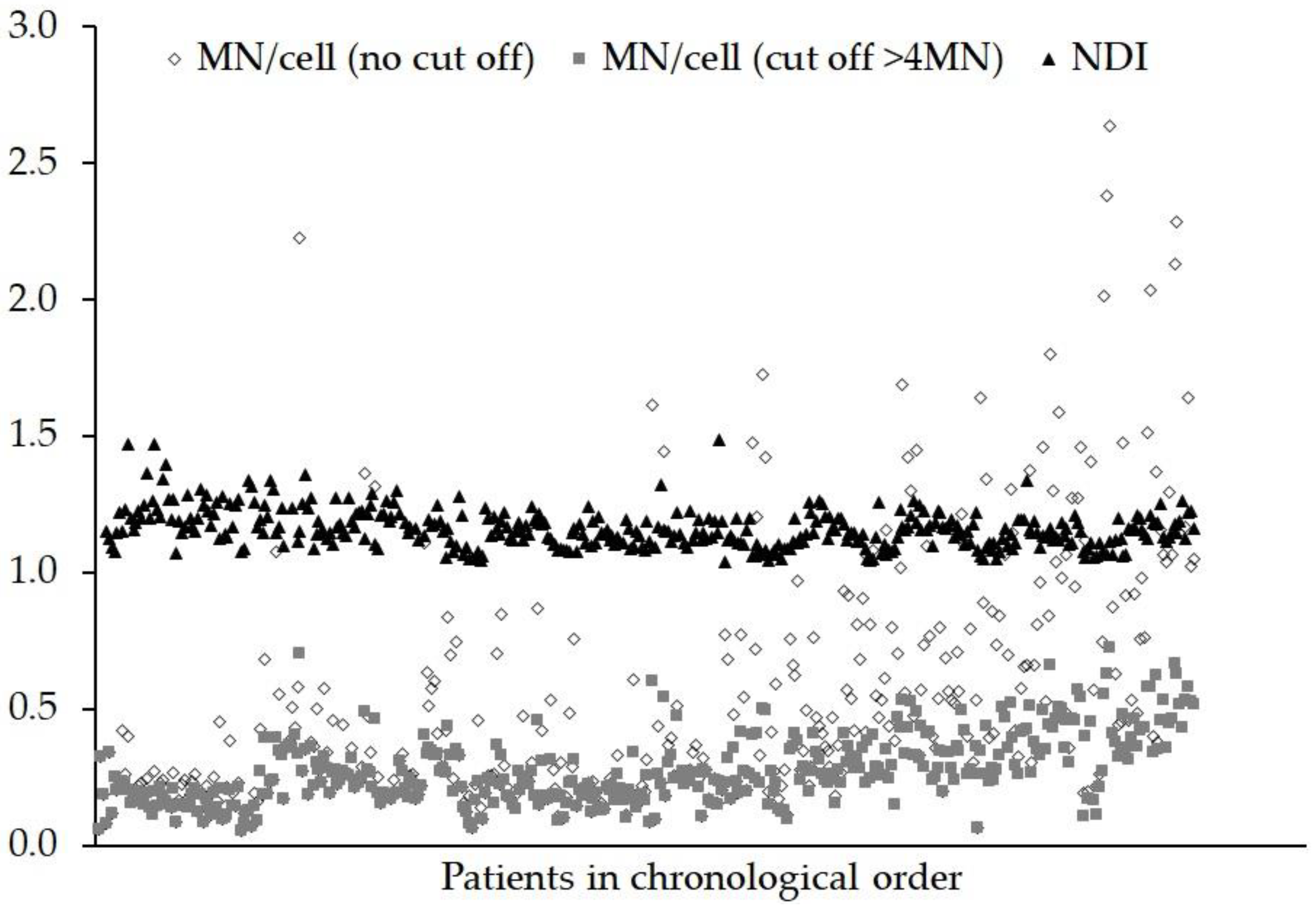
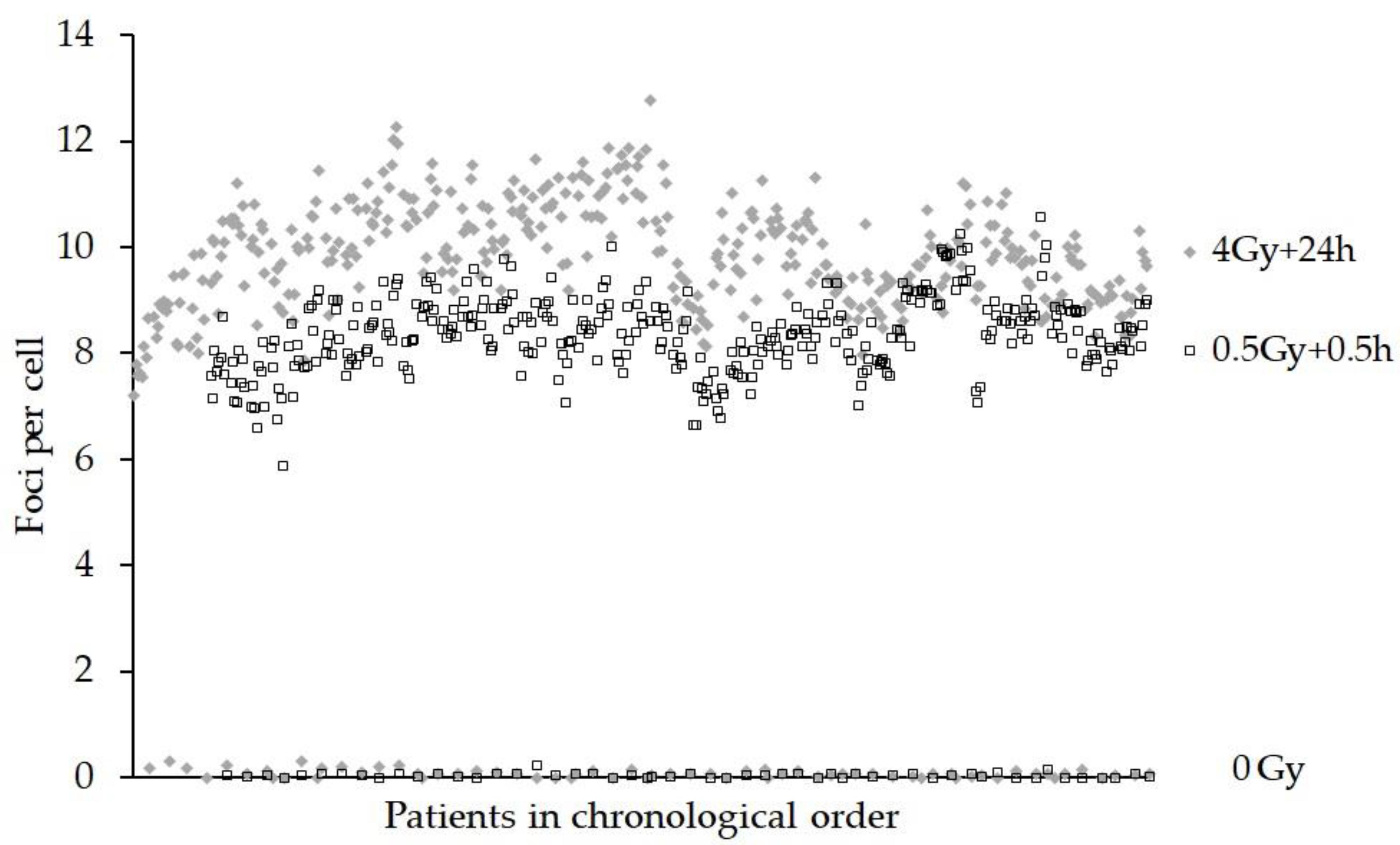
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Obe, G.; Natarajan, A.T. (Eds.) Chromosome Aberrations; Karger: Basel, Switzerland, 2004. [Google Scholar]
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA damage foci Meaning and significance. Environ. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef]
- International Atomic Energy Agency (IAEA). Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies; IAEA: Vienna, Austria, 2011. [Google Scholar]
- Sun, M.; Moquet, J.E.; Barnard, S.; Lloyd, D.C.; Rothkamm, K.; Ainsbury, E.A. Doses in Radiation Accidents Investigated by Chromosomal Aberration Analysis XXV. Review of Cases Investigated, 2006–2015. PHE-CRCE-025. 2016; PHE Publication Gateway Number: 2015730. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/515260/PHE-CRCE-025.pdf (accessed on 12 May 2016).
- Sotnik, N.V.; Azizova, T.V.; Darroudi, F.; Ainsbury, E.A.; Moquet, J.E.; Fomina, J.; Lloyd, D.C.; Edwards, A.A. Verification by the FISH translocation assay of historic doses to Mayak workers from external gamma radiation. Radiat. Environ. Biophys. 2015, 5494, 445–451. [Google Scholar] [CrossRef]
- Chua, M.L.K.; Rothkamm, K. Biomarkers of radiation exposure: Can they predict normal tissue radiosensitivity? Clin. Oncol. 2013, 25, 610–616. [Google Scholar] [CrossRef]
- Rothkamm, K.; Lloyd, D. Established and emerging methods of biological dosimetry. In Comprehensive Biomedical Physics; Brahme, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 7, pp. 289–310. [Google Scholar]
- Borgmann, K.; Röper, B.; El-Awady, R.A.; Brackrock, S.; Bigalke, M.; Dörk, T.; Alberti, W.; Dikomey, E.; Dahm-Daphi, J. Indicators of late normal tissue response after radiotherapy for head and neck cancer: Fibroblasts, lymphocytes, genetics, DNA repair and chromosome aberrations. Radiother. Oncol. 2002, 64, 141–152. [Google Scholar] [CrossRef]
- Chua, M.L.K.; Somaiah, N.; A’Hern, R.; Davies, S.; Gothard, L.; Yarnold, J.; Rothkamm, K. Residual DNA and chromosomal damage in ex vivo irradiated blood lymphocytes correlated with late normal tissue response to breast radiotherapy. Radiother. Oncol. 2011, 99, 362–366. [Google Scholar] [CrossRef]
- Beaton, L.A.; Marro, L.; Samiee, S.; Malone, S.; Grimes, S.; Malone, K.; Wilkins, R.C. Investigating chromosome damage using fluorescent in situ hybridization to identify biomarkers of radiosensitivity in prostate cancer patients. Int. J. Radiat. Biol. 2013, 89, 1087–1093. [Google Scholar] [CrossRef]
- Marková, E.; Somsedíková, A.; Vasilyev, S.; Pobijaková, M.; Lacková, A.; Lukačko, P.; Belyaev, I. DNA repair foci and late apoptosis/necrosis in peripheral blood lymphocytes of breast cancer patients undergoing radiotherapy. Int. J. Radiat. Biol. 2015, 91, 934–945. [Google Scholar]
- Qvarnström, F.; Simonsson, M.; Nyman, J.; Hermansson, I.; Book, M.; Johansson, K.-A.; Turesson, I. Double strand break induction and kinetics indicate preserved hypersensitivity in keratinocytes to subtherapeutic doses for 7 weeks of radiotherapy. Radiother. Oncol. 2017, 122, 163–169. [Google Scholar] [CrossRef]
- Talbot, C.J.; Veldwijk, M.R.; Azria, D.; Batini, C.; Bierbaum, M.; Brengues, M.; Chang-Claude, J.; Johnson, K.; Keller, A.; Smith, S.; et al. Multi-centre technical evaluation of the radiation-induced lymphocyte apoptosis assay as a predictive test for radiotherapy toxicity. Clin. Transl. Radiat. Oncol. 2019, 18, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kulka, U.; Abend, M.; Ainsbury, E.; Badie, C.; Barquinero, J.F.; Barrios, L.; Beinke, C.; Bortolin, E.; Cucu, A.; De Amicis, A.; et al. RENEB–Running the European Network of biological dosimetry and physical retrospective dosimetry. Int. J. Radiat. Biol. 2017, 93, 2–14. [Google Scholar] [CrossRef] [Green Version]
- RENEB Strategic Research Agenda. November 2015. Available online: http://www.reneb.net/wp-content/uploads/2017/10/reneb-sra_november-2015_2.pdf (accessed on 5 July 2016).
- Gregoire, E.; Ainsbury, E.; Barrios, L.; Bassinet, C.; Fattibene, P.; Kulka, U.; Oestreicher, U.; Pantelias, G.; Terzoudi, G.; Trompier, F.; et al. The harmonization process to set up and maintain an operational biological and physical retrospective dosimetry network: QA QM applied to the RENEB network. Int. J. Radiat. Biol. 2017, 93, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Ainsbury, E.; Badie, C.; Barnard, S.; Manning, G.; Moquet, J.; Abend, M.; Antunes, A.C.; Barrios, L.; Bassinet, C.; Beinke, C.; et al. Integration of new biological and physical retrospective dosimetry methods into EU emergency response plans–joint RENEB and EURADOS inter-laboratory comparisons. Int. J. Radiat. Biol. 2017, 93, 99–109. [Google Scholar] [CrossRef]
- FAST Trialists Group; Agrawal, R.K.; Alhasso, A.; Barrett-Lee, P.J.; Bliss, P.; Bloomfield, D.; Bowen, J.; Brunt, A.M.; Donovan, E.; Emson, M.; et al. First results of the randomised UK FAST Trial of radiotherapy hypofractionation for treatment of early breast cancer (CRUKE/04/015). Radiother. Oncol. 2011, 100, 93–100. [Google Scholar]
- Dearnaley, D.; Syndikus, I.; Mossop, H.; Khoo, V.; Birtle, A.; Bloomfield, D.; Graham, J.; Kirkbride, P.; Logue, J.; Malik, Z.; et al. Conventional versus hypofractionated high-dose intensity-modulated radiotherapy for prostate cancer: 5-year outcomes of the randomised, non-inferiority, phase 3 CHHiP trial. Lancet Oncol. 2016, 17, 1047–1060. [Google Scholar] [CrossRef] [Green Version]
- World Health Organisation. Guidance on Regulations for the Transport of Infectious Substances; 2009–2010 WHO/HSE/EPR/2008.10; WHO: Geneva, Switzerland, 2008. [Google Scholar]
- Guogytė, K.; Plieskienė, A.; Ladygienė, R.; Vaisiūnas, Ž.; Sevriukova, O.; Janušonis, V.; Žilukas, J. Assessment of correlation between chromosomal radiosensitivity of peripheral blood lymphocytes after in vitro irradiation and normal tissue side effects for cancer patients undergoing radiotherapy. Genome Integr. 2017, 8, 1. [Google Scholar]
- Rothkamm, K.; Barnard, S.; Ainsbury, E.A.; Al-Hafidh, J.; Barquinero, J.-F.; Lindholm, C.; Moquet, J.; Perälä, M.; Roch-Lefèvre, S.; Scherthan, H.; et al. Manual versus automated γ-H2AX foci analysis across five European laboratories: Can this assay be used for rapid biodosimetry in a large scale radiation accident? Mutat. Res. 2013, 756, 170–173. [Google Scholar] [CrossRef]
- Horn, S.; Barnard, S.; Rothkamm, K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS ONE 2011, 6, e25113. [Google Scholar] [CrossRef]
- Lloyd, D.C.; Purrott, R.J.; Dolphin, G.W.; Bolton, D.; Edwards, A.A.; Corp, M.J. The relationship between chromosome aberrations and low LET radiation dose in human lymphocytes. Int. J. Radiat. Biol. 1975, 28, 75–90. [Google Scholar] [CrossRef]
- Thierens, H.; Vral, A.; Vandevoorde, C.; Vandersickle, V.; de Gelder, V.; Romm, H.; Oestreicher, U.; Rothkamm, K.; Barnard, S.; Ainsbury, E.; et al. Is a semi-automated approach indicated in the application of the automated micronucleus assay for triage purposes? Radiat. Prot. Dosimetry 2014, 159, 87–94. [Google Scholar] [CrossRef]
- Ainsbury, E.A.; Lloyd, D.C. Dose estimation software for radiation biodosimetry. Health Phys. 2010, 98, 290–295. [Google Scholar] [CrossRef]
- Willems, P.; August, L.; Slabbert, J.; Romm, H.; Oestreicher, U.; Thierens, H.; Vral, A. Automated micronucleus (MN) scoring for population triage in case of large scale radiation events. Int. J. Radiat. Biol. 2010, 86, 2–11. [Google Scholar] [CrossRef]
- Lloyd, D.C.; Edward, A.A.; Prosser, J.S. Chromosome aberrations induced in human lymphocytes by in-vitro acute X and gamma radiation. Radiat. Prot. Dosimetry 1986, 15, 83–88. [Google Scholar] [CrossRef]
- Moquet, J.; Barnard, S.; Staynova, A.; Lindholm, C.; Monteiro Gil, O.; Martins, V.; Rößler, U.; Vral, A.; Vandevoorde, C.; Wojewódzka, M.; et al. The second gamma-H2AX assay inter-comparison exercise carried out in the framework of the European biodosimetry network (RENEB). Int. J. Radiat. Biol. 2017, 93, 58–64. [Google Scholar] [CrossRef]
- Oestreicher, U.; Samaga, D.; Ainsbury, E.; Antunes, A.C.; Baeyens, A.; Barrios, L.; Beinke, C.; Beukes, P.; Blakely, W.F.; Cucu, A.; et al. RENEB intercomparisons applying the conventional dicentric chromosome assay (DCA). Int. J. Radiat. Biol. 2017, 93, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, D.C.; Purrott, R.J.; Reeder, E.J. The incidence of unstable chromosomes aberrations in peripheral blood lymphocytes from unirradiated and occupationally exposed people. Mutat. Res. 1980, 72, 523–532. [Google Scholar] [CrossRef]
- Hoffmann, G.R.; Sayer, A.M.; Littlefield, L.G. Higher frequency of chromosome aberrations in late-arising first-division metaphases than in early-arising metaphases after exposure of human lymphocytes to X-rays in G0. Int. J. Radiat. Biol. 2002, 78, 765–772. [Google Scholar] [CrossRef]
- Hone, P.A.; Edwards, A.A.; Lloyd, D.C.; Moquet, J.E. The yield of radiation-induced chromosomal aberrations in first division human lymphocytes depends on the culture time. Int. J. Radiat. Biol. 2005, 81, 523–529. [Google Scholar] [CrossRef]
- Livingston, G.K.; Wilkins, R.C.; Ainsbury, E.A. Pilot website to support international collaboration for dose assessments in a radiation emergency. Radiat. Meas. 2011, 46, 912–915. [Google Scholar] [CrossRef]
- Romm, H.; Ainsbury, E.; Bajinskis, A.; Barnard, S.; Barquinero, J.F.; Barrios, L.; Beinke, C.; Puig-Casanovas, R.; Deperas-Kaminska, M.; Gregoire, E.; et al. Web-based scoring of the dicentric assay, a collaborative biodosimetric scoring strategy for population triage in large scale radiation accidents. Radiat. Environ. Biophys. 2014, 53, 241–254. [Google Scholar] [CrossRef]
- Romm, H.; Ainsbury, E.A.; Barquinero, J.F.; Barrios, L.; Beinke, C.; Cucu, A.; Moreno Domene, M.; Filippi, S.; Monteiro Gil, O.; Gregoire, E.; et al. Web based scoring is useful for validation and harmonisation of scoring criteria within RENEB. Int. J. Radiat. Biol. 2017, 93, 110–117. [Google Scholar] [CrossRef] [Green Version]
- Depuydt, J.; Baeyens, A.; Barnard, S.; Beinke, C.; Benedek, A.; Beukes, P.; Buraczewska, I.; Darroudi, F.; De Sanctis, S.; Dominguez, I.; et al. RENEB intercomparison exercises analyzing micronuclei (Cytokinesis-block Micronucleus Assay). Int. J. Radiat. Biol. 2017, 93, 36–47. [Google Scholar] [CrossRef]
- Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007, 2, 1084–1104. [Google Scholar] [CrossRef] [Green Version]
- Rothkamm, K.; Horn, S. gamma-H2AX as protein biomarker for radiation exposure. Ann. 1st Super Sanità. 2009, 45, 265–271. [Google Scholar]
- Ainsbury, E.A.; Al-Hafidh, J.; Bajinskis, A.; Barnard, S.; Barquinero, J.F.; Beinke, C.; de Gelder, V.; Gregoire, E.; Jaworska, A.; Lindholm, C.; et al. Inter- and intra-laboratory comparison of a multibiodosimetric approach to triage in a simulated, large scale radiation emergency. Int. J. Radiat. Biol. 2014, 90, 193–202. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moquet, J.; Rothkamm, K.; Barnard, S.; Ainsbury, E. Radiation Biomarkers in Large Scale Human Health Effects Studies. J. Pers. Med. 2020, 10, 155. https://doi.org/10.3390/jpm10040155
Moquet J, Rothkamm K, Barnard S, Ainsbury E. Radiation Biomarkers in Large Scale Human Health Effects Studies. Journal of Personalized Medicine. 2020; 10(4):155. https://doi.org/10.3390/jpm10040155
Chicago/Turabian StyleMoquet, Jayne, Kai Rothkamm, Stephen Barnard, and Elizabeth Ainsbury. 2020. "Radiation Biomarkers in Large Scale Human Health Effects Studies" Journal of Personalized Medicine 10, no. 4: 155. https://doi.org/10.3390/jpm10040155
APA StyleMoquet, J., Rothkamm, K., Barnard, S., & Ainsbury, E. (2020). Radiation Biomarkers in Large Scale Human Health Effects Studies. Journal of Personalized Medicine, 10(4), 155. https://doi.org/10.3390/jpm10040155





