Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis
Abstract
:1. Introduction
2. Materials and Methods
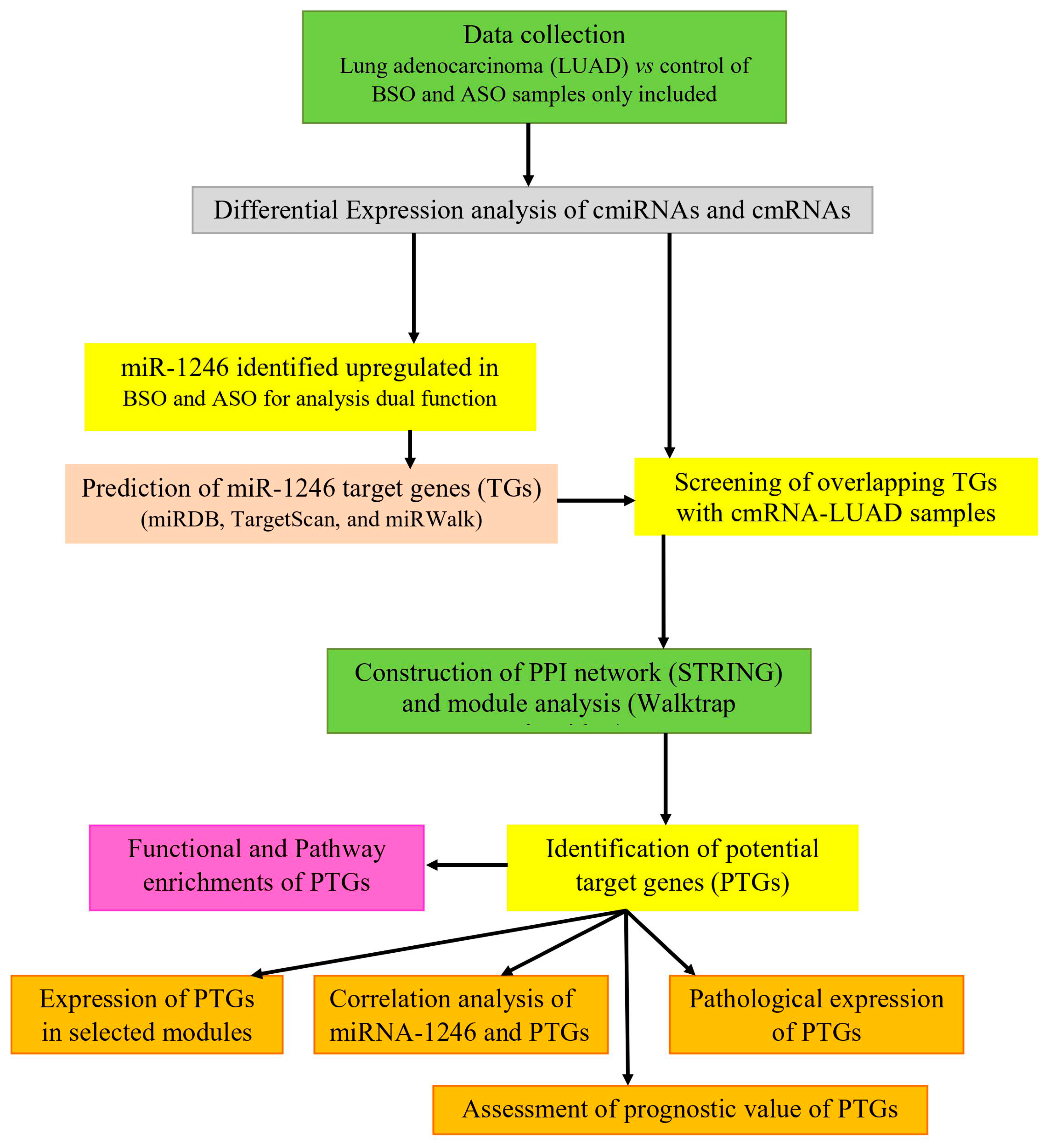
2.1. Data Collection
2.2. Differential Expression of cmiRNAs and cmRNAs
2.3. miR-1246 Target Gene Prediction
2.4. Screening of Overlapping Target Genes
2.5. Construction of PPI Network
2.6. Identification of Modules and Hub Genes
2.7. Functional Enrichment Analysis
2.8. Validation of Potential Target Genes (PTGs)
2.8.1. Expression of PTGs in LUAD
2.8.2. Correlation Analysis of miR-1246 and PTGs
2.8.3. Survival Analysis
2.8.4. Protein Expression Analysis in LUAD
3. Results
3.1. Differentially Expressed cmiRNAs and cmRNAs
3.2. Identification of Overlapping miR-1246 Target Genes
3.3. Functional and Pathway Enrichment of Overlapping miR-1246 Target Genes
3.4. Modules and PTGs Identification
3.5. Function and Pathway Enrichments of PTGs
3.6. Validation of PTGs
3.6.1. Expression of PTGs
3.6.2. Spearman’s Correlation Analysis of PTGs
3.6.3. Prognostic Impact of PTGs
3.6.4. Protein Expression of PTGs
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Subotic, D.D.; Van Schil, P.P.; Grigoriu, B. Radiation therapy for post-operative recurrence: Yes, but only for limited indications. Eur. Respir. J. 2016, 48, 278–279. [Google Scholar] [CrossRef]
- Le, H.B.B.; Zhu, W.Y.Y.; Chen, D.D.; He, J.Y.; Huang, Y.Y.; Liu, X.G.; Zhang, Y.K. Evaluation of dynamic change of serum miR-21 and miR-24 in pre-and post-operative lung carcinoma patients. Med. Oncolog. 2012, 29, 3190–3197. [Google Scholar] [CrossRef]
- Liloglou, T.; Bediaga, N.G.; Brown, B.R.; Field, J.K.; Davies, M.P. Epigenetic biomarkers in lung cancer. Cancer Lett. 2014, 342, 200–212. [Google Scholar] [CrossRef]
- Rothschild, S.I. Epigenetic therapy in lung cancer–role of microRNAs. Front. Oncolog. 2013, 3, 158. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, A.; Bunkar, N.; Aglawe, A.; Pandey, K.C.; Tiwari, R.; Chaudhury, K.; Goryacheva, I.Y.; Mishra, P.K. Epigenetic biomarkers for risk assessment of particulate matter associated lung cancer. Curr. Drug Targets 2018, 19, 1127–1147. [Google Scholar] [CrossRef] [PubMed]
- Afzali, F.; Salimi, M. Unearthing regulatory axes of breast cancer circRNAs networks to find novel targets and fathom pivotal mechanisms. Interdiscip. Sci. 2019, 11, 711–722. [Google Scholar] [CrossRef]
- Han, H.; Zhang, Z.; Yang, X.; Yang, W.; Xue, C.; Cao, X. miR-23b suppresses lung carcinoma cell proliferation through CCNG1. Oncolog. Lett. 2018, 16, 4317–4324. [Google Scholar]
- Feng, M.; Zhao, J.; Wang, L.; Liu, J. Upregulated expression of serum exosomal microRNAs as diagnostic biomarkers of lung adenocarcinoma. Ann. Clin. Lab. Sci. 2018, 48, 712–718. [Google Scholar] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Hou, L.K.; Ma, Y.S.; Han, Y.; Lu, G.X.; Luo, P.; Chang, Z.Y.; Xie, R.; Yang, H.; Chai, L.; Cai, M.; et al. Association of microRNA-33a molecular signature with non-small cell lung cancer diagnosis and prognosis after chemotherapy. PLoS ONE 2017, 12, e0170431. [Google Scholar] [CrossRef] [PubMed]
- Hirono, T.; Jingushi, K.; Nagata, T.; Sato, M.; Minami, K.; Aoki, M.; Takeda, A.H.; Umehara, T.; Egawa, H.; Nakatsuji, Y.; et al. MicroRNA-130b functions as an oncomiRNA in non-small cell lung cancer by targeting tissue inhibitor of metalloproteinase-2. Sci. Rep. 2019, 9, 6956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Dou, Y.; Sui, Z.; Cheng, H.; Liu, X.; Wang, Q.; Gao, P.; Qu, Y.; Xu, M. Upregulated miRNA-182-5p expression in tumor tissue and peripheral blood samples from patients with non-small cell lung cancer is associated with downregulated Caspase 2 expression. Exp. Ther. Med. 2020, 19, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Zahran, A.M.; El-Mahdy, R.I.; Nabil, E.E.; Esmaeel, H.M.; Alkady, O.A.; Elkady, A.; Mohareb, D.A.; Mostafa, M.M.; John, J. Assessment of Circulating miRNA-17 and miRNA-222 Expression Profiles as Non-Invasive Biomarkers in Egyptian Patients with Non-Small-Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Geng, S.; Hu, Y. miR-486–5p inhibits cell proliferation and invasion through repressing GAB2 in non-small cell lung cancer. Oncolog. Lett. 2018, 16, 3525–3530. [Google Scholar] [CrossRef]
- Zhang, J.G.; Guo, J.F.; Liu, D.L.; Liu, Q.; Wang, J.J. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J. Thorac. Oncolog. 2011, 6, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Nohata, N.; Kinoshita, T.; Mutallip, M.; Okamoto, T.; Yoshida, S.; Suzuki, M.; Yoshino, I.; Seki, N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J. Hum. Genet. 2012, 57, 38–45. [Google Scholar] [CrossRef] [Green Version]
- Asakura, K.; Kadota, T.; Matsuzaki, J.; Yoshida, Y.; Yamamoto, Y.; Nakagawa, K.; Takizawa, S.; Aoki, Y.; Nakamura, E.; Miura, J.; et al. A miRNA-based diagnostic model predicts resectable lung cancer in humans with high accuracy. Commun. Biol. 2020, 3, 134. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Yi, H.; Ma, S. Measures for the degree of overlap of gene signatures and applications to TCGA. Brief. Bioinform. 2015, 16, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.Q.; Selvaraj, G.; Kaushik, A.C. Computational Perspective on the Current State of the Methods and New Challenges in Cancer Drug Discovery. Curr. Pharm. Des. 2018, 24, 3725. [Google Scholar] [CrossRef] [Green Version]
- Selvaraj, G.; Kaliamurthi, S.; Kaushik, A.C.; Khan, A.; Wei, Y.K.; Cho, W.C.; Gu, K.; Wei, D.Q. Identification of target gene and prognostic evaluation for lung adenocarcinoma using gene expression meta-analysis, network analysis and neural network algorithms. J. Biomed. Inform. 2018, 86, 120–134. [Google Scholar] [CrossRef]
- Shih, C.L.; Luo, J.D.; Chang, J.W.C.; Chen, T.L.; Chien, Y.T.; Yu, C.J.; Chiou, C.C. Circulating messenger RNA profiling with microarray and next-generation sequencing: Cross-platform comparison. Cancer Genom. Proteom. 2015, 12, 223–230. [Google Scholar]
- Gentleman, R.; Carey, V.; Huber, W.; Irizarry, R.; Dudoit, S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer Science and Business Media: New York, NY, USA, 2006. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2. 0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 25 July 2020).
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Bozhilova, L.V.; Whitmore, A.V.; Wray, J.; Reinert, G.; Deane, C.M. Measuring rank robustness in scored protein interaction networks. BMC Bioinform. 2019, 20, 446. [Google Scholar] [CrossRef] [Green Version]
- Pons, P.; Latapy, M. Computing Communities in Large Networks Using Random Walks. In International Symposium on Computer and Information Sciences; Springer: Berlin/Heidelberg, Germany, October 2005; pp. 284–293. [Google Scholar]
- Kohl, P.; Sachs, F.; Franz, M.R. Cardiac Mechano-Electric Coupling and Arrhythmias; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Jérôme, G.; et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinform. (Oxf. Engl.) 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Medica Biochem. Medica 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Gyorffy, B.; Surowiak, P.; Budczies, J.; Lanczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Björling, E.; Agaton, C.; Al-KhaliliSzigyarto, C.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based onantibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 6352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvaraj, G.; Selvaraj, C.; Wei, D.Q. Computational advances in chronic diseases diagnostics and therapy-I. Curr. Drug Targets 2020, 21, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, G.; Kaliamurthi, S.; Lin, S.; Gu, K.; Wei, D.Q. Prognostic impact of tissue inhibitor of metalloproteinase-1 in non-small cell lung cancer: Systematic review and meta-analysis. Curr. Med. Chem. 2019, 26, 7694. [Google Scholar] [CrossRef] [PubMed]
- Kaliamurthi, S.; Selvaraj, G.; Junaid, M.; Khan, A.; Gu, K.; Wei, D.Q. Cancer immunoinformatics: A promising era in the development of peptide vaccines for human papillomavirus-induced cervical cancer. Curr. Pharm. Des. 2018, 24, 3791–3817. [Google Scholar] [CrossRef] [PubMed]
- Nangraj, A.S.; Selvaraj, G.; Kaliamurthi, S.; Cho, W.C.S.; Aman, C.; Cho, W.C.; Wei, D.Q. Integrated PPI and WGCNA retrieving shared gene signatures between Barrett’s esophagus and esophageal adenocarcinoma. Front. Pharm. 2020, 11, 881. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, I.; Yokota, H.; Ishige, F.; Iwatate, Y.; Takeshita, N.; Nagase, H.; Uno, T.; Matsubara, H. Radiogenomics predicts the expression of microRNA-1246 in the serum of esophageal cancer patients. Sci. Rep. 2020, 10, 2532. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell. Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef]
- Du, P.; Lai, Y.H.; Yao, D.S.; Chen, J.Y.; Ding, N. Downregulation of microRNA-1246 inhibits tumor growth and promotes apoptosis of cervical cancer cells by targeting thrombospondin-2. Oncolog. Lett. 2019, 18, 2491–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, D.; Xu, J.; Wang, J.; Pan, Y.; Fu, J.; Bai, Y.; Zhang, J.; Shao, C. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 2016, 7, 32707. [Google Scholar] [CrossRef] [Green Version]
- Morla, S. Glycosaminoglycans and Glycosaminoglycan Mimetics in Cancer and Inflammation. Int. J. Mol. Sci. 2019, 20, 1963. [Google Scholar] [CrossRef] [Green Version]
- Salanti, A.; Clausen, T.M.; Agerbæk, M.Ø.; Al Nakouzi, N.; Dahlbäck, M.; Oo, H.Z.; Lee, S.; Gustavsson, T.; Rich, J.R.; Hedberg, B.J.; et al. Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein. Cancer Cell 2015, 28, 500–514. [Google Scholar] [CrossRef] [Green Version]
- Brunetti, J.; Depau, L.; Falciani, C.; Gentile, M.; Mandarini, E.; Riolo, G.; Lupetti, P.; Pini, A.; Bracci, L. Insights into the role of sulfated glycans in cancer cell adhesion and migration through use of branched peptide probe. Sci. Rep. 2016, 6, 27174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dastsooz, H.; Cereda, M.; Donna, D.; Oliviero, S. A Comprehensive Bioinformatics Analysis of UBE2C in Cancers. Int. J. Mol. Sci. 2019, 20, 2228. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Powell, C.; Yao, M.; Wu, J.; Dong, Q. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int. J. Biochem. Cell Biol. 2014, 47, 113–117. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, T.; Wei, G.; Wang, Y. Inhibition of microRNA-17/20a suppresses cell proliferation in gastric cancer by modulating UBE2C expression. Oncolog. Rep. 2015, 33, 2529–2536. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Guo, J.; Wu, Y.; Du, J.; Wang, X.; An, J.; Hu, B.; Kong, L.; Di, W.; Wang, W. UBE2C, directly targeted by miR-548e-5p, increases the cellular growth and invasive abilities of cancer cells interacting with the EMT marker protein zinc finger E-box binding homeobox 1/2 in NSCLC. Theranostics 2019, 9, 2036. [Google Scholar] [CrossRef] [PubMed]
- Kwan, S.Y.; Au-Yeung, C.L.; Yeung, T.L.; Rynne-Vidal, A.; Wong, K.K.; Risinger, J.I.; Lin, H.K.; Schmandt, R.E.; Yates, M.S.; Mok, S.C.; et al. Ubiquitin Carboxyl-Terminal Hydrolase L1 (UCHL1) Promotes Uterine Serous Cancer Cell Proliferation and Cell Cycle Progression. Cancers 2020, 12, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; González-Prieto, R.; Zhang, M.; Geurink, P.P.; Kooij, R.; Iyengar, P.V.; van Dinther, M.; Bos, E.; Zhang, X.; Sylvia, E.; et al. Deubiquitinase activity profiling identifies UCHL1 as a candidate oncoprotein that promotes TGFβ-induced breast cancer metastasis. Clin. Cancer Res. 2020, 26, 1460–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Zhao, Y.; Zhang, Y.; AiErken, N.; Shao, N.; Ye, R.; Lin, Y.; Wang, S. TNNT1 facilitates proliferation of breast cancer cells by promoting G1/S phase transition. Life Sci. 2018, 208, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.H.; Yu, S.Y.; Tu, R.S.; Cai, Y.Q. TNNT1, a prognostic indicator in colon adenocarcinoma, regulates cell behaviors and mediates EMT process. Biosci. Biotechnol. Biochem. 2020, 84, 111–117. [Google Scholar] [CrossRef]
- Chapard, C.; Hohl, D.; Huber, M. The TRAF-interacting protein (TRAIP) is a novel E2F target with peak expression in mitosis. Oncotarget 2015, 6, 20933–20945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Geahlen, R.L. The protein-tyrosine kinase Syk interacts with TRAF-interacting protein TRIP in breast epithelial cells. Oncogene 2009, 28, 1348–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.; Zeng, Y.; Chen, Y.; Liu, M.; Chen, S.; Yao, M.; Zhang, P.; Zhong, F.; Jiang, K.; He, S.; et al. TRAIP promotes malignant behaviors and correlates with poor prognosis in liver cancer. Biomed. Pharm. 2020, 124, 109857. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, A.J. Overview of Cell Signaling Pathways in Cancer. In Predictive Biomarkers in Oncology; Springer: Cham, Switzerland, 2019; pp. 167–182. [Google Scholar]









| miRNA_ID | Log2FC | p-Value | miRNA_ID | Log2FC | p-Value |
|---|---|---|---|---|---|
| BSO overexpressed | BSO underexpressed | ||||
| hsa-miR-1246 | 6.28 | 2.79 × 10−110 | hsa-miR-373-5p | −5.92 | 0 |
| hsa-miR-8060 | 5.69 | 6.62 × 10−189 | hsa-miR-1199-5p | −6.05 | 0 |
| hsa-miR-920 | 5.46 | 0 | hsa-miR-208b-5p | −6.07 | 0 |
| hsa-miR-6131 | 5.31 | 9.32 × 10−187 | hsa-miR-6777-5p | −6.07 | 0 |
| hsa-miR-4259 | 5.08 | 9.10 × 10−249 | hsa-miR-4648 | −6.32 | 0 |
| hsa-miR-6849-5p | 4.61 | 2.22 × 10−172 | hsa-miR-4435 | −6.38 | 0 |
| hsa-miR-193a-5p | 4.39 | 4.87 × 10−182 | hsa-miR-4276 | −6.46 | 0 |
| hsa-miR-6717-5p | 4.24 | 2.02 × 10−226 | hsa-miR-6857-5p | −6.49 | 0 |
| hsa-miR-3934-5p | 4.11 | 2.63 × 10−128 | hsa-miR-92a-2-5p | −7.19 | 0 |
| hsa-miR-1343-3p | 3.96 | 0 | hsa-miR-1203 | −7.37 | 0 |
| ASO overexpressed | ASO underexpressed | ||||
| hsa-miR-1246 | 7.09 | 0 | hsa-miR-3184-5p | −8.41 | 0 |
| hsa-miR-1290 | 6.17 | 0 | hsa-miR-1203 | −1.54 | 2.73 × 10−214 |
| hsa-miR-29b-1-5p | 6.03 | 0 | hsa-miR-4730 | −1.60 | 0 |
| hsa-miR-191-5p | 5.75 | 0 | hsa-miR-873-3p | −1.64 | 1.79 × 10−173 |
| hsa-miR-451a | 5.64 | 0 | hsa-miR-92a-2-5p | −1.74 | 0 |
| hsa-miR-103a-3p | 5.17 | 0 | hsa-miR-4276 | −1.89 | 2.65 × 10−242 |
| hsa-miR-4755-3p | 5.09 | 0 | hsa-miR-3184-5p | −2.01 | 0 |
| hsa-miR-6131 | 4.99 | 0 | hsa-miR-4648 | −2.05 | 3.64 × 10−225 |
| hsa-miR-4771 | 4.96 | 0 | hsa-miR-6857-5p | −2.36 | 4.82 × 10−302 |
| hsa-miR-4480 | 4.89 | 0 | hsa-miR-4481 | −2.55 | 1.76 × 10−312 |
| Gene Symbol | Description | Log2FC | p-Value |
|---|---|---|---|
| Overexpressed genes | |||
| BTBD11 | BTB domain containing 11 | 3.108 | 4.69 × 10−4 |
| ZNF683 | Zinc finger protein 683 | 1.991 | 6.82 × 10−3 |
| GPATCH4 | G-patch domain containing 4 | 1.754 | 8.86 × 10−4 |
| EHMT1 | Euchromatic histone lysine methyltransferase 1 | 1.652 | 3.61 × 10−3 |
| RAB6B | Ras-related protein Rab-6B | 1.576 | 9.06 × 10−3 |
| C12orf5 | TP53 induced glycolysis regulatory phosphatase | 1.569 | 1.44 × 10−3 |
| GNLY | Granulysin | 1.569 | 9.71 × 10−3 |
| RPGRIP1 | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 | 1.542 | 3.44 × 10−4 |
| CPT1B | Carnitine palmitoyltransferase I | 1.527 | 4.17 × 10−3 |
| SRI | Sorcin | 1.525 | 1.38 × 10−3 |
| Underexpressed genes | |||
| WISP3 | WNT1-inducible-signaling pathway protein 3 | −1.855 | 5.39 × 10−3 |
| HFE2 | Hemojuvelin | −1.858 | 3.08 × 10−3 |
| LOR | Loricrin | −1.861 | 4.96 × 10−3 |
| SLC26A11 | Sodium-independent sulfate anion transporter | −1.875 | 3.97 × 10−3 |
| DCAF12L2 | DDB1- and CUL4-associated factor 12-like protein 2 | −1.885 | 3.31 × 10−4 |
| DKFZp564N2472 | POM121 transmembrane nucleoporin-like 12 | −1.885 | 4.22 × 10−3 |
| FRG2C | FSHD region gene 2 family member C | −1.921 | 4.13 × 10−4 |
| PRM2 | Protamine 2 | −1.95 | 8.97 × 10−3 |
| PTCH2 | Patched 2 | −2.022 | 4.04 × 10−3 |
| NNAT | Neuronatin | −2.298 | 9.95 × 10−3 |
| Official Symbol | Gene ID | Official Full Name | Chromosome Location | Exon Count | Degree | Betweenness |
|---|---|---|---|---|---|---|
| UBE2C | 11,065 | Ubiquitin conjugating enzyme E2 C | 20q13.12 | 8 | 34 | 7811.25 |
| TBCE | 6905 | Tubulin folding cofactor E | 1q42.3 | 18 | 13 | 39.11 |
| DNAJA3 | 9093 | DNAJ heat shock protein family (Hsp40) member 3 | 16p13.3 | 12 | 12 | 6127.74 |
| PITX2 | 5308 | Paired-like homeodomain transcription factor 2 | 4q25 | 9 | 07 | 4584.14 |
| TGIF1 | 7050 | TGFB induced factor homeobox 1 | 18p11.31 | 12 | 07 | 22.32 |
| TRAIP | 10,293 | TRAF interacting protein | 3p21.31 | 16 | 06 | 1533.11 |
| UCHL1 | 7345 | Ubiquitin C-terminal hydrolase L1 | 4p13 | 9 | 06 | 1537.48 |
| TNNI3 | 7137 | Troponin I3 | 19q13.42 | 8 | 04 | 0.23 |
| TNNT1 | 7138 | Troponin T1 | 19q13.42 | 15 | 04 | 10.91 |
| NRAS | 4893 | Neuroblastoma RAS viral oncogene homolog | 1p13.2 | 7 | 03 | 247.07 |
| RAC3 | 5881 | Rac family small GTPase 3 | 17q25.3 | 6 | 03 | 630.68 |
| EFNA4 | 1945 | Ephrin A4 | 1q21.3 | 4 | 03 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Wei, Y.-K.; Kaliamurthi, S.; Cao, Y.; Nangraj, A.S.; Sui, X.; Chu, D.; Wang, H.; Wei, D.-Q.; Peslherbe, G.H.; et al. Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis. J. Pers. Med. 2020, 10, 162. https://doi.org/10.3390/jpm10040162
Huang S, Wei Y-K, Kaliamurthi S, Cao Y, Nangraj AS, Sui X, Chu D, Wang H, Wei D-Q, Peslherbe GH, et al. Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis. Journal of Personalized Medicine. 2020; 10(4):162. https://doi.org/10.3390/jpm10040162
Chicago/Turabian StyleHuang, Siyuan, Yong-Kai Wei, Satyavani Kaliamurthi, Yanghui Cao, Asma Sindhoo Nangraj, Xin Sui, Dan Chu, Huan Wang, Dong-Qing Wei, Gilles H. Peslherbe, and et al. 2020. "Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis" Journal of Personalized Medicine 10, no. 4: 162. https://doi.org/10.3390/jpm10040162
APA StyleHuang, S., Wei, Y.-K., Kaliamurthi, S., Cao, Y., Nangraj, A. S., Sui, X., Chu, D., Wang, H., Wei, D.-Q., Peslherbe, G. H., Selvaraj, G., & Shi, J. (2020). Circulating miR-1246 Targeting UBE2C, TNNI3, TRAIP, UCHL1 Genes and Key Pathways as a Potential Biomarker for Lung Adenocarcinoma: Integrated Biological Network Analysis. Journal of Personalized Medicine, 10(4), 162. https://doi.org/10.3390/jpm10040162









