Polymorphisms and Gene-Gene Interaction in AGER/IL6 Pathway Might Be Associated with Diabetic Ischemic Heart Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Measurements
2.3. Serum Markers
2.4. Genotyping
2.5. Definition of Diseases and Recommendation Level of Their Risk Factors
2.6. Statistical Analysis
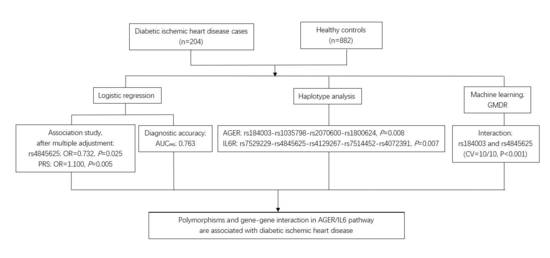
3. Results
3.1. General Characteristics of the Studied Participants
3.2. Association of AGER and IL6R Polymorphisms with Diabetic Ischemic Heart Disease
3.3. Association between Haplotypes and Diabetic Ischemic Heart Disease
3.4. The Effect of Gene-Gene Interactions on Diabetic Ischemic Heart Disease
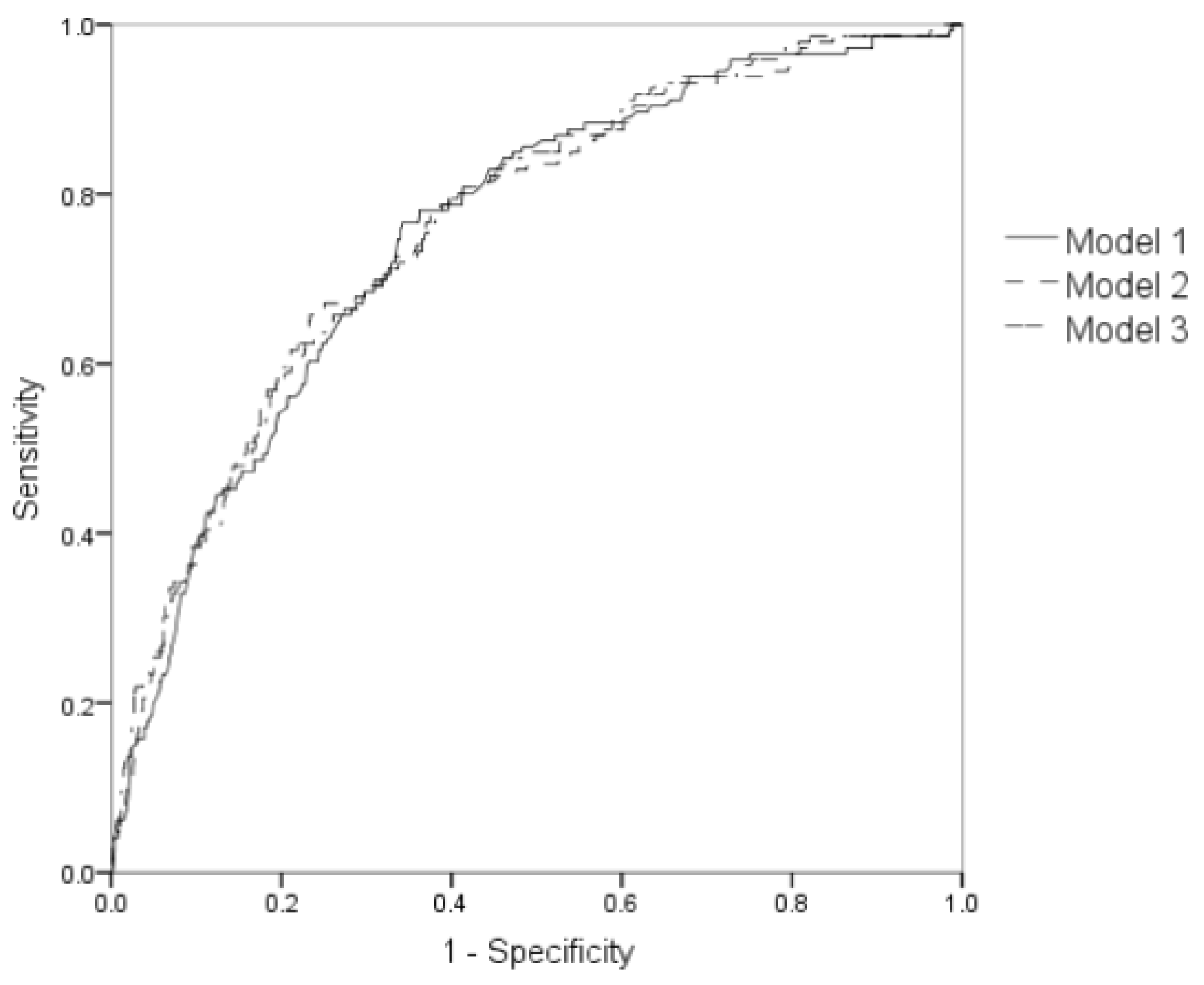
3.5. Sensitivity Analysis and Subgroup Analysis
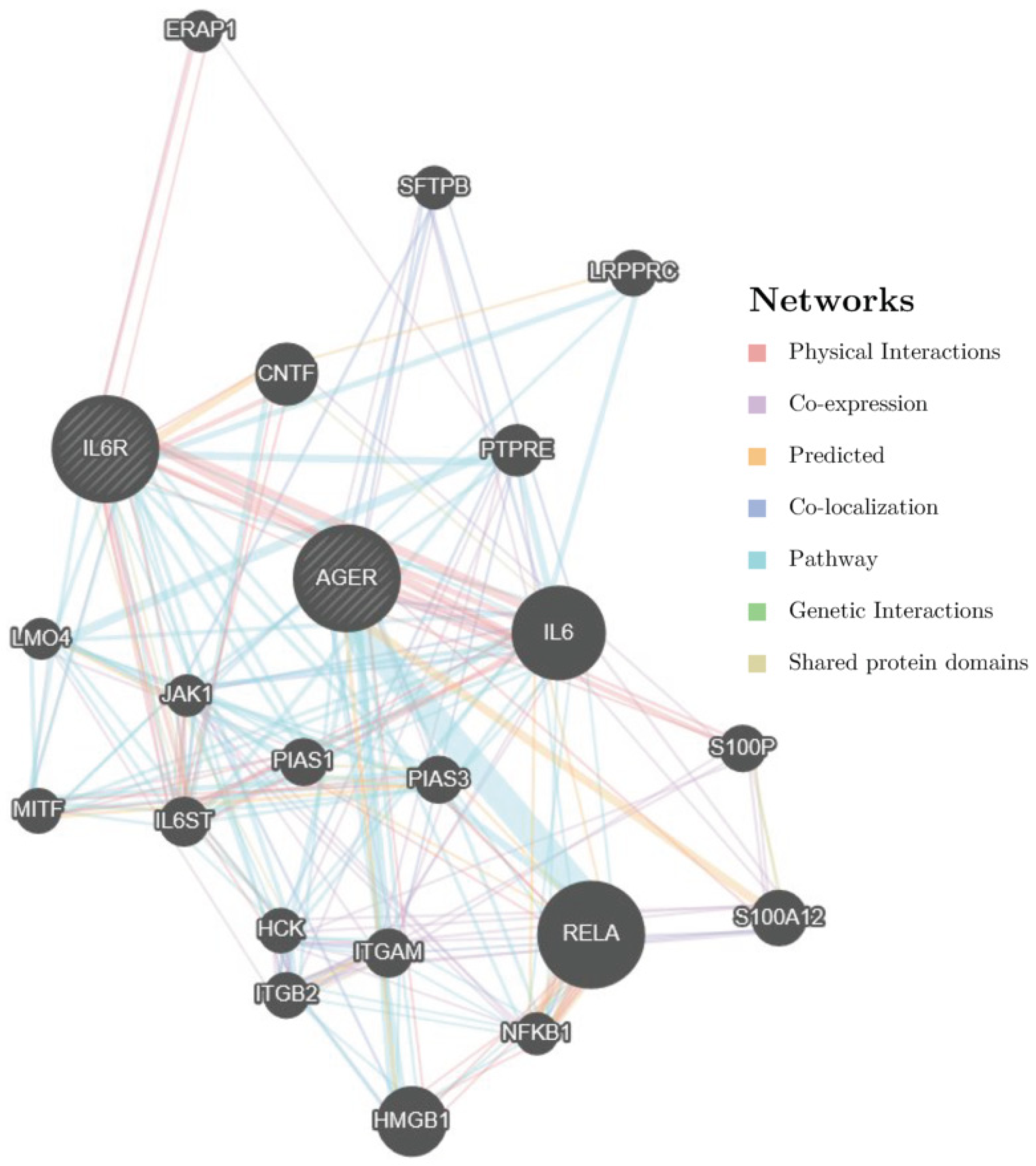
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shah, A.D.; Langenberg, C.; Rapsomaniki, E.; Denaxas, S.; Pujades-Rodriguez, M.; Gale, C.P.; Deanfield, J.; Smeeth, L.; Timmis, A.; Hemingway, H. Type 2 diabetes and incidence of cardiovascular diseases: A cohort study in 1.9 million people. Lancet Diabetes Endocrinol. 2015, 3, 105–113. [Google Scholar] [CrossRef]
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Maugeri, N.; Malato, S.; Femia, E.A.; Pugliano, M.; Campana, L.; Lunghi, F.; Rovere-Querini, P.; Lussana, F.; Podda, G.; Cattaneo, M.; et al. Clearance of circulating activated platelets in polycythemia vera and essential thrombocythemia. Blood 2011, 118, 3359–3366. [Google Scholar] [CrossRef] [PubMed]
- Zegeye, M.M.; Lindkvist, M.; Falker, K.; Kumawat, A.K.; Paramel, G.; Grenegard, M.; Sirsjö, A.; Ljungberg, L.U. Activation of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun. Signal. 2018, 16, 55. [Google Scholar] [CrossRef]
- Grozovsky, R.; Giannini, S.; Falet, H.; Hoffmeister, K.M. Novel mechanisms of platelet clearance and thrombopoietin regulation. Curr. Opin. Hematol. 2015, 22, 445–451. [Google Scholar] [CrossRef]
- Serveaux-Dancer, M.; Jabaudon, M.; Creveaux, I.; Belville, C.; Blondonnet, R.; Gross, C. Pathological Implications of Receptor for Advanced Glycation End-Product (AGER) Gene Polymorphism. Dis. Markers 2019, 2019, 2067353. [Google Scholar] [CrossRef]
- Li, J.; Cai, W.; Zhang, W.; Zhu, W.F.; Liu, Y.; Yue, L.X.; Zhu, L.Y.; Xiao, J.R.; Liu, J.Y.; Xu, J.X. Polymorphism 2184A/G in the AGER gene is not associated with diabetic retinopathy in Han Chinese patients with type 2 diabetes. J. Int. Med. Res. 2016, 44, 520–528. [Google Scholar] [CrossRef]
- Fan, W.Y.; Gu, H.; Yang, X.F.; She, C.Y.; Liu, X.P.; Liu, N.P. Association of candidate gene polymorphisms with diabetic retinopathy in Chinese patients with type 2 diabetes. Int. J. Ophthalmol. 2020, 13, 301–308. [Google Scholar] [CrossRef]
- Peng, F.; Hu, D.; Jia, N.; Li, X.; Li, Y.; Chu, S.; Zhu, D.; Shen, W.; Lin, J.; Niu, W. Association of four genetic polymorphisms of AGER and its circulating forms with coronary artery disease: A meta-analysis. PLoS ONE 2013, 8, e70834. [Google Scholar] [CrossRef]
- Lu, W.; Feng, B. The -374A allele of the RAGE gene as a potential protective factor for vascular complications in type 2 diabetes: A meta-analysis. Tohoku J. Exp. Med. 2010, 220, 291–297. [Google Scholar] [CrossRef]
- Ma, W.Q.; Qu, Q.R.; Zhao, Y.; Liu, N.F. Association of RAGE gene Gly82Ser polymorphism with coronary artery disease and ischemic stroke: A systematic review and meta-analysis. Medicine 2016, 95, e5593. [Google Scholar] [CrossRef] [PubMed]
- Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium; Swerdlow, D.I.; Holmes, M.V.; Kuchenbaecker, K.B.; Engmann, J.E.; Shah, T.; Sofat, R.; Guo, Y.; Chung, C.; Peasey, A.; et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: A mendelian randomisation analysis. Lancet 2012, 379, 1214–1224. [Google Scholar] [PubMed]
- Rafiq, S.; Melzer, D.; Weedon, M.N.; Lango, H.; Saxena, R.; Scott, L.J.; DIAGRAM Consortium; Palmer, C.N.; Morris, A.D.; McCarthy, M.I.; et al. Gene variants influencing measures of inflammation or predisposing to autoimmune and inflammatory diseases are not associated with the risk of type 2 diabetes. Diabetologia 2008, 51, 2205–2213. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Peng, D.Q.; Zhao, S.P.; Nie, S.; Li, J. Gene-gene interaction of PPARgamma and ApoE affects coronary heart disease risk. Int. J. Cardiol. 2003, 92, 257–263. [Google Scholar] [CrossRef]
- Carty, C.L.; Heagerty, P.; Heckbert, S.R.; Enquobahrie, D.A.; Jarvik, G.P.; Davis, S.; Tracy, R.P.; Reiner, A.P. Association of genetic variation in serum amyloid-A with cardiovascular disease and interactions with IL6, IL1RN, IL1beta and TNF genes in the Cardiovascular Health Study. J. Atheroscler. Thromb. 2009, 16, 419–430. [Google Scholar] [CrossRef]
- Carty, C.L.; Heagerty, P.; Heckbert, S.R.; Jarvik, G.P.; Lange, L.A.; Cushman, M.; Tracy, R.P.; Reiner, A.P. Interaction between fibrinogen and IL-6 genetic variants and associations with cardiovascular disease risk in the Cardiovascular Health Study. Ann. Hum. Genet. 2010, 74, 1–10. [Google Scholar] [CrossRef]
- Zhu, Z.; Tong, X.; Zhu, Z.; Liang, M.; Cui, W.; Su, K.; Li, M.D.; Zhu, J. Development of GMDR-GPU for gene-gene interaction analysis and its application to WTCCC GWAS data for type 2 diabetes. PLoS ONE 2013, 8, e61943. [Google Scholar] [CrossRef]
- Xu, H.M.; Xu, L.F.; Hou, T.T.; Luo, L.F.; Chen, G.B.; Sun, X.W.; Lou, X.Y. GMDR: Versatile Software for Detecting Gene-Gene and Gene-Environment Interactions Underlying Complex Traits. Curr. Genom. 2016, 17, 396–402. [Google Scholar] [CrossRef]
- Gajjala, P.R.; Sanati, M.; Jankowski, J. Cellular and Molecular Mechanisms of Chronic Kidney Disease with Diabetes Mellitus and Cardiovascular Diseases as Its Comorbidities. Front. Immunol. 2015, 6, 340. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37, S81–S90. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; Executive Group on behalf of the Joint European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF) Task Force for the Universal Definition of Myocardial Infarction. Fourth Universal Definition of Myocardial Infarction. J. Am. Coll. Cardiol. 2018, 72, 2231–2264. [Google Scholar] [CrossRef]
- Luepker, R.V.; Apple, F.S.; Christenson, R.H.; Crow, R.S.; Fortmann, S.P.; Goff, D.; Goldberg, R.J.; Hand, M.M.; Jaffe, A.S.; Julian, D.G.; et al. Case definitions for acute coronary heart disease in epidemiology and clinical research studies: A statement from the AHA Council on Epidemiology and Prevention; AHA Statistics Committee; World Heart Federation Council on Epidemiology and Prevention; the European Society of Cardiology Working Group on Epidemiology and Prevention; Centers for Disease Control and Prevention; and the National Heart, Lung, and Blood Institute. Circulation 2003, 108, 2543–2549. [Google Scholar]
- Amsterdam, E.A.; Wenger, N.K.; Brindis, R.G.; Casey, D.E., Jr.; Ganiats, T.G.; Holmes, D.R., Jr.; Jaffe, A.S.; Jneid, H.; Kelly, R.F.; Kontos, M.C.; et al. 2014 AHA/ACC Guideline for the Management of Patients with Non-ST-Elevation Acute Coronary Syndromes: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2014, 64, e139–e228. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.S. Joint committee for guideline revision. 2016 Chinese guidelines for the management of dyslipidemia in adults. J. Geriatr. Cardiol. 2018, 15, 1–29. [Google Scholar]
- Stevens, L.A.; Schmid, C.H.; Greene, T.; Zhang, Y.L.; Beck, G.J.; Froissart, M.; Hamm, L.L.; Lewis, J.B.; Mauer, M.; Navis, G.J.; et al. Comparative performance of the CKD Epidemiology Collaboration (CKD-EPI) and the Modification of Diet in Renal Disease (MDRD) Study equations for estimating GFR levels above 60 mL/min/1.73 m2. Am. J. Kidney Dis. 2010, 56, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, Z.J.; Li, Y.C.; Wang, L.M.; Jiang, Y.; Zhao, W.H. Prediction of 10-year risk for ischemic cardiovascular disease in adults aged ≥35 years in China. Zhonghua Liu Xing Bing Xue Za Zhi 2016, 37, 689–693. [Google Scholar] [PubMed]
- Kirkman, M.S.; Mahmud, H.; Korytkowski, M.T. Intensive Blood Glucose Control and Vascular Outcomes in Patients with Type 2 Diabetes Mellitus. Endocrinol. Metab. Clin. N. Am. 2018, 47, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Feng, B.; Xie, G.; Liu, F. Association of AGER gene G82S polymorphism with the severity of coronary artery disease in Chinese Han population. Clin. Endocrinol. 2011, 75, 470–474. [Google Scholar] [CrossRef]
- Maruthur, N.M.; Li, M.; Halushka, M.K.; Astor, B.C.; Pankow, J.S.; Boerwinkle, E.; Coresh, J.; Selvin, E.; Kao, W.H. Genetics of Plasma Soluble Receptor for Advanced Glycation End-Products and Cardiovascular Outcomes in a Community-based Population: Results from the Atherosclerosis Risk in Communities Study. PLoS ONE 2015, 10, e0128452. [Google Scholar]
- Lim, S.C.; Dorajoo, R.; Zhang, X.; Wang, L.; Ang, S.F.; Tan, C.S.H.; Yeoh, L.Y.; Ng, X.W.; Li, N.; Su, C.; et al. Genetic variants in the receptor for advanced glycation end products (RAGE) gene were associated with circulating soluble RAGE level but not with renal function among Asians with type 2 diabetes: A genome-wide association study. Nephrol. Dial. Transpl. 2017, 32, 1697–1704. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Shao, Y.; Lai, W.; Ren, H.; Xu, D. Association of polymorphisms in the RAGE gene with serum CRP levels and coronary artery disease in the Chinese Han population. J. Hum. Genet. 2010, 55, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.H.; Lu, L.; Wang, L.J.; Yan, X.X.; Chen, Q.J.; Zhang, Q.; Zhang, R.Y.; Shen, W.F. RAGE gene polymorphisms are associated with circulating levels of endogenous secretory RAGE but not with coronary artery disease in Chinese patients with type 2 diabetes mellitus. Arch. Med. Res. 2009, 40, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, J.; Zhu, H.; Xia, Y.; Gao, L.; Li, Z.; Jia, N.; Shen, W.; Yang, Y.; Niu, W. An interactive association of advanced glycation end-product receptor gene four common polymorphisms with coronary artery disease in northeastern Han Chinese. PLoS ONE 2013, 8, e76966. [Google Scholar]
- Niu, W.; Qi, Y.; Wu, Z.; Liu, Y.; Zhu, D.; Jin, W. A meta-analysis of receptor for advanced glycation end products gene: Four well-evaluated polymorphisms with diabetes mellitus. Mol. Cell Endocrinol. 2012, 358, 9–17. [Google Scholar] [CrossRef]
- Li, T.; Qin, W.; Liu, Y.; Li, S.; Qin, X.; Liu, Z. Effect of RAGE gene polymorphisms and circulating sRAGE levels on susceptibility to gastric cancer: A case-control study. Cancer Cell Int. 2017, 17, 19. [Google Scholar] [CrossRef]
- Ligthart, S.; Sedaghat, S.; Ikram, M.A.; Hofman, A.; Franco, O.H.; Dehghan, A. EN-RAGE: A novel inflammatory marker for incident coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2695–2699. [Google Scholar] [CrossRef]
- Reichert, S.; Triebert, U.; Santos, A.N.; Hofmann, B.; Schaller, H.G.; Schlitt, A.; Schulz, S. Soluble form of receptor for advanced glycation end products and incidence of new cardiovascular events among patients with cardiovascular disease. Atherosclerosis 2017, 266, 234–239. [Google Scholar] [CrossRef]
- Heier, M.; Margeirsdottir, H.D.; Gaarder, M.; Stensæth, K.H.; Brunborg, C.; Torjesen, P.A.; Seljeflot, I.; Hanssen, K.F.; Dahl-Jørgensen, K. Soluble RAGE and atherosclerosis in youth with type 1 diabetes: A 5-year follow-up study. Cardiovasc. Diabetol. 2015, 14, 126. [Google Scholar] [CrossRef]
- Egaña-Gorroño, L.; López-Díez, R.; Yepuri, G.; Ramirez, L.S.; Reverdatto, S.; Gugger, P.F.; Shekhtman, A.; Ramasamy, R.; Schmidt, A.M. Receptor for Advanced Glycation End Products (RAGE) and Mechanisms and Therapeutic Opportunities in Diabetes and Cardiovascular Disease: Insights from Human Subjects and Animal Models. Front. Cardiovasc. Med. 2020, 7, 37. [Google Scholar] [CrossRef]
- IL6R Genetics Consortium Emerging Risk Factors Collaboration; Sarwar, N.; Butterworth, A.S.; Freitag, D.F.; Gregson, J.; Willeit, P.; Gorman, D.N.; Gao, P.; Saleheen, D.; Rendon, A.; et al. Interleukin-6 receptor pathways in coronary heart disease: A collaborative meta-analysis of 82 studies. Lancet 2012, 379, 1205–1213. [Google Scholar]
- Chen, Z.; Qian, Q.; Tang, C.; Ding, J.; Feng, Y.; Ma, G. Association of two variants in the interleukin-6 receptor gene and premature coronary heart disease in a Chinese Han population. Mol. Biol. Rep. 2013, 40, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Teng, X.; Gu, H.; Liu, H.; Zhou, Z.; Zhao, Y.; Hu, S.; Zheng, Z. Interleukin-6 receptor rs7529229 T/C polymorphism is associated with left main coronary artery disease phenotype in a Chinese population. Int. J. Mol. Sci. 2014, 15, 5623–5633. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Tokoro, F.; Matsuoka, R.; Arai, M.; Noda, T.; Watanabe, S.; Horibe, H.; Fujimaki, T.; Oguri, M.; Kato, K.; et al. Association of genetic variants with dyslipidemia. Mol. Med. Rep. 2015, 12, 5429–5436. [Google Scholar] [CrossRef][Green Version]
- Horibe, H.; Fujimaki, T.; Oguri, M.; Kato, K.; Matsuoka, R.; Abe, S.; Tokoro, F.; Arai, M.; Noda, T.; Watanabe, S.; et al. Association of a polymorphism of the interleukin 6 receptor gene with chronic kidney disease in Japanese individuals. Nephrology 2015, 20, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Kraakman, M.J.; Lee, M.K.; Al-Sharea, A.; Dragoljevic, D.; Barrett, T.J.; Montenont, E.; Basu, D.; Heywood, S.; Kammoun, H.L.; Flynn, M.; et al. Neutrophil-derived S100 calcium-binding proteins A8/A9 promote reticulated thrombocytosis and atherogenesis in diabetes. J. Clin. Investig. 2017, 127, 2133–2147. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Liu, J.; Lau, C.W.; Huang, Y. IL-6 in diabetes and cardiovascular complications. Br. J. Pharmacol. 2014, 171, 3595–3603. [Google Scholar] [CrossRef]
- Society, C.D. Guidelines for the prevention and control of type 2 diabetes in China (2017 Edition). Chin. J. Pract. Int. Med. 2018, 38, 292–344. [Google Scholar]
- Scheen, A.J. Cardiovascular Effects of New Oral Glucose-Lowering Agents: DPP-4 and SGLT-2 Inhibitors. Circ. Res. 2018, 122, 1439–1459. [Google Scholar] [CrossRef]
- Olsson, S.; Jood, K. Genetic variation in the receptor for advanced glycation end-products (RAGE) gene and ischaemic stroke. Eur. J. Neurol. 2013, 20, 991–993. [Google Scholar] [CrossRef]
- Wang, Z.T.; Wang, L.Y.; Wang, L.; Cheng, S.; Fan, R.; Zhou, J.; Zhong, J. Association between RAGE gene polymorphisms and ulcerative colitis susceptibility: A case-control study in a Chinese Han population. Genet. Mol. Res. 2015, 14, 19242. [Google Scholar] [CrossRef]
- Kang, P.; Tian, C.; Jia, C. Association of RAGE gene polymorphisms with type 2 diabetes mellitus, diabetic retinopathy and diabetic nephropathy. Gene 2012, 500, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Niu, W.; Yu, T.; Dong, F.; Huang, K.; Duan, R.; Qumu, S.; Lu, M.; Li, Y.; Yang, T.; et al. Association of RAGE gene multiple variants with the risk for COPD and asthma in northern Han Chinese. Aging 2019, 11, 3220–3237. [Google Scholar] [CrossRef] [PubMed]
- Wadén, J.M.; Dahlström, E.H.; Elonen, N.; Thorn, L.M.; Wadén, J.; Sandholm, N.; Forsblom, C.; Groop, P.H.; FinnDiane Study Group. Soluble receptor for AGE in diabetic nephropathy and its progression in Finnish individuals with type 1 diabetes. Diabetologia 2019, 62, 1268–1274. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Yoo, S.D.; Chon, J.; Yun, D.H.; Kim, H.S.; Park, H.J.; Kim, S.K.; Chung, J.H.; Kang, J.K.; Lee, S.A. Interleukin-6 Receptor Polymorphisms Contribute to the Neurological Status of Korean Patients with Ischemic Stroke. J. Korean Med. Sci. 2016, 31, 430–434. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Yang, R.; Li, X.Y.; Ye, C.; He, B.C.; Lin, T.; Xu, X.Q.; Zheng, L.L.; Luo, W.T.; Cai, L. Single nucleotide polymorphisms of the NF-κB and STAT3 signaling pathway genes predict lung cancer prognosis in a Chinese Han population. Cancer Genet. 2015, 208, 310–318. [Google Scholar] [CrossRef]
- Key, K.V.; Mudd-Martin, G.; Moser, D.K.; Rayens, M.K.; Morford, L.A. Inflammatory Genotype Moderates the Association Between Anxiety and Systemic Inflammation in Adults at Risk for Cardiovascular Disease. J. Cardiovasc. Nurs. 2022, 37, 64–72. [Google Scholar] [CrossRef]
- Arguinano, A.A.; Naderi, E.; Ndiaye, N.C.; Stathopoulou, M.; Dadé, S.; Alizadeh, B.; Visvikis-Siest, S. IL6R haplotype rs4845625*T/rs4537545*C is a risk factor for simultaneously high CRP, LDL and ApoB levels. Genes Immun. 2017, 18, 163–169. [Google Scholar] [CrossRef]
- Tabassum, R.; Mahendran, Y.; Dwivedi, O.P.; Chauhan, G.; Ghosh, S.; Marwaha, R.K.; Tandon, N.; Bharadwaj, D. Common variants of IL6, LEPR, and PBEF1 are associated with obesity in Indian children. Diabetes 2012, 61, 626–631. [Google Scholar] [CrossRef]
- Van Dongen, J.; Jansen, R.; Smit, D.; Hottenga, J.J.; Mbarek, H.; Willemsen, G.; Kluft, C.; Penninx, B.W.; Ferreira, M.A.; Boomsma, D.I.; et al. The contribution of the functional IL6R polymorphism rs2228145, eQTLs and other genome-wide SNPs to the heritability of plasma sIL-6R levels. Behav. Genet. 2014, 44, 368–382. [Google Scholar] [CrossRef]
- Walston, J.D.; Matteini, A.M.; Nievergelt, C.; Lange, L.A.; Fallin, D.M.; Barzilai, N.; Ziv, E.; Pawlikowska, L.; Kwok, P.; Cummings, S.R.; et al. Inflammation and stress-related candidate genes, plasma interleukin-6 levels, and longevity in older adults. Exp. Gerontol. 2009, 44, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Naitza, S.; Porcu, E.; Steri, M.; Taub, D.D.; Mulas, A.; Xiao, X.; Strait, J.; Dei, M.; Lai, S.; Busonero, F.; et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012, 8, e1002480. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, S.; Frayling, T.M.; Murray, A.; Hurst, A.; Stevens, K.; Weedon, M.N.; Henley, W.; Ferrucci, L.; Bandinelli, S.; Corsi, A.M.; et al. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun. 2007, 8, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Webb, T.R.; Erdmann, J.; Stirrups, K.E.; Stitziel, N.O.; Masca, N.G.; Jansen, H.; Kanoni, S.; Nelson, C.P.; Ferrario, P.G.; König, I.R.; et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated with Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 823–836. [Google Scholar] [CrossRef]
- Christiansen, M.K.; Larsen, S.B.; Nyegaard, M.; Neergaard-Petersen, S.; Ajjan, R.; Würtz, M.; Grove, E.L.; Hvas, A.M.; Jensen, H.K.; Kristensen, S.D. Coronary artery disease-associated genetic variants and biomarkers of inflammation. PLoS ONE 2017, 12, e0180365. [Google Scholar]
- Gigante, B.; Strawbridge, R.J.; Velasquez, I.M.; Golabkesh, Z.; Silveira, A.; Goel, A.; Baldassarre, D.; Veglia, F.; Tremoli, E.; Clarke, R.; et al. Analysis of the role of interleukin 6 receptor haplotypes in the regulation of circulating levels of inflammatory biomarkers and risk of coronary heart disease. PLoS ONE 2015, 10, e0119980. [Google Scholar]

| Controls | T2DM + IHD | p Value | |
|---|---|---|---|
| Age (years) 1 | 64.00 ± 11.25 | 65 ± 11.00 | 0.126 |
| Male (n, %) | 488 (55.3) | 119 (58.3) | 0.436 |
| SBP (mmHg) 1 | 136.00 (24.00) | 130.00 (19.50) | <0.001 ** |
| DBP (mmHg) 1 | 79.00 (14.00) | 80 (12.00) | 0.001 ** |
| BMI (kg/m2) 1 | 25.64 (4.46) | 25.36 (3.53) | 0.578 |
| TC (mmol/L) 1 | 5.12 (1.41) | 4.68 (1.53) | <0.001 ** |
| HDLC (mmol/L) 1 | 1.34 (0.48) | 1.22 (0.45) | <0.001 ** |
| LDLC (mmol/L) 1 | 3.02 (1.17) | 2.68 (1.04) | <0.001 ** |
| TG (mmol/L) 1 | 1.40 (0.90) | 2.35 (1.57) | <0.001 ** |
| FPG (mmol/L) 1 | 5.60 (0.93) | 7.07 (3.53) | <0.001 ** |
| Current smoking (n, %) | 194 (22.0) | 18 (8.8) | <0.001 ** |
| Current drinking (n, %) | 295 (33.5) | 32 (15.7) | <0.001 ** |
| AGEs (mmol/L) 1 | 31.05 (15.94) | 38.07 (16.82) | <0.001 ** |
| IL-6 (mmol/L) 1 | 133.08 (49.32) | 136.83 (40.18) | 0.353 |
| Genotype | Crude OR 1 (95%CI) | Crude p Value | Adjusted OR¤ (95%CI) | Adjusted p Value | |
|---|---|---|---|---|---|
| rs184003 | GG | Ref | Ref | Ref | Ref |
| GT | 1.435 (1.019, 2.020) | 0.039 * | 1.223 (0.797, 1.878) | 0.357 | |
| TT | 2.525 (1.092, 5.837) | 0.030 * | 1.651 (0.580, 4.702) | 0.348 | |
| additive | 1.491 (1.125, 1.976) | 0.005 ** | 1.247 (0.880, 1.767) | 0.215 | |
| dominant | 1.518 (1.093, 2.017) | 0.012 * | 1.265 (0.839, 1.905) | 0.261 | |
| recessive | 2.282 (0.993, 5.241) | 0.046 | 1.571 (0.555, 4.449) | 0.395 | |
| rs2070600 | CC | Ref | Ref | Ref | Ref |
| CT | 0.713 (0.496, 1.024) | 0.067 | 0.843 (0.550, 1.294) | 0.435 | |
| TT | 0.536 (0.237, 1.211) | 0.134 | 0.611 (0.206, 1.807) | 0.373 | |
| additive | 0.721 (0.542, 0.960) | 0.025 * | 0.819 (0.578, 1.162) | 0.264 | |
| dominant | 0.684 (0.485, 0.964) | 0.030 * | 1.399 (0.914, 2.140) | 0.122 | |
| recessive | 0.587 (0.261, 1.320) | 0.198 | 2.204 (0.493, 9.851) | 0.301 | |
| rs4845625 | CC | Ref | Ref | Ref | Ref |
| CT | 0.692 (0.483, 0.991) | 0.045 * | 0.619 (0.398, 0.961) | 0.033 | |
| TT | 0.503 (0.318, 0.795) | 0.003 ** | 0.542 (0.318, 0.924) | 0.025 | |
| additive | 0.707 (0.563, 0.888) | 0.003 ** | 0.732 (0.557, 0.961) | 0.025 | |
| dominant | 0.632 (0.448, 0.889) | 0.008 ** | 0.594 (0.392, 0.902) | 0.014 | |
| recessive | 0.644 (0.434, 0.955) | 0.028 | 0.757 (0.481, 1.191) | 0.229 |
| Haplotypes | F_U 1 | F_A¤ | Chi-Square | OR (95%CI) | p Value | |
|---|---|---|---|---|---|---|
| Block 1 2 | Omnibus test | - | - | 11.750 | 0.008 ** | |
| C-A-C-T | 0.162 | 0.170 | 0.162 | 1.049 (0.830, 1.327) | 0.687 | |
| C-G-T-A | 0.202 | 0.150 | 5.575 | 0.743 (0.580, 0.951) | 0.018 * | |
| A-G-C-A | 0.140 | 0.197 | 8.229 | 1.407 (1.114, 1.777) | 0.004 ** | |
| C-G-C-A | 0.497 | 0.482 | 0.247 | 0.970 (0.859, 1.094) | 0.620 | |
| Block 2 3 | Omnibus test | - | - | 11.99 | 0.007 ** | |
| T-T-C-C-T | 0.093 | 0.100 | 0.227 | 1.075 (0.798, 1.449) | 0.634 | |
| C-C-T-T-C | 0.387 | 0.431 | 2.639 | 1.114 (0.978, 1.268) | 0.104 | |
| T-C-C-T-C | 0.095 | 0.131 | 4.551 | 1.379 (1.026, 1.853) | 0.033 * | |
| T-T-C-T-C | 0.426 | 0.338 | 10.32 | 0.793 (0.689, 0.914) | 0.001 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Xie, Y.; Zhao, Q.; Peng, W.; Guo, C.; Zhang, J.; Zhang, L. Polymorphisms and Gene-Gene Interaction in AGER/IL6 Pathway Might Be Associated with Diabetic Ischemic Heart Disease. J. Pers. Med. 2022, 12, 392. https://doi.org/10.3390/jpm12030392
Liu K, Xie Y, Zhao Q, Peng W, Guo C, Zhang J, Zhang L. Polymorphisms and Gene-Gene Interaction in AGER/IL6 Pathway Might Be Associated with Diabetic Ischemic Heart Disease. Journal of Personalized Medicine. 2022; 12(3):392. https://doi.org/10.3390/jpm12030392
Chicago/Turabian StyleLiu, Kuo, Yunyi Xie, Qian Zhao, Wenjuan Peng, Chunyue Guo, Jie Zhang, and Ling Zhang. 2022. "Polymorphisms and Gene-Gene Interaction in AGER/IL6 Pathway Might Be Associated with Diabetic Ischemic Heart Disease" Journal of Personalized Medicine 12, no. 3: 392. https://doi.org/10.3390/jpm12030392
APA StyleLiu, K., Xie, Y., Zhao, Q., Peng, W., Guo, C., Zhang, J., & Zhang, L. (2022). Polymorphisms and Gene-Gene Interaction in AGER/IL6 Pathway Might Be Associated with Diabetic Ischemic Heart Disease. Journal of Personalized Medicine, 12(3), 392. https://doi.org/10.3390/jpm12030392