Race-Specific Genetic Profiles of Homologous Recombination Deficiency in Multiple Cancers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohorts and Patients
2.2. The Determination of Pathogenicity for Somatic Variant Calls
2.3. Groupwise Association Test, Outlier Detection Analysis, and Variant Annotation
2.4. HRD Score Calculation
2.5. Statistical Analysis
2.6. Survival Analysis
3. Results
3.1. Distribution of the TCGA Pan-Cancer Atlas Cohort across Racial Groups
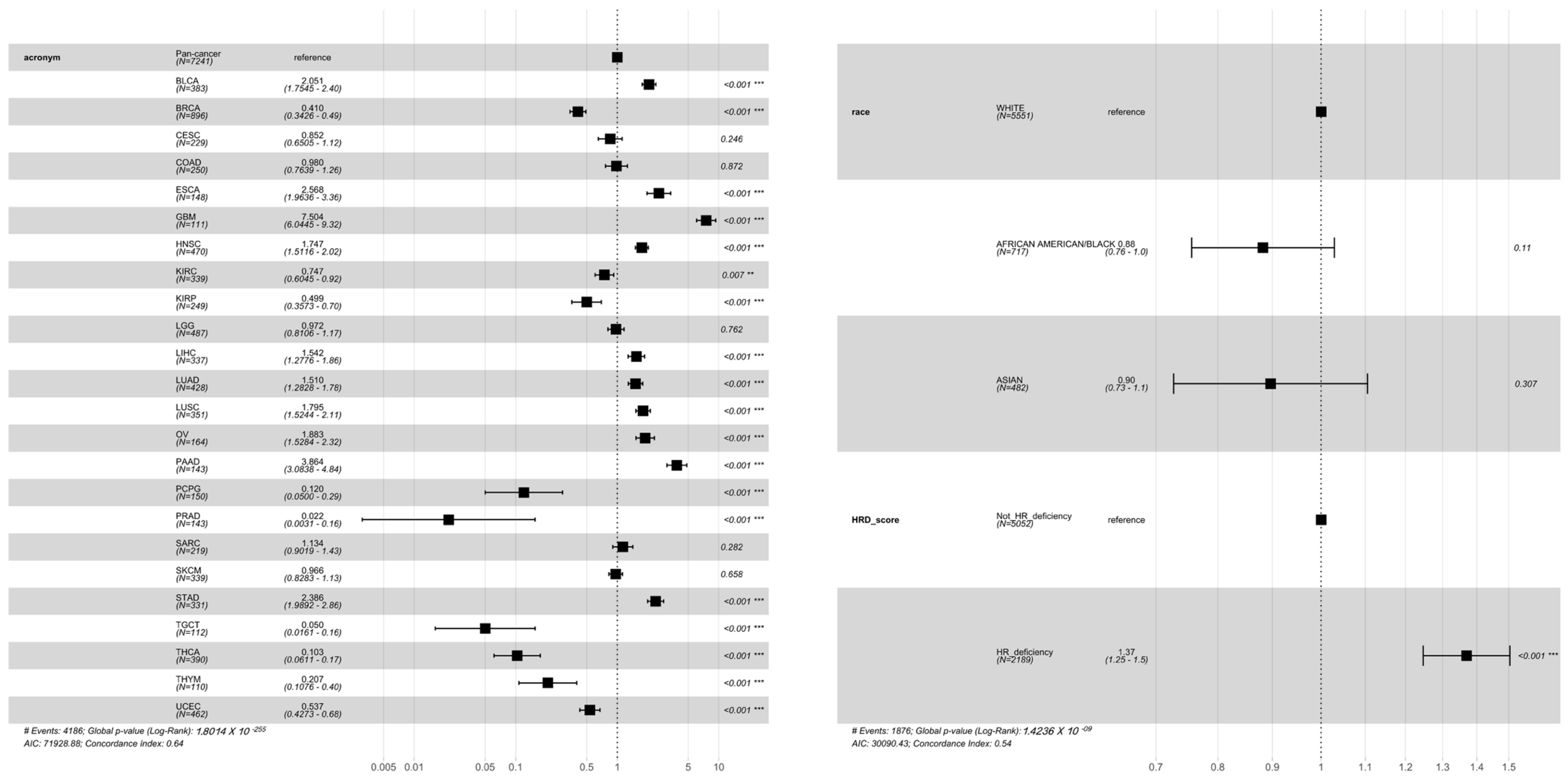
3.2. Association of HRD Scores with Survival across Cancer Types
3.3. Synergistic Effects between Genome-Wide HRD and Global TMB among Cancers
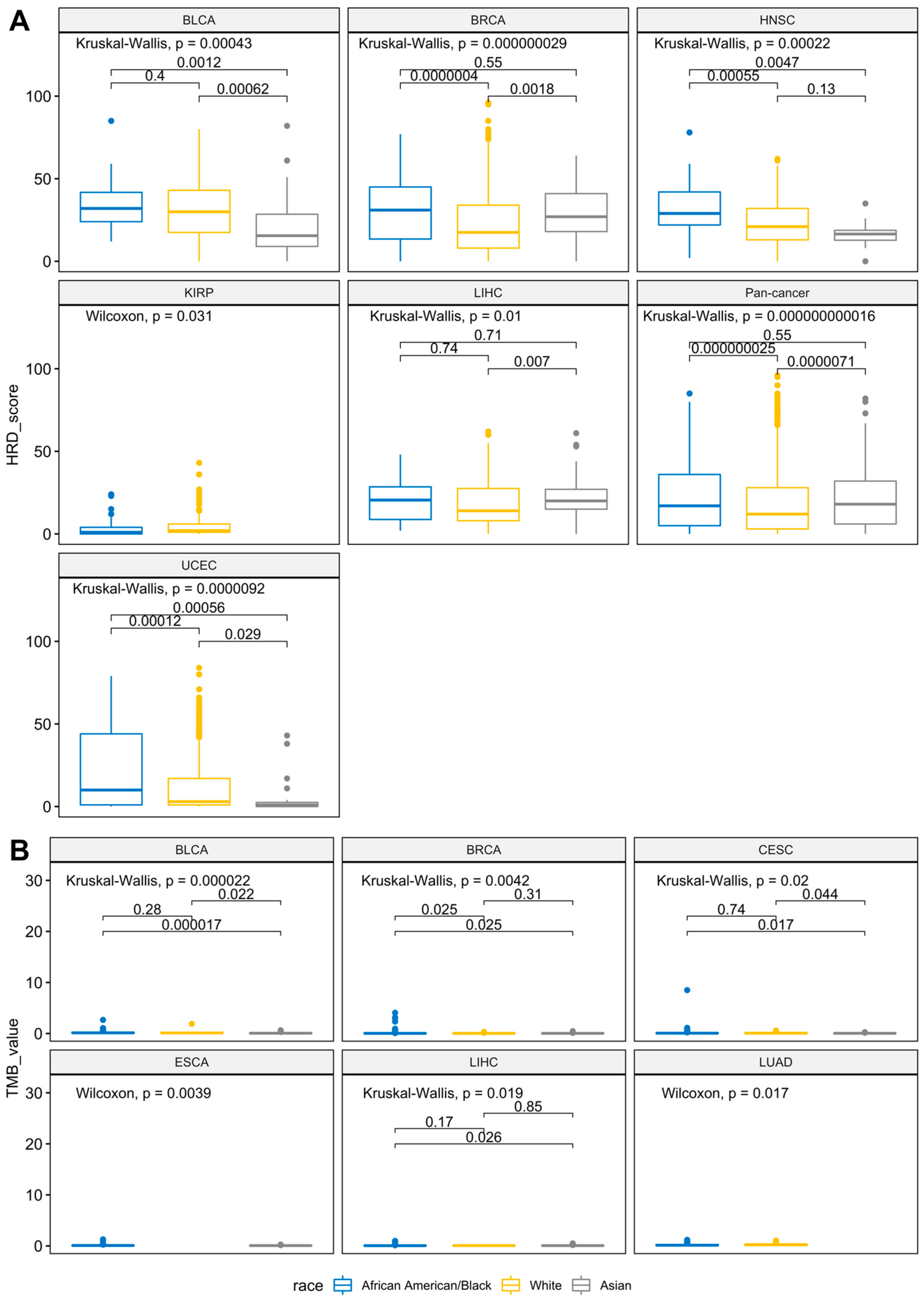
3.4. Racial Differences of Genome-Wide HRD and Global TMB across Cancers
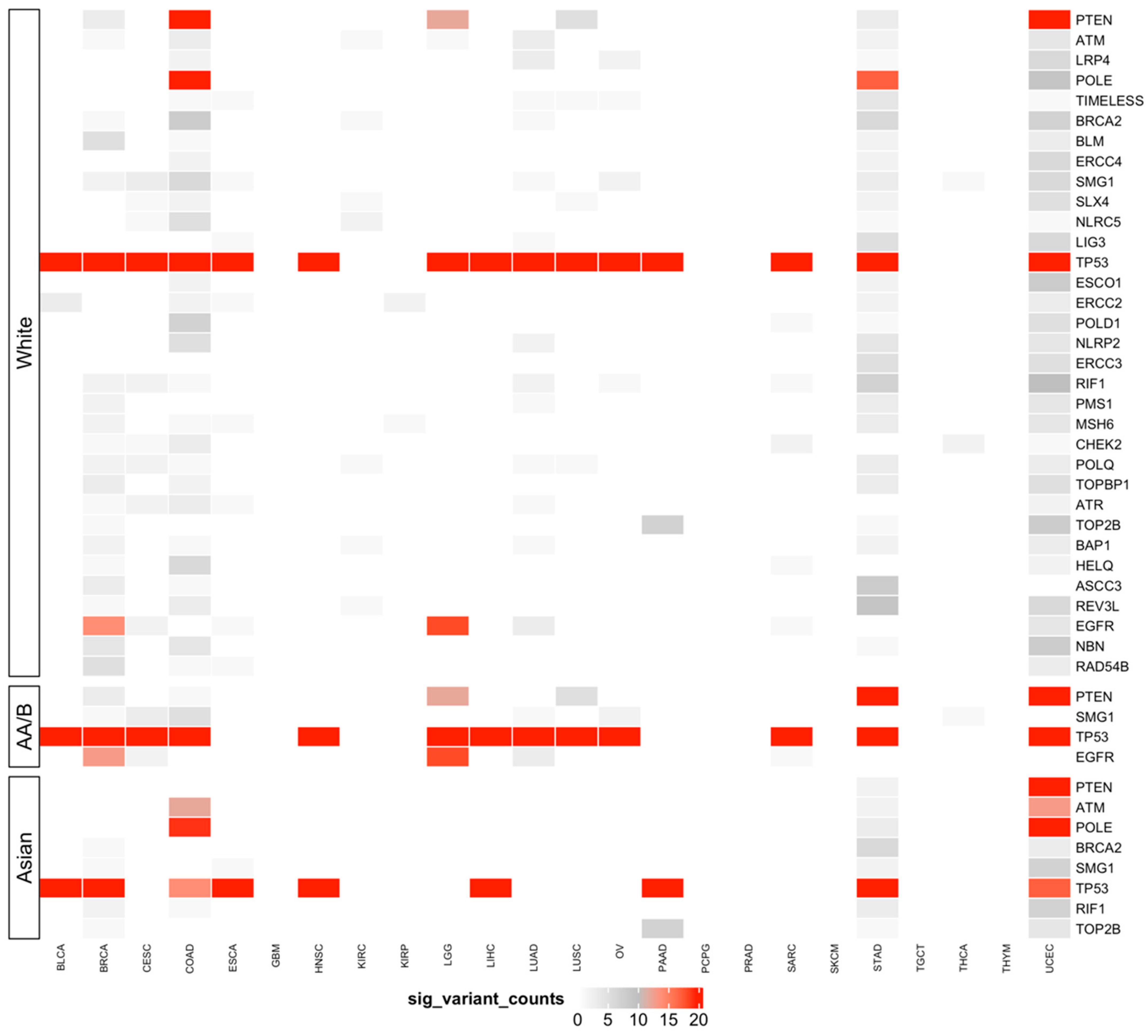
3.5. Predisposing HRD-Related Genes That Are Specific to Race
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef]
- Thompson, L.H.; Schild, D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. Fundamen. Mol. Mech. Mutagenes. 2001, 477, 131–153. [Google Scholar] [CrossRef]
- Abkevich, V.; Timms, K.; Hennessy, B.; Potter, J.; Carey, M.; Meyer, L.; Smith-McCune, K.; Broaddus, R.; Lu, K.; Chen, J. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 2012, 107, 1776–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkbak, N.J.; Wang, Z.C.; Kim, J.-Y.; Eklund, A.C.; Li, Q.; Tian, R.; Bowman-Colin, C.; Li, Y.; Greene-Colozzi, A.; Iglehart, J.D. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012, 2, 366–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, T.; Manié, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, J.E.; Gourley, C.; Powell, C.B.; High, H.; Shapira-Frommer, R.; Castonguay, V.; De Greve, J.; Atkinson, T.; Yap, T.A.; Sandhu, S. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: A multi-institutional study. Clin. Cancer Res. 2013, 19, 5485–5493. [Google Scholar] [CrossRef] [Green Version]
- Audeh, M.W.; Carmichael, J.; Penson, R.T.; Friedlander, M.; Powell, B.; Bell-McGuinn, K.M.; Scott, C.; Weitzel, J.N.; Oaknin, A.; Loman, N. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 2010, 376, 245–251. [Google Scholar] [CrossRef]
- Tutt, A.; Robson, M.; Garber, J.E.; Domchek, S.M.; Audeh, M.W.; Weitzel, J.N.; Friedlander, M.; Arun, B.; Loman, N.; Schmutzler, R.K. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 2010, 376, 235–244. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N. DNA-repair defects and olaparib in metastatic prostate cancer. N. Eng. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef]
- Yarchoan, M.; Myzak, M.C.; Johnson, B.A., III; De Jesus-Acosta, A.; Le, D.T.; Jaffee, E.M.; Azad, N.S.; Donehower, R.C.; Zheng, L.; Oberstein, P.E. Olaparib in combination with irinotecan, cisplatin, and mitomycin C in patients with advanced pancreatic cancer. Oncotarget 2017, 8, 44073. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2016, 16, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Ellis, L.; Canchola, A.J.; Spiegel, D.; Ladabaum, U.; Haile, R.; Gomez, S.L. Racial and ethnic disparities in cancer survival: The contribution of tumor, sociodemographic, institutional, and neighborhood characteristics. J. Clin. Oncol. 2018, 36, 25. [Google Scholar] [CrossRef]
- Siegel, R.; Miller, K.; Fuchs, H.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, B.C.; Dotto, G.-P. Racial differences in cancer susceptibility and survival: More than the color of the skin? Trends Cancer 2017, 3, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Wallace, T.A.; Martin, D.N.; Ambs, S. Interactions among genes, tumor biology and the environment in cancer health disparities: Examining the evidence on a national and global scale. Carcinogenesis 2011, 32, 1107–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, B.; Olopade, O.I. A perfect storm: How tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J. Clin. 2015, 65, 221–238. [Google Scholar] [CrossRef] [Green Version]
- Conti, D.V.; Darst, B.F.; Moss, L.C.; Saunders, E.J.; Sheng, X.; Chou, A.; Schumacher, F.R.; Al Olama, A.A.; Benlloch, S.; Dadaev, T. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021, 53, 65–75. [Google Scholar] [CrossRef]
- Sinha, S.; Mitchell, K.A.; Zingone, A.; Bowman, E.; Sinha, N.; Schäffer, A.A.; Lee, J.S.; Ruppin, E.; Ryan, B.M. Higher prevalence of homologous recombination deficiency in tumors from African Americans versus European Americans. Nat. Cancer 2020, 1, 112–121. [Google Scholar] [CrossRef] [Green Version]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [Green Version]
- Syed, Y.Y. Oncotype DX Breast Recurrence Score®: A Review of its Use in Early-Stage Breast Cancer. Mol. Diagn. Ther. 2020, 24, 621–632. [Google Scholar] [CrossRef]
- Slodkowska, E.A.; Ross, J.S. MammaPrint™ 70-gene signature: Another milestone in personalized medical care for breast cancer patients. Expert Rev. Mol. Diagn. 2009, 9, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Balasubramaniam, S.; Zhang, H.; Berman, T.; Narayan, P.; Suzman, D.; Bloomquist, E.; Tang, S.; Gong, Y.; Sridhara, R. FDA approval summary: Olaparib monotherapy or in combination with Bevacizumab for the maintenance treatment of patients with advanced ovarian cancer. Oncologist 2021, 26, e164–e172. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Takahama, T.; Sakai, K.; Shimizu, S.; Watanabe, S.; Kawakami, H.; Tanaka, K.; Sato, C.; Hayashi, H.; Nonagase, Y. Clinical Application of the FoundationOne CDx Assay to Therapeutic Decision-Making for Patients with Advanced Solid Tumors. Oncologist 2021, 26, e588–e596. [Google Scholar] [CrossRef]
- Tung, N.M.; Garber, J.E. BRCA 1/2 testing: Therapeutic implications for breast cancer management. Br. J. Cancer 2018, 119, 141–152. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N. Eng. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Absher, D.M.; Tang, H.; Southwick, A.M.; Casto, A.M.; Ramachandran, S.; Cann, H.M.; Barsh, G.S.; Feldman, M.; Cavalli-Sforza, L.L. Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008, 319, 1100–1104. [Google Scholar] [CrossRef]
- Barbujani, G.; Colonna, V. Human genome diversity: Frequently asked questions. Trends Genet. 2010, 26, 285–295. [Google Scholar] [CrossRef]
- Phan, V.H.; Tan, C.; Rittau, A.; Xu, H.; McLachlan, A.J.; Clarke, S.J. An update on ethnic differences in drug metabolism and toxicity from anti-cancer drugs. Expert Opin. Drug Metab. Toxicol. 2011, 7, 1395–1410. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, J.M.; Hill, L.A.; Deharo, S.; Irwin, G.; Boyle, D.; Keating, K.E.; Raji, O.Y.; McDyer, F.A.; O’Brien, E.; Bylesjo, M. Identification and validation of an anthracycline/cyclophosphamide–based chemotherapy response assay in breast cancer. J. Natl. Cancer Inst. 2014, 106, djt335. [Google Scholar] [CrossRef] [Green Version]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Wu, M.C.; Lin, X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012, 13, 762–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blumhagen, R.Z.; Schwartz, D.A.; Langefeld, C.D.; Fingerlin, T.E. Identification of Influential Variants in Significant Aggregate Rare Variant Tests. Hum. Hered. 2020, 85, 11–23. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef] [Green Version]
- Iacocca, M.A.; Chora, J.R.; Carrié, A.; Freiberger, T.; Leigh, S.E.; Defesche, J.C.; Kurtz, C.L.; DiStefano, M.T.; Santos, R.D.; Humphries, S.E. ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum. Mutat. 2018, 39, 1631–1640. [Google Scholar] [CrossRef]
- Sztupinszki, Z.; Diossy, M.; Krzystanek, M.; Reiniger, L.; Csabai, I.; Favero, F.; Birkbak, N.J.; Eklund, A.C.; Syed, A.; Szallasi, Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer 2018, 4, 1–4. [Google Scholar] [CrossRef]
- Kassambara, A.; Kassambara, M.A. Package ‘ggpubr’. 2020. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 2 June 2021).
- Ahlmann-Eltze, C.; Patil, I. ggsignif: R Package for Displaying Significance Brackets for ‘ggplot2’. 2021. Available online: https://cran.r-project.org/web/packages/ggsignif/vignettes/intro.html (accessed on 2 June 2021).
- Da Costa, A.A.; Do Canto, L.M.; Larsen, S.J.; Ribeiro, A.R.G.; Stecca, C.E.; Petersen, A.H.; Aagaard, M.M.; De Brot, L.; Baumbach, J.; Baiocchi, G. Genomic profiling in ovarian cancer retreated with platinum based chemotherapy presented homologous recombination deficiency and copy number imbalances of CCNE1 and RB1 genes. BMC Cancer 2019, 19, 422. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T.M.; Lumley, T. Package ‘survival’. R Top Doc 2015, 128, 28–33. [Google Scholar]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S.; Package ‘survminer’. Drawing Survival Curves Using ‘ggplot2’ (R Package Version 03 1). 2017. Available online: https://cran.r-project.org/web/packages/survminer/index.html (accessed on 2 June 2021).
- Jiang, Y.; Dang, S.; Yang, L.; Han, Y.; Zhang, Y.; Mu, T.; Chen, S.; Kong, F.-M. Association between homologous recombination deficiency and tumor mutational burden in lung cancer. J. Clin. Oncol. 2020, 38. [Google Scholar] [CrossRef]
- Raymond, C.; Hernandez, J.; Brobey, R.; Wang, Y.; Potts, K.; Garg, K.; Lim, L.; DiPasquo, D.; Li, M.; Dehghani, M. Detection of HRD gene mutations and copy number changes in cfDNA from prostate cancer patients. J. Clin. Oncol. 2017, 35. [Google Scholar] [CrossRef]
- Chao, A.; Lai, C.-H.; Wang, T.-H.; Jung, S.-M.; Lee, Y.-S.; Chang, W.-Y.; Yang, L.-Y.; Ku, F.-C.; Huang, H.-J.; Chao, A.-S. Genomic scar signatures associated with homologous recombination deficiency predict adverse clinical outcomes in patients with ovarian clear cell carcinoma. J. Mol. Med. 2018, 96, 527–536. [Google Scholar] [CrossRef]
- Patel, J.N.; Braicu, I.; Timms, K.M.; Solimeno, C.; Tshiaba, P.; Reid, J.; Lanchbury, J.S.; Darb-Esfahani, S.; Ganapathi, M.K.; Sehouli, J. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br. J. Cancer 2018, 119, 1060–1066. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ung, M.H.; Cantor, S.; Cheng, C. Computational investigation of homologous recombination DNA repair deficiency in sporadic breast cancer. Sci. Rep. 2017, 7, 15742. [Google Scholar] [CrossRef] [Green Version]
- Byun, J.S.; Singhal, S.K.; Park, S.; Yi, D.I.; Yan, T.; Caban, A.; Jones, A.; Mukhopadhyay, P.; Gil, S.M.; Hewitt, S.M. Racial differences in the association between luminal master regulator gene expression levels and breast cancer survival. Clin. Cancer Res. 2020, 26, 1905–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, J.; Sicotte, H.; Fan, J.-B.; Humphray, S.; Cunningham, J.M.; Kalli, K.R.; Oberg, A.L.; Hart, S.N.; Li, Y.; Davila, J.I. TP53 mutations, tetraploidy and homologous recombination repair defects in early stage high-grade serous ovarian cancer. Nucleic Acids Res. 2015, 43, 6945–6958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Cui, L.; Zhang, B.; Zhao, X.; Xu, B. Efficacy of platinum-based chemotherapy in advanced triple-negative breast cancer in association with homologous recombination deficiency. J. Clin. Oncol. 2021, 39. [Google Scholar] [CrossRef]
- Vallböhmer, D.; Lenz, H.-J. Epidermal growth factor receptor as a target for chemotherapy. Clin. Colorectal Cancer 2005, 5, S19–S27. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.-G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIß-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef]
- Jin, M.H.; Oh, D.-Y. ATM in DNA repair in cancer. Pharmacol. Ther. 2019, 203, 107391. [Google Scholar] [CrossRef] [PubMed]
- Shahriyari, L.; Abdel-Rahman, M.; Cebulla, C. BAP1 expression is prognostic in breast and uveal melanoma but not colon cancer and is highly positively correlated with RBM15B and USP19. PLoS ONE 2019, 14, e0211507. [Google Scholar] [CrossRef] [Green Version]
- Helgason, H.; Rafnar, T.; Olafsdottir, H.S.; Jonasson, J.G.; Sigurdsson, A.; Stacey, S.N.; Jonasdottir, A.; Tryggvadottir, L.; Alexiusdottir, K.; Haraldsson, A. Loss-of-function variants in ATM confer risk of gastric cancer. Nat. Genet. 2015, 47, 906–910. [Google Scholar] [CrossRef]
- Cai, H.; Jing, C.; Chang, X.; Ding, D.; Han, T.; Yang, J.; Lu, Z.; Hu, X.; Liu, Z.; Wang, J. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J. Trans. Med. 2019, 17, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cybulski, C.; Kluźniak, W.; Huzarski, T.; Wokołorczyk, D.; Kashyap, A.; Rusak, B.; Stempa, K.; Gronwald, J.; Szymiczek, A.; Bagherzadeh, M. The spectrum of mutations predisposing to familial breast cancer in Poland. Int. J. Cancer 2019, 145, 3311–3320. [Google Scholar] [CrossRef]
- Doherty, J.A.; Weiss, N.S.; Fish, S.; Fan, W.; Loomis, M.M.; Sakoda, L.C.; Rossing, M.A.; Zhao, L.P.; Chen, C. Polymorphisms in nucleotide excision repair genes and endometrial cancer risk. Cancer Epidemiol. Prev. Biomark. 2011, 20, 1873–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpińska-Kaczmarczyk, K.; Lewandowska, M.; Ławniczak, M.; Białek, A.; Urasińska, E. Expression of mismatch repair proteins in early and advanced gastric cancer in Poland. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 2886. [Google Scholar] [CrossRef] [Green Version]
- Nemtsova, M.V.; Kalinkin, A.I.; Kuznetsova, E.B.; Bure, I.V.; Alekseeva, E.A.; Bykov, I.I.; Khorobrykh, T.V.; Mikhaylenko, D.S.; Tanas, A.S.; Kutsev, S.I. Clinical relevance of somatic mutations in main driver genes detected in gastric cancer patients by next-generation DNA sequencing. Sci. Rep. 2020, 10, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, D.C. Median absolute deviation. In Encyclopedia of Statistics in Behavioral Science; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Sherman, R.M.; Forman, J.; Antonescu, V.; Puiu, D.; Daya, M.; Rafaels, N.; Boorgula, M.P.; Chavan, S.; Vergara, C.; Ortega, V.E. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 2019, 51, 30–35. [Google Scholar] [CrossRef]

| Asian | African American/Black | White | Total | |
|---|---|---|---|---|
| BLCA | 42 | 22 | 319 | 383 |
| BRCA | 59 | 159 | 678 | 896 |
| CESC | 18 | 27 | 184 | 229 |
| COAD | 11 | 55 | 184 | 250 |
| ESCA | 44 | 0 | 104 | 148 |
| GBM | 0 | 0 | 111 | 111 |
| HNSC | 10 | 46 | 414 | 470 |
| KIRC | 0 | 52 | 287 | 339 |
| KIRP | 0 | 59 | 190 | 249 |
| LGG | 0 | 21 | 466 | 487 |
| LIHC | 150 | 16 | 171 | 337 |
| LUAD | 0 | 52 | 376 | 428 |
| LUSC | 0 | 26 | 325 | 351 |
| OV | 0 | 18 | 146 | 164 |
| PAAD | 11 | 0 | 132 | 143 |
| PCPG | 0 | 19 | 131 | 150 |
| PRAD | 0 | 0 | 143 | 143 |
| SARC | 0 | 17 | 202 | 219 |
| SKCM | 0 | 0 | 339 | 339 |
| STAD | 75 | 11 | 245 | 331 |
| TGCT | 0 | 0 | 112 | 112 |
| THCA | 49 | 26 | 315 | 390 |
| THYM | 12 | 0 | 98 | 110 |
| UCEC | 20 | 101 | 341 | 462 |
| Total | 501 (6.92%) | 727 (10.04%) | 6013 (83.04%) | 7241 (100%) |
| Positive Correlation | Inverse Correlation |
|---|---|
| OV (R = 0.31) | COAD (R = −0.24) |
| LUSC (R = 0.33) | UCEC (R = −0.32) |
| BLCA (R = 0.38) | |
| PAAD (R = 0.54) | |
| LUAD (R = 0.49) | |
| SARC (R = 0.5) | |
| BRCA (R = 0.54) | |
| THYM (R = 0.32) | |
| KIRC (R = 0.25) | |
| HNSC (R = 0.23) | |
| PRAD (R = 0.39) | |
| LGG (R = 0.34) | |
| PCPG (R = 0.27) | |
| Pan-cancer (R = 0.26) |
| White | African American/Black | Asian | |
|---|---|---|---|
| BLCA | 18 | 51 | 11 |
| BRCA | 135 | 97 | 56 |
| CESC | 48 | 41 | 2 |
| COAD | 200 | 9 | 160 |
| ESCA | 52 | 0 | 52 |
| HNSC | 13 | 20 | 14 |
| KIRC | 27 | 27 | 0 |
| KIRP | 9 | 9 | 0 |
| LGG | 17 | 17 | 0 |
| LIHC | 6 | 13 | 4 |
| LUAD | 118 | 118 | 0 |
| LUSC | 52 | 52 | 0 |
| OV | 18 | 18 | 0 |
| PAAD | 57 | 0 | 57 |
| SARC | 25 | 25 | 0 |
| STAD | 206 | 20 | 191 |
| THCA | 10 | 3 | 7 |
| THYM | 2 | 0 | 2 |
| UCEC | 891 | 43 | 213 |
| Cancer | Gene | Population | Previous Reports | Identified Variant (GRCh38) | REVEL Score 1 | Clinvar 2 | |
|---|---|---|---|---|---|---|---|
| Same Population | Other Population | ||||||
| BRCA | BAP1 | White | Shahriyari et al. (2019) | 3:52442072-T/C | 0.829 | pathogenic | |
| African American/Black | |||||||
| BLM | White | Cybulski et al. (2019) | 15:91312388-C/T | 0.677 | uncertain significance | ||
| African American/Black | |||||||
| OV | ERCC5 | White | Doherty et al. (2011) | 3:103525633-G/T | 0.939 | pathogenic | |
| African American/Black | Doherty et al. (2011) | ||||||
| STAD | ATM | White | Helgason et al. (2015) | 11:108141997-C/T | 0.739 | uncertain significance | |
| Asian | Cai et al. (2015) | ||||||
| MSH6 | White | Karpińska-Kaczmarczyk et al. (2016) | 2:48028049-G/A | 0.857 | uncertain significance | ||
| Asian | |||||||
| PTEN | African American/Black | Nemtsova et al. (2020) | 10:89692905-G/A | 0.976 | likely pathogenic, pathogenic | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsiao, Y.-W.; Lu, T.-P. Race-Specific Genetic Profiles of Homologous Recombination Deficiency in Multiple Cancers. J. Pers. Med. 2021, 11, 1287. https://doi.org/10.3390/jpm11121287
Hsiao Y-W, Lu T-P. Race-Specific Genetic Profiles of Homologous Recombination Deficiency in Multiple Cancers. Journal of Personalized Medicine. 2021; 11(12):1287. https://doi.org/10.3390/jpm11121287
Chicago/Turabian StyleHsiao, Yi-Wen, and Tzu-Pin Lu. 2021. "Race-Specific Genetic Profiles of Homologous Recombination Deficiency in Multiple Cancers" Journal of Personalized Medicine 11, no. 12: 1287. https://doi.org/10.3390/jpm11121287
APA StyleHsiao, Y.-W., & Lu, T.-P. (2021). Race-Specific Genetic Profiles of Homologous Recombination Deficiency in Multiple Cancers. Journal of Personalized Medicine, 11(12), 1287. https://doi.org/10.3390/jpm11121287






