Detecting Blastocyst Components by Artificial Intelligence for Human Embryological Analysis to Improve Success Rate of In Vitro Fertilization
Abstract
:1. Introduction
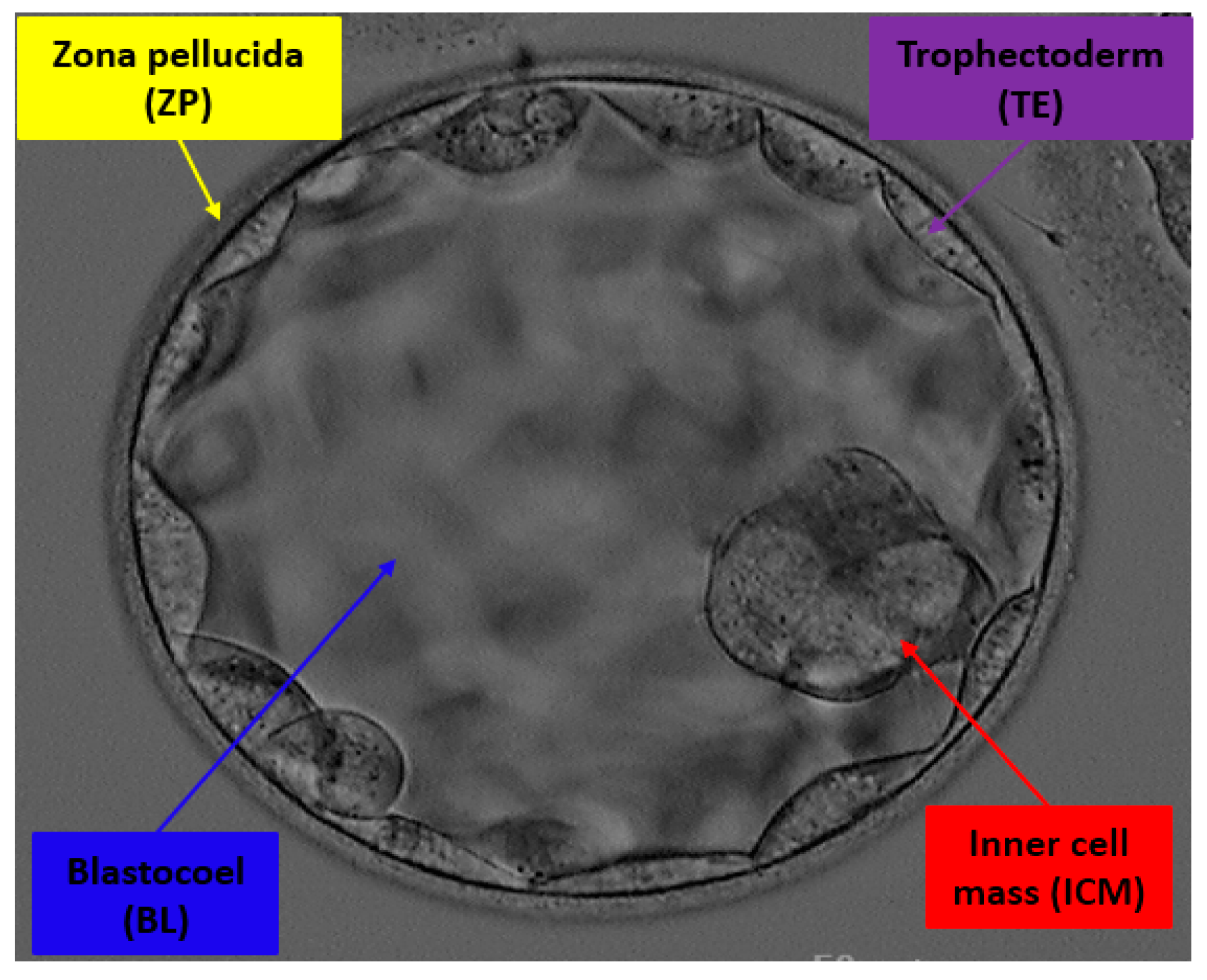
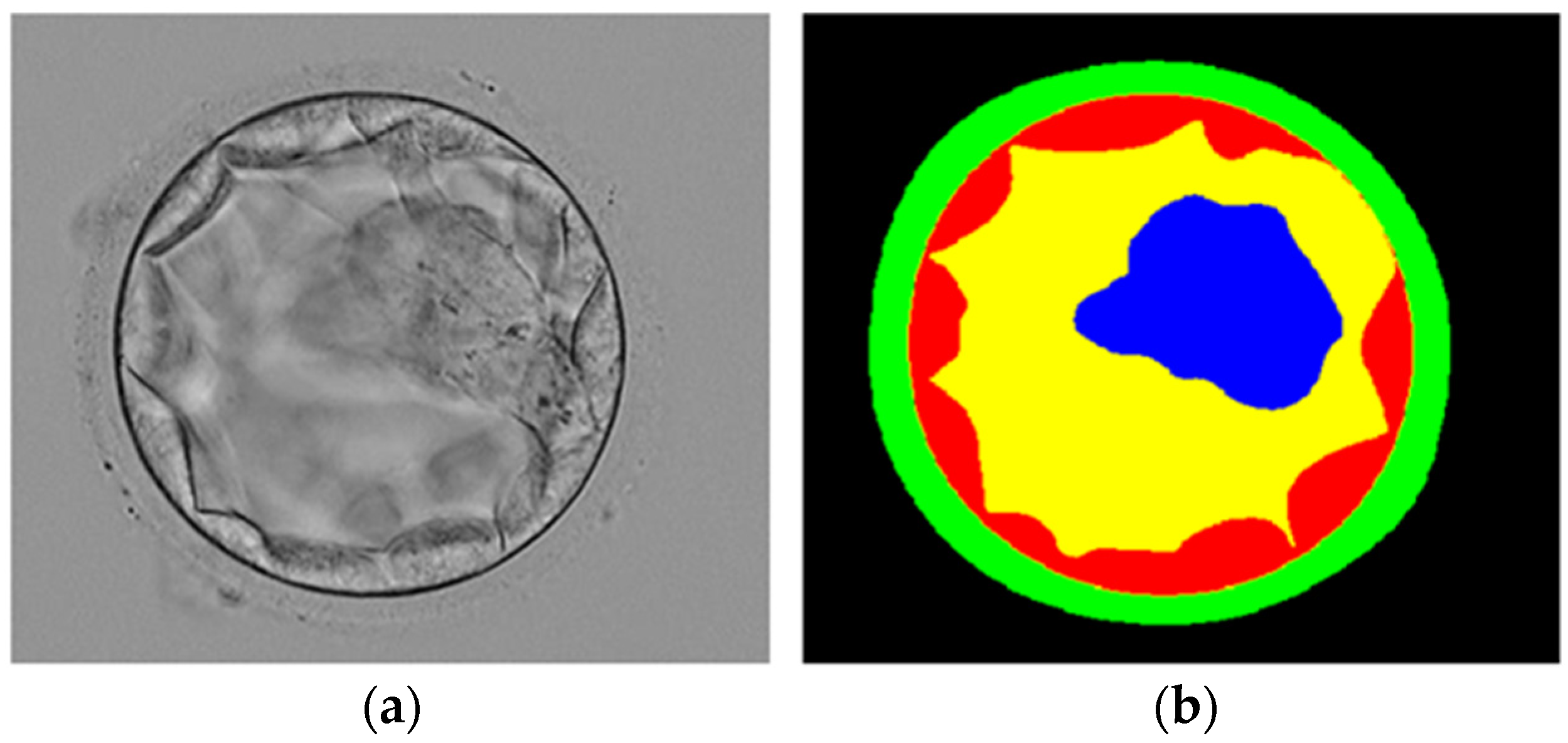
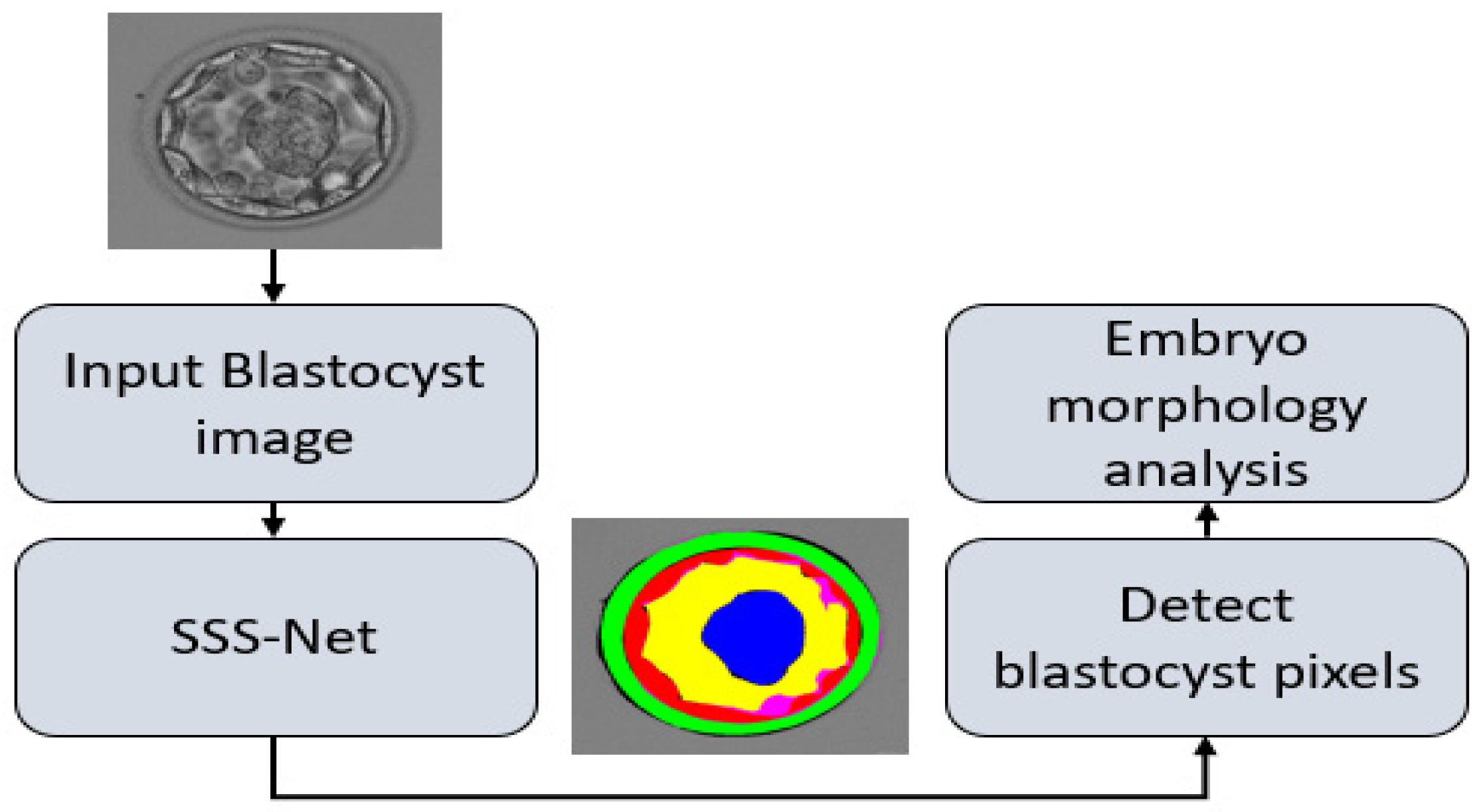
- Multiclass semantic segmentation architecture that segments ZP, TE, BL, and ICM from the background without preprocessing.
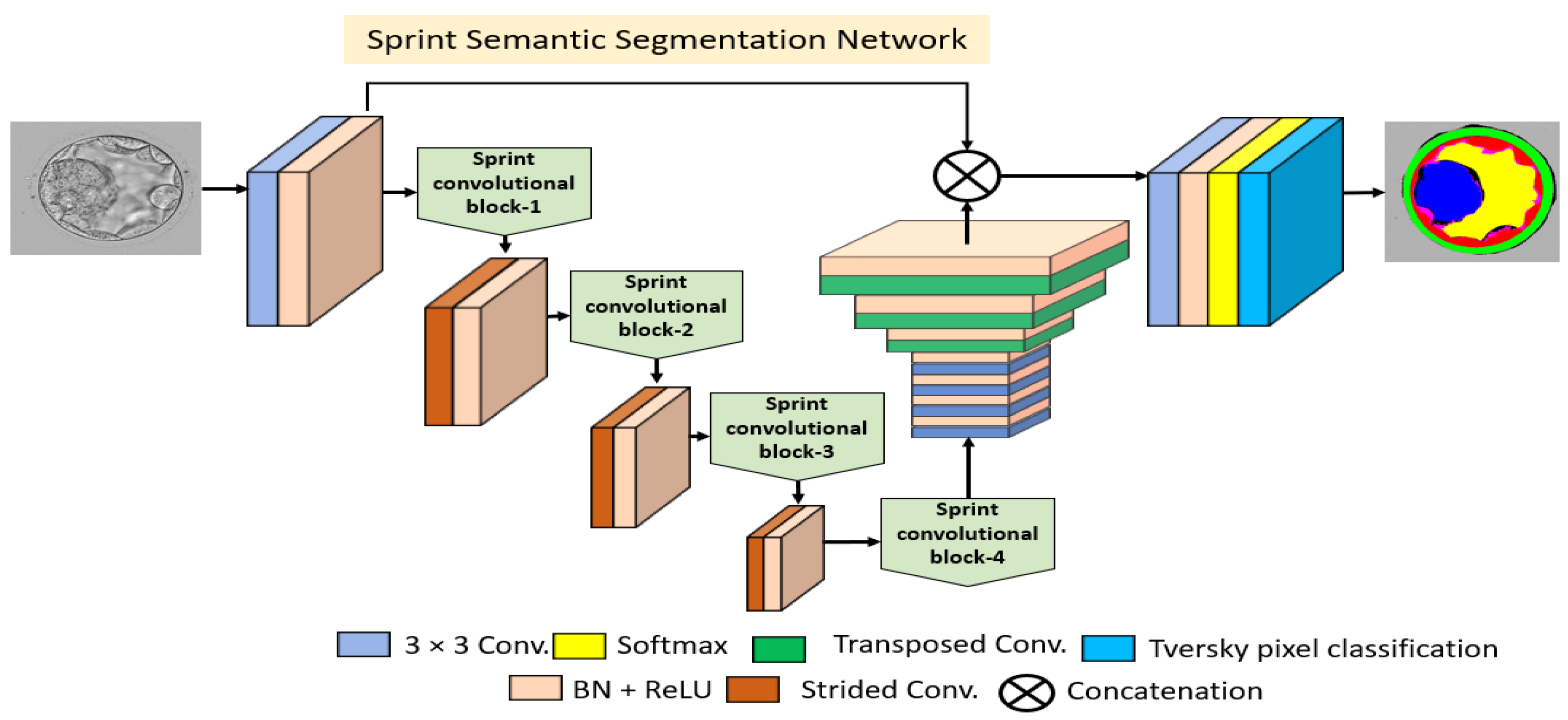
- SCB uses asymmetric kernel-based convolutions in combination with depth-wise separable convolutions to reduce floating-point operations. Low-cost shallow architecture with an overall 4.04 million trainable parameters and 28 Giga floating-point operations per second (GFLOPS).
- The SSS-Net provides high segmentation performance, and the output of the network can be used to observe morphometric properties of the blastocyst components for embryological analysis and blastocyst viability assessment.
- Our trained networks and codes are publicly available for comparison [35].
2. Material and Methods
2.1. Datasets
2.2. Method
2.2.1. Summary of Proposed Method
2.2.2. Structure of Proposed Encoder Block
2.2.3. Structure of Proposed Decoder Block
2.2.4. Experimental Environment and Data Augmentation
2.2.5. Ablation Study
3. Results
3.1. Evaluation of Proposed Method
3.2. Comparison of Proposed Method with Existing Methods
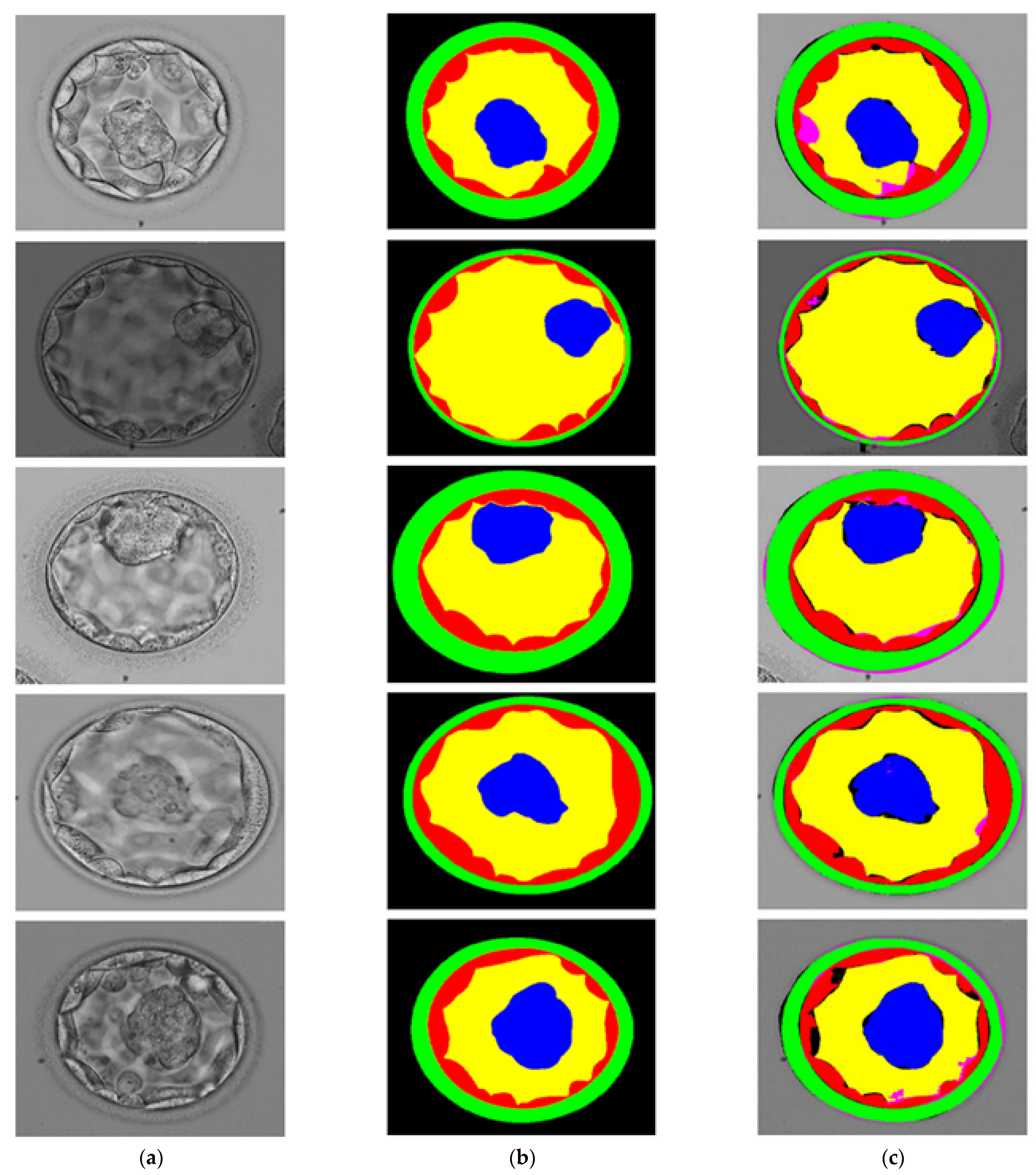
3.3. Visual Results of Proposed Method for Blastocyst Component Detection
4. Discussion
4.1. Principal Findings
4.2. Limitations and Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Purkayastha, N.; Sharma, H. Prevalence and Potential Determinants of Primary Infertility in India: Evidence from Indian Demographic Health Survey. Clin. Epidemiol. Glob. Health 2021, 9, 162–170. [Google Scholar] [CrossRef]
- Lepore, M.; Petruzziello, A. A Situation-Aware DSS to Support Assisted Reproductive Technology Outcome Prediction. In Proceedings of the IEEE Conference on Cognitive and Computational Aspects of Situation Management, Tallin, Estonia, 14–21 May 2021; pp. 103–107. [Google Scholar]
- Galic, I.; Swanson, A.; Warren, C.; Negris, O.; Bozen, A.; Brown, D.; Lawson, A.; Jain, T. Infertility in the Midwest: Perceptions and Attitudes of Current Treatment. Am. J. Obstet. Gynecol. 2021, 225, 61.e1–61.e11. [Google Scholar] [CrossRef] [PubMed]
- Zaninovic, N.; Rosenwaks, Z. Artificial Intelligence in Human in Vitro Fertilization and Embryology. Fertil. Steril. 2020, 114, 914–920. [Google Scholar] [CrossRef]
- Forman, E.J.; Hong, K.H.; Ferry, K.M.; Tao, X.; Taylor, D.; Levy, B.; Treff, N.R.; Scott, R.T. In Vitro Fertilization with Single Euploid Blastocyst Transfer: A Randomized Controlled Trial. Fertil. Steril. 2013, 100, 100–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, M.T.; Nguyen, T.T.T.; Nguyen, T.V.; Dang, H.N.T.; Nguyen, Q.H.V. Blastocyst Transfer after Extended Culture of Cryopreserved Cleavage Embryos Improves in Vitro Fertilization Cycle Outcomes. Cryobiology 2021, 100, 26–31. [Google Scholar] [CrossRef]
- Cutting, R. Single Embryo Transfer for All. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 53, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, S.; Lv, Q. Single Blastocyst Stage versus Single Cleavage Stage Embryo Transfer Following Fresh Transfer: A Systematic Review and Meta-Analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021, 267, 11–17. [Google Scholar] [CrossRef]
- Pribenszky, C.; Nilselid, A.-M.; Montag, M. Time-Lapse Culture with Morphokinetic Embryo Selection Improves Pregnancy and Live Birth Chances and Reduces Early Pregnancy Loss: A Meta-Analysis. Reprod. Biomed. Online 2017, 35, 511–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bori, L.; Dominguez, F.; Fernandez, E.I.; Del Gallego, R.; Alegre, L.; Hickman, C.; Quiñonero, A.; Nogueira, M.F.G.; Rocha, J.C.; Meseguer, M. An Artificial Intelligence Model Based on the Proteomic Profile of Euploid Embryos and Blastocyst Morphology: A Preliminary Study. Reprod. Biomed. Online 2021, 42, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, N.; Xu, Z.; Chen, L.; Zhao, X.; Zeng, H.; Jiang, Y.; Sun, H. Effects of Abnormal Zona Pellucida on Fertilization and Pregnancy in IVF/ICSI-ET. J. Reprod. Contracept. 2015, 26, 73–80. [Google Scholar] [CrossRef]
- Ozgur, K.; Berkkanoglu, M.; Bulut, H.; Donmez, L.; Isikli, A.; Coetzee, K. Blastocyst Age, Expansion, Trophectoderm Morphology, and Number Cryopreserved Are Variables Predicting Clinical Implantation in Single Blastocyst Frozen Embryo Transfers in Freeze-Only-IVF. J. Assist. Reprod. Genet. 2021, 38, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.; Palini, S.; Vento, M.E.; La Ferlita, A.; Lo Faro, M.J.; Caroppo, E.; Borzì, P.; Falzone, L.; Barbagallo, D.; Ragusa, M.; et al. Identification of Extracellular Vesicles and Characterization of MiRNA Expression Profiles in Human Blastocoel Fluid. Sci. Rep. 2019, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Y.; Yu, Y.; Zhang, X.-W. Overall Blastocyst Quality, Trophectoderm Grade, and Inner Cell Mass Grade Predict Pregnancy Outcome in Euploid Blastocyst Transfer Cycles. Chin. Med. J. 2018, 131, 1261–1267. [Google Scholar] [CrossRef]
- Sciorio, R.; Meseguer, M. Focus on Time-Lapse Analysis: Blastocyst Collapse and Morphometric Assessment as New Features of Embryo Viability. Reprod. Biomed. Online 2021, 43, 821–832. [Google Scholar] [CrossRef]
- Harada, Y.; Maeda, T.; Fukunaga, E.; Shiba, R.; Okano, S.; Kinutani, M.; Horiuchi, T. Selection of High-Quality and Viable Blastocysts Based on Timing of Morula Compaction and Blastocyst Formation. Reprod. Med. Biol. 2020, 19, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Loewke, K.E.; Bossert, N.L.; Behr, B.; De Jonge, C.J.; Baer, T.M.; Pera, R.A.R. Non-Invasive Imaging of Human Embryos before Embryonic Genome Activation Predicts Development to the Blastocyst Stage. Nat. Biotechnol. 2010, 28, 1115–1121. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Au, J.; Saeedi, P.; Havelock, J. Automatic Segmentation of Trophectoderm in Microscopic Images of Human Blastocysts. IEEE Trans. Biomed. Eng. 2015, 62, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Yee, D.; Au, J.; Havelock, J. Automatic Identification of Human Blastocyst Components via Texture. IEEE Trans. Biomed. Eng. 2017, 64, 2968–2978. [Google Scholar] [CrossRef] [PubMed]
- Filho, E.S.; Noble, J.A.; Poli, M.; Griffiths, T.; Emerson, G.; Wells, D. A Method for Semi-Automatic Grading of Human Blastocyst Microscope Images. Hum. Reprod. 2012, 27, 2641–2648. [Google Scholar] [CrossRef] [Green Version]
- Kheradmand, S.; Saeedi, P.; Bajic, I. Human Blastocyst Segmentation Using Neural Network. In Proceedings of the IEEE Canadian Conference on Electrical and Computer Engineering, Vancouver, BC, Canada, 15–19 May 2016; pp. 1–4. [Google Scholar]
- Kheradmand, S.; Singh, A.; Saeedi, P.; Au, J.; Havelock, J. Inner Cell Mass Segmentation in Human HMC Embryo Images Using Fully Convolutional Network. In Proceedings of the IEEE International Conference on Image Processing, Beijing, China, 17–20 September 2017; pp. 1752–1756. [Google Scholar]
- Rad, R.M.; Saeedi, P.; Au, J.; Havelock, J. Multi-Resolutional Ensemble of Stacked Dilated U-Net for Inner Cell Mass Segmentation in Human Embryonic Images. In Proceedings of the 25th IEEE International Conference on Image Processing, Athens, Greece, 7–10 October 2018; pp. 3518–3522. [Google Scholar]
- Rad, R.M.; Saeedi, P.; Au, J.; Havelock, J. BLAST-NET: Semantic Segmentation of Human Blastocyst Components via Cascaded Atrous Pyramid and Dense Progressive Upsampling. In Proceedings of the IEEE International Conference on Image Processing, Taipei, Taiwan, 22–25 September 2019; pp. 1865–1869. [Google Scholar]
- Rad, R.M.; Saeedi, P.; Au, J.; Havelock, J. Human Blastocyst’s Zona Pellucida Segmentation via Boosting Ensemble of Complementary Learning. Inform. Med. Unlocked 2018, 13, 112–121. [Google Scholar] [CrossRef]
- Huang, T.T.F.; Kosasa, T.; Walker, B.; Arnett, C.; Huang, C.T.F.; Yin, C.; Harun, Y.; Ahn, H.J.; Ohta, A. Deep Learning Neural Network Analysis of Human Blastocyst Expansion from Time-Lapse Image Files. Reprod. Biomed. Online 2021, 42, 1075–1085. [Google Scholar] [CrossRef]
- Rad, R.M.; Saeedi, P.; Au, J.; Havelock, J. Trophectoderm Segmentation in Human Embryo Images via Inceptioned U-Net. Med. Image Anal. 2020, 62, 101612. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, C.; Zhang, D.; Chen, L.; Sun, H. A Deep Learning Framework Design for Automatic Blastocyst Evaluation With Multifocal Images. IEEE Access 2021, 9, 18927–18934. [Google Scholar] [CrossRef]
- Raef, B.; Ferdousi, R. A Review of Machine Learning Approaches in Assisted Reproductive Technologies. Acta Inf. Med. 2019, 27, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.M.O.; Francisquini, C.D.D.S. Artificial Intelligence as an Ally to Human Reproduction and Embryology. JBRA Assist. Reprod. 2021, 25, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Zaninovic, N.; Rocha, C.J.; Zhan, Q.; Toschi, M.; Malmsten, J.; Nogueira, M.; Meseguer, M.; Rosenwaks, Z.; Hickman, C. Application of Artificial Intelligence Technology to Increase the Efficacy of Embryo Selection and Prediction of Live Birth Using Human Blastocysts Cultured in a Time-Lapse Incubator. Fertil. Steril. 2018, 110, e372–e373. [Google Scholar] [CrossRef]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the Diagnosis of Diabetic and Hypertensive Retinopathy Using Artificial Intelligence-Based Semantic Segmentation. J. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arsalan, M.; Owais, M.; Mahmood, T.; Choi, J.; Park, K.R. Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases. J. Clin. Med. 2020, 9, 871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, J.; Cardoso, J.S.; Soares, F. Offline Computer-Aided Diagnosis for Glaucoma Detection Using Fundus Images Targeted at Mobile Devices. Comput. Methods Programs Biomed. 2020, 192, 105341. [Google Scholar] [CrossRef]
- SSS-Net. Available online: http://dm.dgu.edu/link.html (accessed on 16 May 2018).
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Iglovikov, V.; Shvets, A. TernausNet: U-Net with VGG11 Encoder Pre-Trained on ImageNet for Image Segmentation. arXiv 2018, arXiv:1801.05746. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, J.; Wang, T.; Borji, A.; Wei, G.; Lu, H. A Multistage Refinement Network for Salient Object Detection. IEEE Trans. Image Process. 2020, 29, 3534–3545. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky Loss Function for Image Segmentation Using 3D Fully Convolutional Deep Networks. In Proceedings of the Machine Learning in Medical Imaging, Quebec City, QC, Canada, 10 September 2017; pp. 379–387. [Google Scholar]
- NVIDIA GeForce RTX 3080 Family. Available online: https://www.nvidia.com/en-us/geforce/graphics-cards/30-series/rtx-3080-3080ti/ (accessed on 3 December 2021).
- MathWorks Introduces Release 2021a of MATLAB and Simulink. Available online: https://ch.mathworks.com/company/newsroom/mathworks-introduces-release-2021a-of-matlab-and-simulink.html (accessed on 3 December 2021).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations From Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE international Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]

| Method | No. of Parameters | Mean JI | Model Size | GFLOPS |
|---|---|---|---|---|
| SSS-Net (Residual) | 4.04 M | 85.93 | 15.0 MB | 28 |
| SSS-Net (Dense) | 4.04 M | 86.34 | 14.5 MB | 28 |
| Method | No. of Parameters | ZP | TE | BL | ICM | Background | Mean JI |
|---|---|---|---|---|---|---|---|
| UNet-Baseline [36] | 31.03 M | 79.32 | 75.06 | 79.41 | 79.03 | 94.04 | 81.37 |
| TernausNet U-Net [37] | 10 M | 80.24 | 76.16 | 78.61 | 77.58 | 94.50 | 81.42 |
| PSP-Net [38] | 35 M | 80.57 | 74.83 | 79.26 | 78.28 | 94.60 | 81.51 |
| DeepLab V3 [39] | 40 M | 80.84 | 73.98 | 78.35 | 80.60 | 94.49 | 81.65 |
| Blast-Net [24] | 25 M | 81.15 | 76.52 | 80.79 | 81.07 | 94.74 | 82.85 |
| SSS-Net Residual (Proposed) | 4.04 M | 82.88 | 77.40 | 88.39 | 84.94 | 96.03 | 85.93 |
| SSS-Net Dense (Proposed) | 4.04 M | 84.51 | 78.15 | 88.68 | 84.50 | 95.82 | 86.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arsalan, M.; Haider, A.; Choi, J.; Park, K.R. Detecting Blastocyst Components by Artificial Intelligence for Human Embryological Analysis to Improve Success Rate of In Vitro Fertilization. J. Pers. Med. 2022, 12, 124. https://doi.org/10.3390/jpm12020124
Arsalan M, Haider A, Choi J, Park KR. Detecting Blastocyst Components by Artificial Intelligence for Human Embryological Analysis to Improve Success Rate of In Vitro Fertilization. Journal of Personalized Medicine. 2022; 12(2):124. https://doi.org/10.3390/jpm12020124
Chicago/Turabian StyleArsalan, Muhammad, Adnan Haider, Jiho Choi, and Kang Ryoung Park. 2022. "Detecting Blastocyst Components by Artificial Intelligence for Human Embryological Analysis to Improve Success Rate of In Vitro Fertilization" Journal of Personalized Medicine 12, no. 2: 124. https://doi.org/10.3390/jpm12020124
APA StyleArsalan, M., Haider, A., Choi, J., & Park, K. R. (2022). Detecting Blastocyst Components by Artificial Intelligence for Human Embryological Analysis to Improve Success Rate of In Vitro Fertilization. Journal of Personalized Medicine, 12(2), 124. https://doi.org/10.3390/jpm12020124






