Searching for New Molecular Targets for Oral Squamous Cell Carcinoma with a View to Clinical Implementation of Precision Medicine
Abstract
:1. Introduction
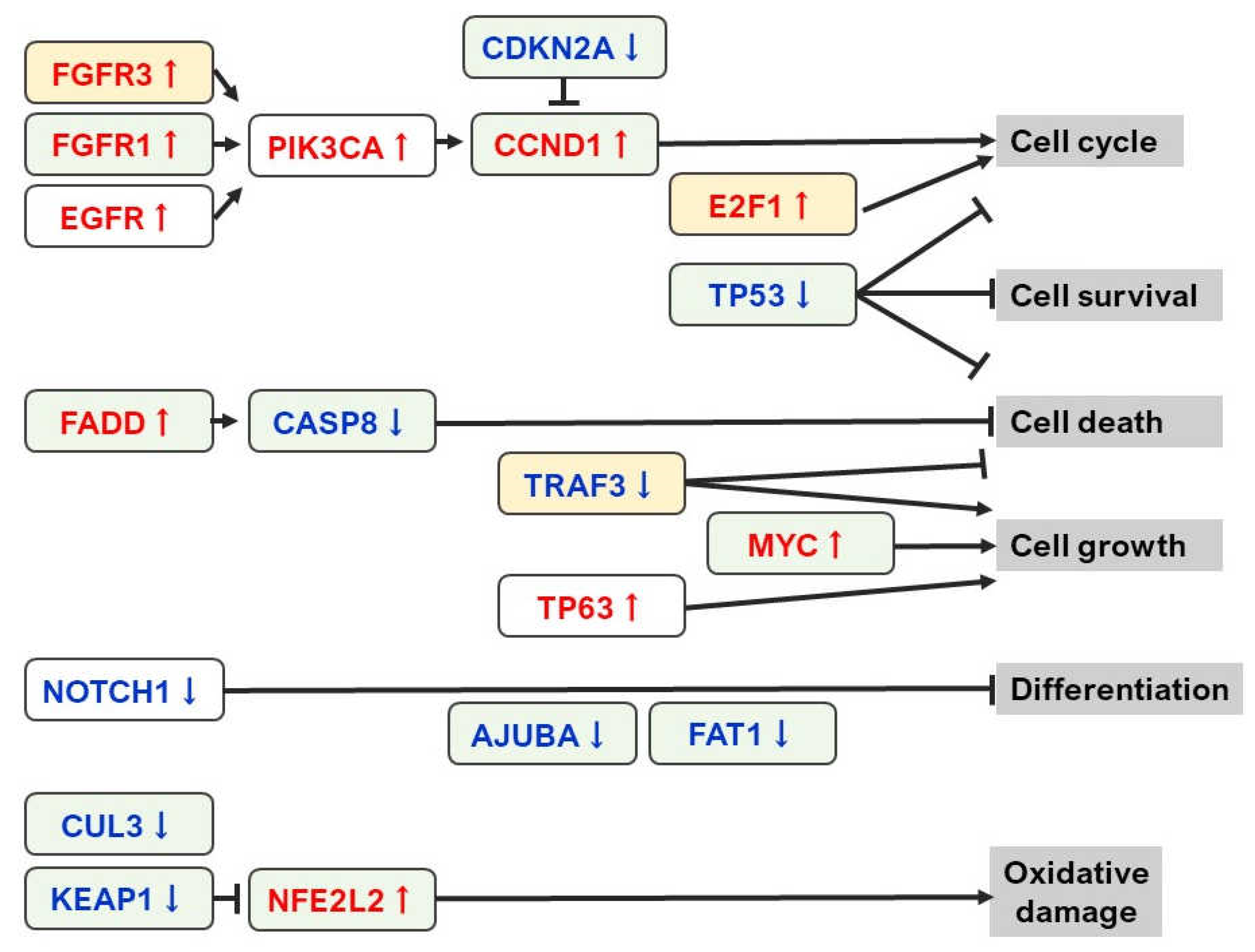
2. Current Status of NGS Analysis in OSCC
3. Tumor Biomarker and Liquid Biopsy
3.1. cfDNA
3.2. CTC
3.3. miRNA
4. Candidate for Novel Secretory Molecular Markers in OSCC
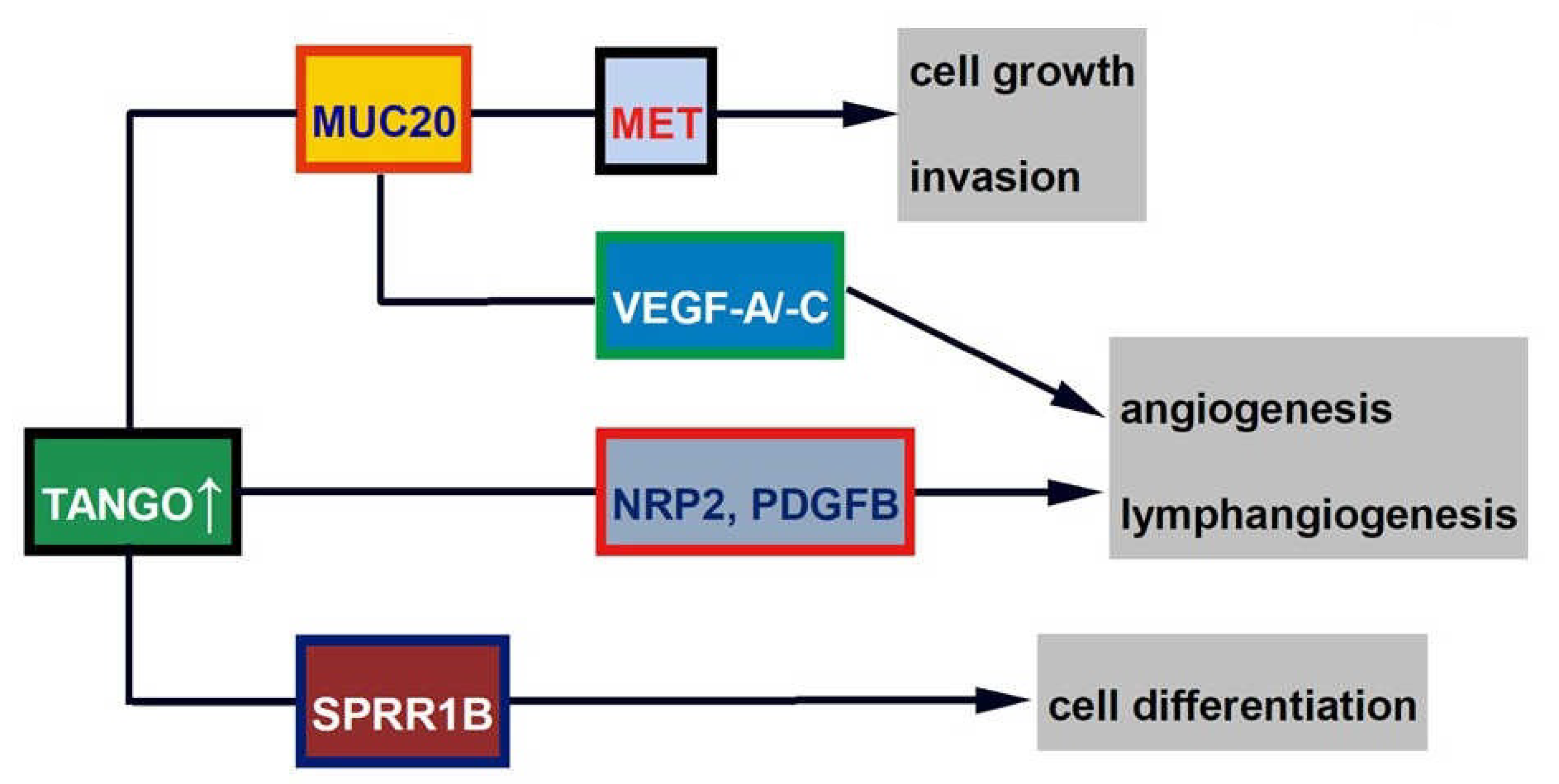
4.1. TANGO and Related Molecules (MUC20 and SRPX2)
4.1.1. TANGO
4.1.2. MUC20
4.1.3. SPRR1B
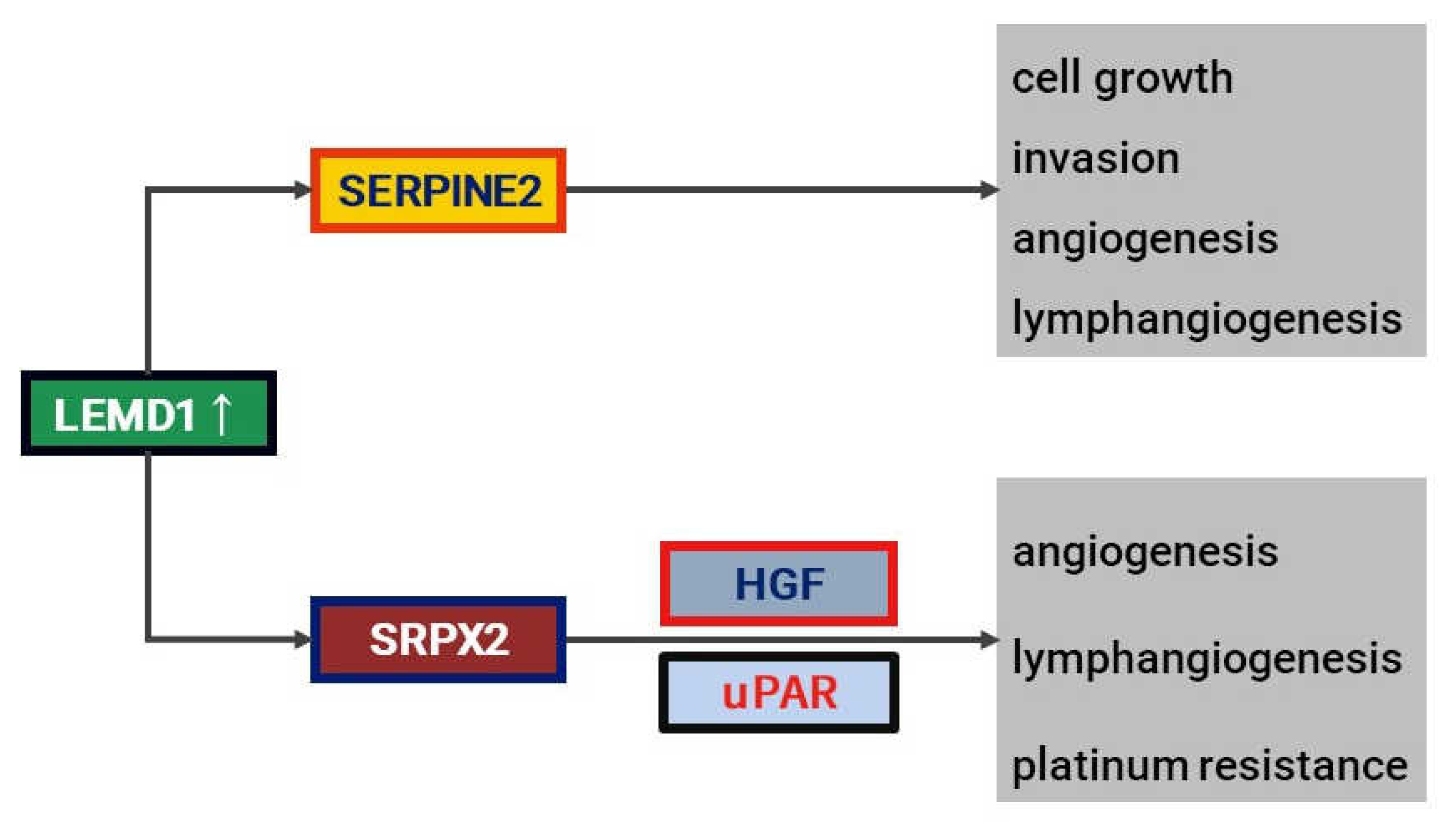
4.2. LEMD1 and Ralated Molecules (SRPX2 and SERPINE2)
4.2.1. LEMD1
4.2.2. SRPX2
4.2.3. SERPINE2
5. Candidate for New Non-Secretory Markers in OSCC
5.1. PXDN
5.2. miR-29b-1-5p
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.; Mahajan, A.; Pradeep, G.L.; Prakash, N.; Patil, S.; Khan, S.M. Expression of matrix metalloproteinase-9 in histological grades of oral squamous cell carcinoma: An immunohistochemical study. J. Oral Maxillofac. Pathol. 2021, 25, 239–246. [Google Scholar] [PubMed]
- Rizzo, J.D.; Curtis, R.E.; Socié, G.; Sobocinski, K.A.; Gilbert, E.; Landgren, O.; Travis, L.B.; Travis, W.D.; Flowers, M.E.; Friedman, D.L.; et al. Solid cancers after allogeneic hematopoietic cell transplantation. Blood 2009, 113, 1175–1183. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Sasahira, T.; Kirita, T. Hallmarks of cancer-related newly prognostic factors of oral squamous cell carcinoma. Int. J. Mol. Sci. 2018, 19, 2413. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, N.; Sento, S.; Yoshizawa, Y.; Sasabe, E.; Kudo, Y.; Yamamoto, T. Current trends and future prospects of molecular targeted therapy in head and neck squamous cell carcinoma. Int. J. Mol. Sci. 2020, 22, 240. [Google Scholar] [CrossRef]
- Akagi, K.; Oki, E.; Taniguchi, H.; Nakatani, K.; Aoki, D.; Kuwata, T.; Yoshino, T. Real-world data on microsatellite instability status in various unresectable or metastatic solid tumors. Cancer Sci. 2021, 112, 1105–1113. [Google Scholar] [CrossRef]
- Hause, R.J.; Pritchard, C.C.; Shendure, J.; Salipante, S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016, 22, 1342–1350. [Google Scholar] [CrossRef]
- Jiang, T.; Wang, G.; Liu, Y.; Feng, L.; Wang, M.; Liu, J.; Chen, Y.; Ouyang, L. Development of small-molecule tropomyosin receptor kinase (TRK) inhibitors for NTRK fusion cancers. Acta Pharm. Sin. B 2021, 11, 355–372. [Google Scholar] [CrossRef]
- Sicklick, J.K.; Kato, S.; Okamura, R.; Schwaederle, M.; Hahn, M.E.; Williams, C.B.; De, P.; Krie, A.; Piccioni, D.E.; Miller, V.A.; et al. Molecular profiling of cancer patients enables personalized combination therapy: The I-PREDICT study. Nat. Med. 2019, 25, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Matsubara, J.; Quy, P.N.; Fukuyama, K.; Nomura, M.; Funakoshi, T.; Doi, K.; Sakamori, Y.; Yoshioka, M.; Yokoyama, A.; et al. Comprehensive genomic profiling for patients with chemotherapy-naïve advanced cancer. Cancer Sci. 2021, 112, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Sunami, K.; Ichikawa, H.; Kubo, T.; Kato, M.; Fujiwara, Y.; Shimomura, A.; Koyama, T.; Kakishima, H.; Kitami, M.; Matsushita, H.; et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 2019, 110, 1480–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunami, K.; Naito, Y.; Aimono, E.; Amano, T.; Ennishi, D.; Kage, H.; Kanai, M.; Komine, K.; Koyama, T.; Maeda, T.; et al. The initial assessment of expert panel performance in core hospitals for cancer genomic medicine in Japan. Int. J. Clin. Oncol. 2021, 26, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Network, C.G.A. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Baumeister, P.; Reiter, M.; Welz, C.; Becker, S.; Betz, C.; Harréus, U. Surgically treated oropharyngeal cancer: Risk factors and tumor characteristics. J. Cancer Res. Clin. Oncol. 2014, 140, 1011–1019. [Google Scholar] [CrossRef]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Cohen, R.B.; Jones, C.U.; Sur, R.K.; Raben, D.; Baselga, J.; Spencer, S.A.; Zhu, J.; et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010, 11, 21–28. [Google Scholar] [CrossRef]
- Gillison, M.L.; Trotti, A.M.; Harris, J.; Eisbruch, A.; Harari, P.M.; Adelstein, D.J.; Jordan, R.C.K.; Zhao, W.; Sturgis, E.M.; Burtness, B.; et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): A randomised, multicentre, non-inferiority trial. Lancet 2019, 393, 40–50. [Google Scholar] [CrossRef]
- Saito, Y.; Hayashi, R.; Iida, Y.; Mizumachi, T.; Fujii, T.; Matsumoto, F.; Beppu, T.; Yoshida, M.; Shinomiya, H.; Kamiyama, R.; et al. Optimization of therapeutic strategy for p16-positive oropharyngeal squamous cell carcinoma: Multi-institutional observational study based on the national Head and Neck Cancer Registry of Japan. Cancer 2020, 126, 4177–4187. [Google Scholar] [CrossRef]
- Walter, V.; Yin, X.; Wilkerson, M.D.; Cabanski, C.R.; Zhao, N.; Du, Y.; Ang, M.K.; Hayward, M.C.; Salazar, A.H.; Hoadley, K.A.; et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE 2013, 8, e56823. [Google Scholar] [CrossRef]
- Keck, M.K.; Zuo, Z.; Khattri, A.; Stricker, T.P.; Brown, C.D.; Imanguli, M.; Rieke, D.; Endhardt, K.; Fang, P.; Brägelmann, J.; et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin. Cancer Res. 2015, 21, 870–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; David, O.; Cabay, R.J.; Valyi-Nagy, K.; Macias, V.; Zhong, R.; Wenig, B.; Feldman, L.; Weichselbaum, R.; Spiotto, M.T. Molecular classification of lymph node metastases subtypes predict for survival in head and neck cancer. Clin. Cancer Res. 2019, 25, 1795–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasahira, T.; Kurihara, M.; Nakashima, C.; Kirita, T.; Kuniyasu, H. LEM domain containing 1 promotes oral squamous cell carcinoma invasion and endothelial transmigration. Br. J. Cancer 2016, 115, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.Y.; Yi, C.; Chung, H.R.; Wang, D.J.; Chang, W.C.; Lee, S.Y.; Lin, C.T.; Yang, Y.C.; Yang, W.C. Potential biomarkers in saliva for oral squamous cell carcinoma. Oral Oncol. 2010, 46, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Zviran, A.; Schulman, R.C.; Shah, M.; Hill, S.T.K.; Deochand, S.; Khamnei, C.C.; Maloney, D.; Patel, K.; Liao, W.; Widman, A.J.; et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020, 26, 1114–1124. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [Green Version]
- Baby, N.T.; Abdullah, A.; Kannan, S. The scope of liquid biopsy in the clinical management of oral cancer. Int. J. Oral Maxillofac. Surg. 2021; in press. [Google Scholar] [CrossRef]
- Perdomo, S.; Avogbe, P.H.; Foll, M.; Abedi-Ardekani, B.; Facciolla, V.L.; Anantharaman, D.; Chopard, P.; Calvez-Kelm, F.L.; Vilensky, M.; Polesel, J.; et al. Circulating tumor DNA detection in head and neck cancer: Evaluation of two different detection approaches. Oncotarget 2017, 8, 72621–72632. [Google Scholar] [CrossRef] [Green Version]
- Schröck, A.; Leisse, A.; de Vos, L.; Gevensleben, H.; Dröge, F.; Franzen, A.; Wachendörfer, M.; Schröck, F.; Ellinger, J.; Teschke, M.; et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: An observational prospective cohort study. Clin. Chem. 2017, 63, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Lousada-Fernandez, F.; Rapado-Gonzalez, O.; Lopez-Cedrun, J.L.; Lopez-Lopez, R.; Muinelo-Romay, L.; Suarez-Cunqueiro, M.M. Liquid biopsy in oral cancer. Int. J. Mol. Sci. 2018, 19, 1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hristozova, T.; Konschak, R.; Stromberger, C.; Fusi, A.; Liu, Z.; Weichert, W.; Stenzinger, A.; Budach, V.; Keilholz, U.; Tinhofer, I. The presence of circulating tumor cells (CTCs) correlates with lymph node metastasis in nonresectable squamous cell carcinoma of the head and neck region (SCCHN). Ann. Oncol. 2011, 22, 1878–1885. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Costa, J.P.; de Carvalho, A.F.; da Silveira, G.G.; Amaya, P.; Wu, Y.; Park, K.J.; Gigliola, M.P.; Lustberg, M.; Buim, M.E.; Ferreira, E.N.; et al. Gene expression patterns through oral squamous cell carcinoma development: PD-L1 expression in primary tumor and circulating tumor cells. Oncotarget 2015, 6, 20902–20920. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.M.; Wu, M.H.; Chang, P.H.; Lin, H.C.; Liao, C.D.; Wu, S.M.; Hung, T.M.; Lin, C.Y.; Chang, T.C.; Tzu-Tsen, Y.; et al. The change in circulating tumor cells before and during concurrent chemoradiotherapy is associated with survival in patients with locally advanced head and neck cancer. Head Neck 2019, 41, 2676–2687. [Google Scholar] [CrossRef]
- Tada, H.; Takahashi, H.; Kawabata-Iwakawa, R.; Nagata, Y.; Uchida, M.; Shino, M.; Ida, S.; Mito, I.; Matsuyama, T.; Chikamatsu, K. Molecular phenotypes of circulating tumor cells and efficacy of nivolumab treatment in patients with head and neck squamous cell carcinoma. Sci. Rep. 2020, 10, 21573. [Google Scholar] [CrossRef]
- Grisanti, S.; Almici, C.; Consoli, F.; Buglione, M.; Verardi, R.; Bolzoni-Villaret, A.; Bianchetti, A.; Ciccarese, C.; Mangoni, M.; Ferrari, L.; et al. Circulating tumor cells in patients with recurrent or metastatic head and neck carcinoma: Prognostic and predictive significance. PLoS ONE 2014, 9, e103918. [Google Scholar] [CrossRef]
- Miyamoto, D.T.; Zheng, Y.; Wittner, B.S.; Lee, R.J.; Zhu, H.; Broderick, K.T.; Desai, R.; Fox, D.B.; Brannigan, B.W.; Trautwein, J.; et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015, 349, 1351–1356. [Google Scholar] [CrossRef] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Rabinowits, G.; Bowden, M.; Flores, L.M.; Verselis, S.; Vergara, V.; Jo, V.Y.; Chau, N.; Lorch, J.; Hammerman, P.S.; Thomas, T.; et al. Comparative analysis of microRNA expression among benign and malignant tongue tissue and plasma of patients with tongue cancer. Front. Oncol. 2017, 7, 191. [Google Scholar] [CrossRef] [Green Version]
- Arndt, S.; Bosserhoff, A.K. TANGO is a tumor suppressor of malignant melanoma. Int. J. Cancer 2006, 119, 2812–2820. [Google Scholar] [CrossRef] [PubMed]
- Arndt, S.; Bosserhoff, A.K. Reduced expression of TANGO in colon and hepatocellular carcinomas. Oncol. Rep. 2007, 18, 885–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoll, R.; Bosserhoff, A. Extracellular SH3 domain containing proteins--features of a new protein family. Curr. Protein Pept. Sci. 2008, 9, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Arndt, S.; Melle, C.; Mondal, K.; Klein, G.; von Eggeling, F.; Bosserhoff, A.K. Interactions of TANGO and leukocyte integrin CD11c/CD18 regulate the migration of human monocytes. J. Leukoc. Biol. 2007, 82, 1466–1472. [Google Scholar] [CrossRef]
- Sasahira, T.; Kirita, T.; Yamamoto, K.; Ueda, N.; Kurihara, M.; Matsushima, S.; Bhawal, U.K.; Bosserhoff, A.K.; Kuniyasu, H. Transport and Golgi organisation protein 1 is a novel tumour progressive factor in oral squamous cell carcinoma. Eur. J. Cancer 2014, 50, 2142–2151. [Google Scholar] [CrossRef]
- Sasahira, T.; Bosserhoff, A.K.; Kirita, T. The importance of melanoma inhibitory activity gene family in the tumor progression of oral cancer. Pathol. Int. 2018, 68, 278–286. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara-Shimomura, M.; Shimomura, H.; Bosserhoff, A.K.; Kirita, T. Identification of oral squamous cell carcinoma markers MUC2 and SPRR1B downstream of TANGO. J. Cancer Res. Clin. Oncol. 2021, 147, 1659–1672. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, L.; Wei, P.; Chi, Y.; Li, D.; Wang, Q.; Ni, S.; Tan, C.; Sheng, W.; Sun, M.; et al. Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. J. Transl. Med. 2013, 11, 151. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.T.; Kuo, T.C.; Liao, Y.Y.; Lin, M.C.; Tien, Y.W.; Huang, M.C. Silencing of MUC20 suppresses the malignant character of pancreatic ductal adenocarcinoma cells through inhibition of the HGF/MET pathway. Oncogene 2018, 37, 6041–6053. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Shyu, M.K.; Wang, S.W.; Chou, C.H.; Huang, M.J.; Lin, T.C.; Chen, S.T.; Lin, H.H.; Huang, M.C. MUC20 promotes aggressive phenotypes of epithelial ovarian cancer cells via activation of the integrin β1 pathway. Gynecol. Oncol. 2016, 140, 131–137. [Google Scholar] [CrossRef]
- Shen, L.Y.; Wang, H.; Dong, B.; Yan, W.P.; Lin, Y.; Shi, Q.; Chen, K.N. Possible prediction of the response of esophageal squamous cell carcinoma to neoadjuvant chemotherapy based on gene expression profiling. Oncotarget 2016, 7, 4531–4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, W.; Nakanishi, G.; Arata, J.; Jetten, A.M. Differential expression of human cornifin alpha and beta in squamous differentiating epithelial tissues and several skin lesions. J. Investig. Dermatol. 1997, 108, 200–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, S.P.; Vuong, H.; Adiseshaiah, P. Interplay between proximal and distal promoter elements is required for squamous differentiation marker induction in the bronchial epithelium: Role for ESE-1, Sp1, and AP-1 proteins. J. Biol. Chem. 2003, 278, 21378–21387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hippo, Y.; Yashiro, M.; Ishii, M.; Taniguchi, H.; Tsutsumi, S.; Hirakawa, K.; Kodama, T.; Aburatani, H. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001, 61, 889–895. [Google Scholar] [PubMed]
- Ludovini, V.; Bianconi, F.; Siggillino, A.; Piobbico, D.; Vannucci, J.; Metro, G.; Chiari, R.; Bellezza, G.; Puma, F.; Della Fazia, M.A.; et al. Gene identification for risk of relapse in stage I lung adenocarcinoma patients: A combined methodology of gene expression profiling and computational gene network analysis. Oncotarget 2016, 7, 30561–30574. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Nadelman, C.; Henry, G.; Fan, J.; Muellenhoff, M.; Medina, E.; Gratch, N.S.; Chen, M.; Han, J.; Woodley, D. The p38-MAPK/SAPK pathway is required for human keratinocyte migration on dermal collagen. J. Investig. Dermatol. 2001, 117, 1601–1611. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.H.; Son, Y.S.; Lee, D.Y.; Chung, E.K.; Hur, K.C.; Hong, S.I.; Fuchs, E. Calcipotriol (MC 903), a synthetic derivative of vitamin D3 stimulates differentiation of squamous carcinoma cell line in the raft culture. Anticancer Res. 1996, 16, 337–347. [Google Scholar]
- Yuki, D.; Lin, Y.M.; Fujii, Y.; Nakamura, Y.; Furukawa, Y. Isolation of LEM domain-containing 1, a novel testis-specific gene expressed in colorectal cancers. Oncol. Rep. 2004, 12, 275–280. [Google Scholar] [CrossRef]
- Gjerstorff, M.F.; Andersen, M.H.; Ditzel, H.J. Oncogenic cancer/testis antigens: Prime candidates for immunotherapy. Oncotarget 2015, 6, 15772–15787. [Google Scholar] [CrossRef] [Green Version]
- Sia, G.M.; Clem, R.L.; Huganir, R.L. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science 2013, 342, 987–991. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Arao, T.; Tamura, D.; Aomatsu, K.; Furuta, K.; Matsumoto, K.; Kaneda, H.; Kudo, K.; Fujita, Y.; Kimura, H.; et al. SRPX2 is a novel chondroitin sulfate proteoglycan that is overexpressed in gastrointestinal cancer. PLoS ONE 2012, 7, e27922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Øster, B.; Linnet, L.; Christensen, L.L.; Thorsen, K.; Ongen, H.; Dermitzakis, E.T.; Sandoval, J.; Moran, S.; Esteller, M.; Hansen, T.F.; et al. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int. J. Cancer 2013, 132, 2303–2315. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, S.R.; Xu, H.X.; Wang, W.Q.; Li, S.; Li, T.J.; Ni, Q.X.; Yu, X.J.; Liu, L.; Wu, C.T. SRPX2 and RAB31 are effective prognostic biomarkers in pancreatic cancer. J. Cancer 2019, 10, 2670–2678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasahira, T.; Kurihara-Shimomura, M.; Nishiguchi, Y.; Shimomura, H.; Kirita, T. Sushi repeat containing protein X-linked 2 is a downstream signal of LEM domain containing 1 and acts as a tumor-promoting factor in oral squamous cell carcinoma. Int. J. Mol. Sci. 2020, 21, 3655. [Google Scholar] [CrossRef]
- Miljkovic-Licina, M.; Hammel, P.; Garrido-Urbani, S.; Bradfield, P.F.; Szepetowski, P.; Imhof, B.A. Sushi repeat protein X-linked 2, a novel mediator of angiogenesis. FASEB J. 2009, 23, 4105–4116. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Lv, W.; Lv, H.; Zhang, R.; Jiang, Y. Genome-wide haplotype association analysis identifies SERPINB9, SERPINE2, GAK, and HSP90B1 as novel risk genes for oral squamous cell carcinoma. Tumour Biol. 2016, 37, 1845–1851. [Google Scholar] [CrossRef]
- Xu, D.; McKee, C.M.; Cao, Y.; Ding, Y.; Kessler, B.M.; Muschel, R.J. Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res. 2010, 70, 6988–6998. [Google Scholar] [CrossRef] [Green Version]
- McKee, C.M.; Xu, D.; Cao, Y.; Kabraji, S.; Allen, D.; Kersemans, V.; Beech, J.; Smart, S.; Hamdy, F.; Ishkanian, A.; et al. Protease nexin 1 inhibits hedgehog signaling in prostate adenocarcinoma. J. Clin. Investig. 2012, 122, 4025–4036. [Google Scholar] [CrossRef] [Green Version]
- Pagliara, V.; Adornetto, A.; Mammì, M.; Masullo, M.; Sarnataro, D.; Pietropaolo, C.; Arcone, R. Protease nexin-1 affects the migration and invasion of C6 glioma cells through the regulation of urokinase plasminogen activator and matrix metalloproteinase-9/2. Biochim. Biophys. Acta 2014, 1843, 2631–2644. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara-Shimomura, M.; Shimomura, H.; Kirita, T. SERPINE2 is an oral cancer-promoting factor that induces angiogenesis and lymphangiogenesis. Int. J. Clin. Oncol. 2021, 26, 1831–1839. [Google Scholar] [CrossRef]
- Sasahira, T.; Kirita, T.; Kurihara, M.; Yamamoto, K.; Bhawal, U.K.; Bosserhoff, A.K.; Kuniyasu, H. MIA-dependent angiogenesis and lymphangiogenesis are closely associated with progression, nodal metastasis and poor prognosis in tongue squamous cell carcinoma. Eur. J. Cancer 2010, 46, 2285–2294. [Google Scholar] [CrossRef]
- Kerbel, R.S. Tumor angiogenesis. N. Engl. J. Med. 2008, 358, 2039–2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mpekris, F.; Baish, J.W.; Stylianopoulos, T.; Jain, R.K. Role of vascular normalization in benefit from metronomic chemotherapy. Proc. Natl. Acad. Sci. USA 2017, 114, 1994–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voutouri, C.; Kirkpatrick, N.D.; Chung, E.; Mpekris, F.; Baish, J.W.; Munn, L.L.; Fukumura, D.; Stylianopoulos, T.; Jain, R.K. Experimental and computational analyses reveal dynamics of tumor vessel cooption and optimal treatment strategies. Proc. Natl. Acad. Sci. USA 2019, 116, 2662–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Zhang, W.; Wang, Y.; Zou, T.; Zhang, B.; Xu, Y.; Pang, T.; Hu, Q.; Chen, M.; Wang, L.; et al. Hypoxia-induced miR-214 expression promotes tumour cell proliferation and migration by enhancing the Warburg effect in gastric carcinoma cells. Cancer Lett. 2018, 414, 44–56. [Google Scholar] [CrossRef]
- Dougan, J.; Hawsawi, O.; Burton, L.J.; Edwards, G.; Jones, K.; Zou, J.; Nagappan, P.; Wang, G.; Zhang, Q.; Danaher, A.; et al. Proteomics-metabolomics combined approach identifies peroxidasin as a protector against metabolic and oxidative stress in prostate cancer. Int. J. Mol. Sci. 2019, 20, 3046. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Yang, X.; Jin, C.; Li, L.; Cui, Q.; Guo, Y.; Dong, Y.; Yang, X.; Guo, L.; Zhang, M. Identification and verification of differentially expressed microRNAs and their target genes for the diagnosis of esophageal cancer. Oncol. Lett. 2018, 16, 3642–3650. [Google Scholar] [CrossRef]
- Zheng, Y.Z.; Liang, L. High expression of PXDN is associated with poor prognosis and promotes proliferation, invasion as well as migration in ovarian cancer. Ann. Diagn. Pathol. 2018, 34, 161–165. [Google Scholar] [CrossRef]
- Kurihara-Shimomura, M.; Sasahira, T.; Shimomura, H.; Kirita, T. Peroxidan plays a tumor-promoting role in oral squamous cell carcinoma. Int. J. Mol. Sci. 2020, 21, 5416. [Google Scholar] [CrossRef]
- Kon, S.; Ishibashi, K.; Katoh, H.; Kitamoto, S.; Shirai, T.; Tanaka, S.; Kajita, M.; Ishikawa, S.; Yamauchi, H.; Yako, Y.; et al. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat. Cell Biol. 2017, 19, 530–541. [Google Scholar] [CrossRef]
- Gao, P.; Zhang, H.; Dinavahi, R.; Li, F.; Xiang, Y.; Raman, V.; Bhujwalla, Z.M.; Felsher, D.W.; Cheng, L.; Pevsner, J.; et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell 2007, 12, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, E.A.; Thompson, I.M., Jr.; Tangen, C.M.; Crowley, J.J.; Lucia, M.S.; Goodman, P.J.; Minasian, L.M.; Ford, L.G.; Parnes, H.L.; Gaziano, J.M.; et al. Vitamin E and the risk of prostate cancer: The Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 2011, 306, 1549–1556. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S.; Tuveson, D.A. The promise and perils of antioxidants for cancer patients. N. Engl. J. Med. 2014, 371, 177–178. [Google Scholar] [CrossRef]
- Piskounova, E.; Agathocleous, M.; Murphy, M.M.; Hu, Z.; Huddlestun, S.E.; Zhao, Z.; Leitch, A.M.; Johnson, T.M.; DeBerardinis, R.J.; Morrison, S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015, 527, 186–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [Green Version]
- Drago-Ferrante, R.; Pentimalli, F.; Carlisi, D.; De Blasio, A.; Saliba, C.; Baldacchino, S.; Degaetano, J.; Debono, J.; Caruana-Dingli, G.; Grech, G.; et al. Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget 2017, 8, 28939–28958. [Google Scholar] [CrossRef] [Green Version]
- Milevskiy, M.J.G.; Sandhu, G.K.; Wronski, A.; Korbie, D.; Brewster, B.L.; Shewan, A.; Edwards, S.L.; French, J.D.; Brown, M.A. MiR-29b-1-5p is altered in BRCA1 mutant tumours and is a biomarker in basal-like breast cancer. Oncotarget 2018, 9, 33577–33588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Zhang, Q.; Cheng, W.; Zhang, Z.; Wang, J.; Ge, J. Effect of miR-29b-1* and miR-29c knockdown on cell growth of the bladder cancer cell line T24. J. Int. Med. Res. 2013, 41, 1803–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, M.P.; Tong, P.; Diao, L.; Cardnell, R.J.; Gibbons, D.L.; William, W.N.; Skoulidis, F.; Parra, E.R.; Rodriguez-Canales, J.; Wistuba, I.I.; et al. A Patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin. Cancer Res. 2016, 22, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Kurihara-Shimomura, M.; Sasahira, T.; Shimomura, H.; Nakashima, C.; Kirita, T. The oncogenic activity of miR-29b-1-5p induces the epithelial-mesenchymal transition in oral squamous cell carcinoma. J. Clin. Med. 2019, 8, 273. [Google Scholar] [CrossRef] [Green Version]
- Sasahira, T.; Kurihara, M.; Bhawal, U.K.; Ueda, N.; Shimomoto, T.; Yamamoto, K.; Kirita, T.; Kuniyasu, H. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br. J. Cancer 2012, 107, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Righini, C.A.; de Fraipont, F.; Timsit, J.F.; Faure, C.; Brambilla, E.; Reyt, E.; Favrot, M.C. Tumor-specific methylation in saliva: A promising biomarker for early detection of head and neck cancer recurrence. Clin. Cancer Res. 2007, 13, 1179–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, Y.; Wan, Y.; Vagenas, D.; Ovchinnikov, D.A.; Perry, C.F.; Davis, M.J.; Punyadeera, C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer 2016, 16, 749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef] [Green Version]
| HPV-Positive Cancer | HPV-Negative Cancer | |
|---|---|---|
| Risk factor | Oral sex | Smoking, alcohol abuse |
| Age | Under 60 years | Above 60 years |
| Surrogate marker | p16 | p53 |
| Susceptible site | Oropharynx | Various sites |
| Outcome | Good | Bad |
| Basal | Mesenchymal | Atypical | Classical | |
|---|---|---|---|---|
| Differentiation | Well, moderate | Moderate, poor | Moderate | Moderate, poor |
| Predominant site | Oral cavity | Oral cavity | Oropharynx | Larynx |
| Lymph node metastability | Low | Moderate | High | Low |
| 7p gain | Yes | Yes | No | Yes |
| CDKN2A loss | Occasionally | Rarely | Rarely | Frequently |
| SOX2 expression | Low | Low | High | High |
| TP63 expression | High | Moderate | Moderate | Moderate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasahira, T.; Kurihara-Shimomura, M.; Shimojjukoku, Y.; Shima, K.; Kirita, T. Searching for New Molecular Targets for Oral Squamous Cell Carcinoma with a View to Clinical Implementation of Precision Medicine. J. Pers. Med. 2022, 12, 413. https://doi.org/10.3390/jpm12030413
Sasahira T, Kurihara-Shimomura M, Shimojjukoku Y, Shima K, Kirita T. Searching for New Molecular Targets for Oral Squamous Cell Carcinoma with a View to Clinical Implementation of Precision Medicine. Journal of Personalized Medicine. 2022; 12(3):413. https://doi.org/10.3390/jpm12030413
Chicago/Turabian StyleSasahira, Tomonori, Miyako Kurihara-Shimomura, Yudai Shimojjukoku, Kaori Shima, and Tadaaki Kirita. 2022. "Searching for New Molecular Targets for Oral Squamous Cell Carcinoma with a View to Clinical Implementation of Precision Medicine" Journal of Personalized Medicine 12, no. 3: 413. https://doi.org/10.3390/jpm12030413






