Evaluation of Cytokines as Robust Diagnostic Biomarkers for COVID-19 Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Patient Selection
2.2. Biological Samples
2.3. Cytokines and Chemokines Analysis and Interpretation
2.4. Variables
2.5. Ethics Approval
2.6. Statistical Analysis
3. Results
3.1. Demographics
3.2. Discovery Cohort
3.3. Validation Cohort
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 7 December 2020).
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet Lond. Engl. 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- CDC Coronavirus Disease 2019 (COVID-19). Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html (accessed on 7 December 2020).
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical Course and Outcomes of Critically Ill Patients with SARS-CoV-2 Pneumonia in Wuhan, China: A Single-Centered, Retrospective, Observational Study. Lancet Respir. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Ponti, G.; Maccaferri, M.; Ruini, C.; Tomasi, A.; Ozben, T. Biomarkers Associated with COVID-19 Disease Progression. Crit. Rev. Clin. Lab. Sci. 2020, 57, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, Biochemical and Immune Biomarker Abnormalities Associated with Severe Illness and Mortality in Coronavirus Disease 2019 (COVID-19): A Meta-Analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, S.; Kay, F.U.; Abbara, S.; Bhalla, S.; Chung, J.H.; Chung, M.; Henry, T.S.; Kanne, J.P.; Kligerman, S.; Ko, J.P.; et al. Radiological Society of North America Expert Consensus Statement on Reporting Chest CT Findings Related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA—Secondary Publication. J. Thorac. Imaging 2020, 35, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.Y.F.; Lam, H.Y.S.; Fong, A.H.-T.; Leung, S.T.; Chin, T.W.-Y.; Lo, C.S.Y.; Lui, M.M.-S.; Lee, J.C.Y.; Chiu, K.W.-H.; Chung, T.W.-H.; et al. Frequency and Distribution of Chest Radiographic Findings in Patients Positive for COVID-19. Radiology 2020, 296, E72–E78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickel, C.H.; Bingisser, R. Mimics and Chameleons of COVID-19. Swiss Med. Wkly. 2020, 150. [Google Scholar] [CrossRef] [PubMed]
- Lascarrou, J.-B.; Colin, G.; Le Thuaut, A.; Serck, N.; Ohana, M.; Sauneuf, B.; Geri, G.; Mesland, J.-B.; Ribeyre, G.; Hussenet, C.; et al. Predictors of Negative First SARS-CoV-2 RT-PCR despite Final Diagnosis of COVID-19 and Association with Outcome. Sci. Rep. 2021, 11, 2388. [Google Scholar] [CrossRef]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef]
- Kinloch, N.N.; Ritchie, G.; Brumme, C.J.; Dong, W.; Dong, W.; Lawson, T.; Jones, R.B.; Montaner, J.S.G.; Leung, V.; Romney, M.G.; et al. Suboptimal Biological Sampling as a Probable Cause of False-Negative COVID-19 Diagnostic Test Results. J. Infect. Dis. 2020, 222, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Harahwa, T.A.; Lai Yau, T.H.; Lim-Cooke, M.-S.; Al-Haddi, S.; Zeinah, M.; Harky, A. The Optimal Diagnostic Methods for COVID-19. Diagn. Berl. Ger. 2020, 7, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Halalau, A.; Imam, Z.; Karabon, P.; Mankuzhy, N.; Shaheen, A.; Tu, J.; Carpenter, C. External Validation of a Clinical Risk Score to Predict Hospital Admission and In-Hospital Mortality in COVID-19 Patients. Ann. Med. 2021, 53, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Wynants, L.; Calster, B.V.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.A.; Debray, T.P.A.; et al. Prediction Models for Diagnosis and Prognosis of Covid-19: Systematic Review and Critical Appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; He, Y.; Yang, H.; Yu, H.; Wang, T.; Chen, Z.; Yao, R.; Liang, Z. Development and Validation a Nomogram for Predicting the Risk of Severe COVID-19: A Multi-Center Study in Sichuan, China. PLoS ONE 2020, 15, e0233328. [Google Scholar] [CrossRef]
- Haimovich, A.D.; Ravindra, N.G.; Stoytchev, S.; Young, H.P.; Wilson, F.P.; van Dijk, D.; Schulz, W.L.; Taylor, R.A. Development and Validation of the Quick COVID-19 Severity Index: A Prognostic Tool for Early Clinical Decompensation. Ann. Emerg. Med. 2020, 76, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Yang, P.; Xie, Y.; Woodruff, H.C.; Rao, X.; Guiot, J.; Frix, A.-N.; Louis, R.; Moutschen, M.; Li, J.; et al. Development of a Clinical Decision Support System for Severity Risk Prediction and Triage of COVID-19 Patients at Hospital Admission: An International Multicentre Study. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 Are Disease Severity Predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef]
- Meizlish, M.L.; Pine, A.B.; Bishai, J.D.; Goshua, G.; Nadelmann, E.R.; Simonov, M.; Chang, C.-H.; Zhang, H.; Shallow, M.; Bahel, P.; et al. A Neutrophil Activation Signature Predicts Critical Illness and Mortality in COVID-19. Blood Adv. 2021, 5, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and Immunologic Features in Severe and Moderate Coronavirus Disease 2019. J. Clin. Investig. 2020. [Google Scholar] [CrossRef] [Green Version]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False Negative of RT-PCR and Prolonged Nucleic Acid Conversion in COVID-19: Rather than Recurrence. J. Med. Virol. 2020, 92, 1755–1756. [Google Scholar] [CrossRef] [Green Version]
- Ducray, V.; Vlachomitrou, A.S.; Bouscambert-Duchamp, M.; Si-Mohamed, S.; Gouttard, S.; Mansuy, A.; Wickert, F.; Sigal, A.; Gaymard, A.; Talbot, F.; et al. Chest CT for Rapid Triage of Patients in Multiple Emergency Departments during COVID-19 Epidemic: Experience Report from a Large French University Hospital. Eur. Radiol. 2020. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus Disease (COVID-19): RRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961. [Google Scholar] [CrossRef]
- Sun, Y.; Koh, V.; Marimuthu, K.; Ng, O.T.; Young, B.; Vasoo, S.; Chan, M.; Lee, V.J.M.; De, P.P.; Barkham, T.; et al. Epidemiological and Clinical Predictors of COVID-19. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 786–792. [Google Scholar] [CrossRef] [Green Version]
- Wan, S.; Yi, Q.; Fan, S.; Lv, J.; Zhang, X.; Guo, L.; Lang, C.; Xiao, Q.; Xiao, K.; Yi, Z.; et al. Relationships among Lymphocyte Subsets, Cytokines, and the Pulmonary Inflammation Index in Coronavirus (COVID-19) Infected Patients. Br. J. Haematol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qin, L.; Zhang, P.; Li, K.; Liang, L.; Sun, J.; Xu, B.; Dai, Y.; Li, X.; Zhang, C.; et al. Longitudinal COVID-19 Profiling Associates IL-1RA and IL-10 with Disease Severity and RANTES with Mild Disease. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal Characteristics of Lymphocyte Responses and Cytokine Profiles in the Peripheral Blood of SARS-CoV-2 Infected Patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Liu, C.; Su, L.; Zhang, D.; Fan, J.; Yang, Y.; Xiao, M.; Xie, J.; Xu, Y.; et al. IP-10 and MCP-1 as Biomarkers Associated with Disease Severity of COVID-19. Mol. Med. 2020, 26, 97. [Google Scholar] [CrossRef]
- Cannas, A.; Calvo, L.; Chiacchio, T.; Cuzzi, G.; Vanini, V.; Lauria, F.N.; Pucci, L.; Girardi, E.; Goletti, D. IP-10 Detection in Urine Is Associated with Lung Diseases. BMC Infect. Dis. 2010, 10, 333. [Google Scholar] [CrossRef] [Green Version]
- Rhoades, E.R.; Cooper, A.M.; Orme, I.M. Chemokine Response in Mice Infected with Mycobacterium Tuberculosis. Infect. Immun. 1995, 63, 3871–3877. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Le, T.Q.; Kurihara, N.; Chida, J.; Cisse, Y.; Yano, M.; Kido, H. Influenza Virus-Cytokine-Protease Cycle in the Pathogenesis of Vascular Hyperpermeability in Severe Influenza. J. Infect. Dis. 2010, 202, 991–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Shen, C.; Li, J.; Yuan, J.; Wei, J.; Huang, F.; Wang, F.; Li, G.; Li, Y.; Xing, L.; et al. Plasma IP-10 and MCP-3 Levels Are Highly Associated with Disease Severity and Predict the Progression of COVID-19. J. Allergy Clin. Immunol. 2020, 146, 119–127.e4. [Google Scholar] [CrossRef] [PubMed]
- Agostini, C.; Facco, M.; Siviero, M.; Carollo, D.; Galvan, S.; Cattelan, A.M.; Zambello, R.; Trentin, L.; Semenzato, G. CXC Chemokines IP-10 and Mig Expression and Direct Migration of Pulmonary CD8+/CXCR3+ T Cells in the Lungs of Patients with HIV Infection and T-Cell Alveolitis. Am. J. Respir. Crit. Care Med. 2000, 162, 1466–1473. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, P.; Zhong, Y.; Zhao, Z.; Xing, L.; Zhao, Y.; Zou, Z.; Zhang, Y.; Li, C.; Li, T.; et al. Monoclonal Antibody against CXCL-10/IP-10 Ameliorates Influenza A (H1N1) Virus Induced Acute Lung Injury. Cell Res. 2013, 23, 577–580. [Google Scholar] [CrossRef]

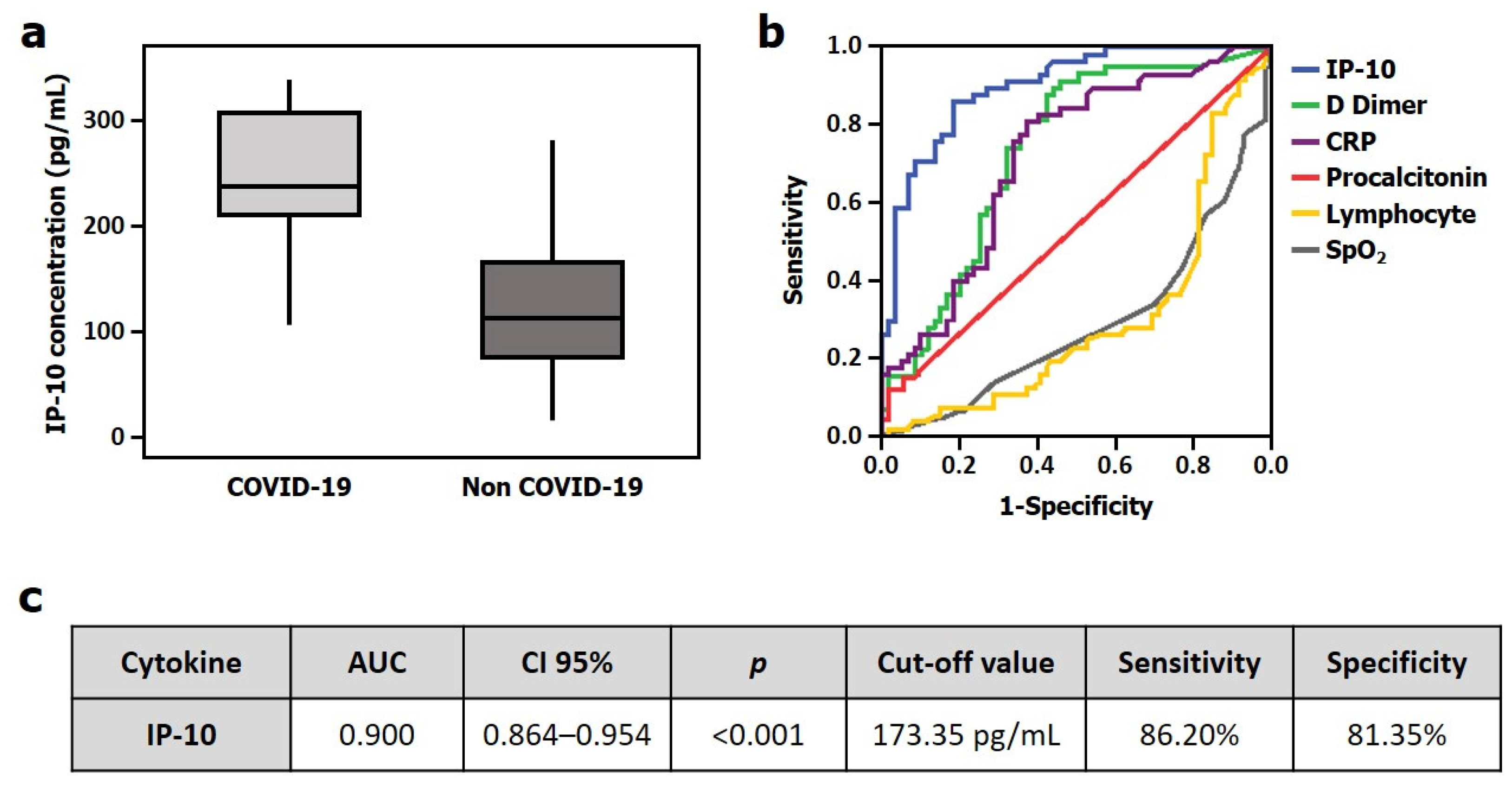

| Discovery Cohort | Validation Cohort | |||||
|---|---|---|---|---|---|---|
| COVID-19 (n = 108) | NON-COVID-19 (n = 28) | p | COVID-19 (n = 58) | NON-COVID-19 (n = 59) | p | |
| Age (years; median (IQR)) | 69 (20) | 71 (37) | 0.296 | 77.5 (19) | 59 (36) | <0.001 |
| Male (%(n)) | 55.3 (73) | 57.1 (16) | 0.722 | 44.8 (26) | 44.1 (26) | 0.934 |
| -Comorbidities (%(n)) | ||||||
| Use of tobacco | 8.3 (9) | 10.7 (3) | 0.692 | 1.7 (1) | 10.2 (6) | 0.154 |
| Use of alcohol | 2.8 (3) | 7.1 (2) | 0.274 | 1.7 (1) | 5.1 (3) | 0.317 |
| Hypertension | 46.3 (50) | 50 (14) | 0.492 | 50 (29) | 28.8 (17) | 0.019 |
| Cardicac disease | 13. 0 (14) | 21.4 (6) | 0.815 | 13.8 (8) | 8.5 (5) | 0.360 |
| Diabetes | 17.6 (19) | 14.3 (4) | 0.677 | 5.2 (3) | 11.9 (7) | 0.195 |
| Neurological disease | 2.8 (3) | 3.6 (1) | 0.974 | 6.9 (4) | 6.8 (4) | 0.980 |
| Liver disease | 1.9 (2) | 3.6 (1) | 0.581 | 0 (0) | 3.4 (2) | 0.157 |
| Obesity | 9.3 (10) | 10.7 (3) | 0.770 | 10.3 (6) | 5.1 (3) | 0.286 |
| Lung disease | 16.8 (18) | 14.3 (4) | 0.944 | 6.9 (4) | 10.2 (6) | 0.527 |
| Kidney disease | 2.8 (3) | 3.6 (1) | 0.825 | 0 (0) | 0 (0) | - |
| -Laboratory (median (IQR)) | ||||||
| Glycaemia (mg/dL) | 123 (82) | 94.6 (12) | 0.051 | 109 (28) | 109 (24) | 0.391 |
| Creatinine (mg/dL) | 0.84 (0.36) | 0.8 (0.35) | 0.495 | 0.87 (0.63) | 0.78 (0.35) | 0.030 |
| Total bilirubin (mg/dL) | 0.5 (0.34) | 0.3 (0.4) | 0.06 | 0.27 (0.2) | 0.27 (0.21) | 0.696 |
| Leukocytes (×109/L) | 6.68 (3.13) | 6.87 (2.15) | <0.001 | 8.11 (6.80) | 7.76 (4.35) | 0.482 |
| Lymphocytes (×109/L) | 1.09 (0.90) | 2.25 (0.94) | <0.001 | 1.14 (0.67) | 1.6 (0.81) | <0.001 |
| Neutrophil (×109/L) | 4.46 (3.22) | 3.76 (1.34) | <0.001 | 5.18 (5.13) | 4.69 (3.59) | 0.821 |
| Platelet (×109/L) | 218 (108) | 250 (580) | 0.005 | 203 (67) | 203 (64) | 0.839 |
| D-dimer (ng/mL) | 742 (1267) | 255 (106) | <0.001 | 258 (976) | 992 (1860) | <0.001 |
| CRP (mg/L) | 80 (120) | 10 (3) | <0.001 | 32.9 (99.75) | 3.3 (50.8) | <0.001 |
| Procalcitonin (ng/mL) | 0.13 (0.26) | 0.11 (0.1) | 0.323 | 0.01 (0) | 0.01 (0) | 0.185 |
| -Hospital meters (median (IQR)) | ||||||
| Length of hospital stay | 12 (13) | 4.50 (3) | <0.001 | 11.5 (15.5) | 7 (1) | 0.320 |
| -Mortality (%(n)) | ||||||
| 28-day mortality | 18.5 (20) | 0 (0) | <0.001 | 24.1 (14) | 0 (0) | <0.001 |
| CYTOKINE | AUC | CI 95% | p | |
|---|---|---|---|---|
| 1 | IP-10 | 0.962 | 0.933–0.992 | <0.001 |
| 2 | IL1RA | 0.866 | 0.795–0.936 | <0.001 |
| 3 | PDGFBB | 0.866 | 0.800–0.931 | <0.001 |
| 4 | MCP1 | 0.839 | 0.767–0.911 | <0.001 |
| 5 | IL17a | 0.800 | 0.718–0.882 | <0.001 |
| 6 | IL15 | 0.792 | 0.703–0.880 | <0.001 |
| 7 | IL1b | 0.790 | 0.710–0.869 | <0.001 |
| 8 | HGF | 0.787 | 0.700–0.874 | <0.001 |
| 9 | IL18 | 0.787 | 0.701–0.873 | <0.001 |
| 10 | IL7 | 0.758 | 0.681–0.836 | <0.001 |
| 11 | IL2 | 0.723 | 0.639–0.807 | <0.001 |
| 12 | VEGFA | 0.705 | 0.591–0.818 | 0.001 |
| 13 | RANTES | 0.704 | 0.603–0.806 | 0.001 |
| 14 | VEGFD | 0.671 | 0.538–0.804 | 0.005 |
| 15 | IP1b | 0.666 | 0.548–0.748 | 0.060 |
| 16 | IL6 | 0.659 | 0.572–0.746 | 0.045 |
| 17 | INF | 0.329 | 0.234–0.423 | 0.048 |
| 18 | IL4 | 0.321 | 0.223–0.420 | 0.050 |
| OR | CI 95% | p | ||
|---|---|---|---|---|
| COVID-19 disease | IP-10 > 173.35 pg/mL | 25.573 | 8.127–80.469 | <0.001 |
| Age | 1.011 | 0.977–1.047 | 0.528 | |
| Hypertension | 0.806 | 0.211–3.081 | 0.753 | |
| Creatinine | 1.507 | 0.608–3.739 | 0.376 | |
| Lymphocytes | 0.999 | 0.999–1.000 | 0.235 | |
| D Dimer | 1.000 | 1.000–1.000 | 0.191 | |
| CRP | 1.001 | 0.993–1.008 | 0.897 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamayo-Velasco, Á.; Peñarrubia-Ponce, M.J.; Álvarez, F.J.; Gonzalo-Benito, H.; de la Fuente, I.; Martín-Fernández, M.; Eiros, J.M.; Martínez-Paz, P.; Miramontes-González, J.P.; Fiz-López, A.; et al. Evaluation of Cytokines as Robust Diagnostic Biomarkers for COVID-19 Detection. J. Pers. Med. 2021, 11, 681. https://doi.org/10.3390/jpm11070681
Tamayo-Velasco Á, Peñarrubia-Ponce MJ, Álvarez FJ, Gonzalo-Benito H, de la Fuente I, Martín-Fernández M, Eiros JM, Martínez-Paz P, Miramontes-González JP, Fiz-López A, et al. Evaluation of Cytokines as Robust Diagnostic Biomarkers for COVID-19 Detection. Journal of Personalized Medicine. 2021; 11(7):681. https://doi.org/10.3390/jpm11070681
Chicago/Turabian StyleTamayo-Velasco, Álvaro, María Jesús Peñarrubia-Ponce, Francisco Javier Álvarez, Hugo Gonzalo-Benito, Ignacio de la Fuente, Marta Martín-Fernández, José María Eiros, Pedro Martínez-Paz, José Pablo Miramontes-González, Aida Fiz-López, and et al. 2021. "Evaluation of Cytokines as Robust Diagnostic Biomarkers for COVID-19 Detection" Journal of Personalized Medicine 11, no. 7: 681. https://doi.org/10.3390/jpm11070681
APA StyleTamayo-Velasco, Á., Peñarrubia-Ponce, M. J., Álvarez, F. J., Gonzalo-Benito, H., de la Fuente, I., Martín-Fernández, M., Eiros, J. M., Martínez-Paz, P., Miramontes-González, J. P., Fiz-López, A., Arribas-Rodríguez, E., Cal-Sabater, P., Aller, R., Dueñas, C., Heredia-Rodríguez, M., Tamayo, E., Bernardo, D., & Gómez-Sánchez, E. (2021). Evaluation of Cytokines as Robust Diagnostic Biomarkers for COVID-19 Detection. Journal of Personalized Medicine, 11(7), 681. https://doi.org/10.3390/jpm11070681








