Deciphering of Adult Glioma Vulnerabilities through Expression Pattern Analysis of GABA, Glutamate and Calcium Neurotransmitter Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Extraction
2.2. GABA, Glutamate and Calcium Pathway Gene Extraction
2.3. Gene Expression Normalization
2.4. Unsupervised Clustering
2.5. Differential Gene Expression Analysis
2.6. miRNAs Interactome Profiling
2.7. Statistical Analysis
2.8. Snakemake Pipepeline Creation
3. Results
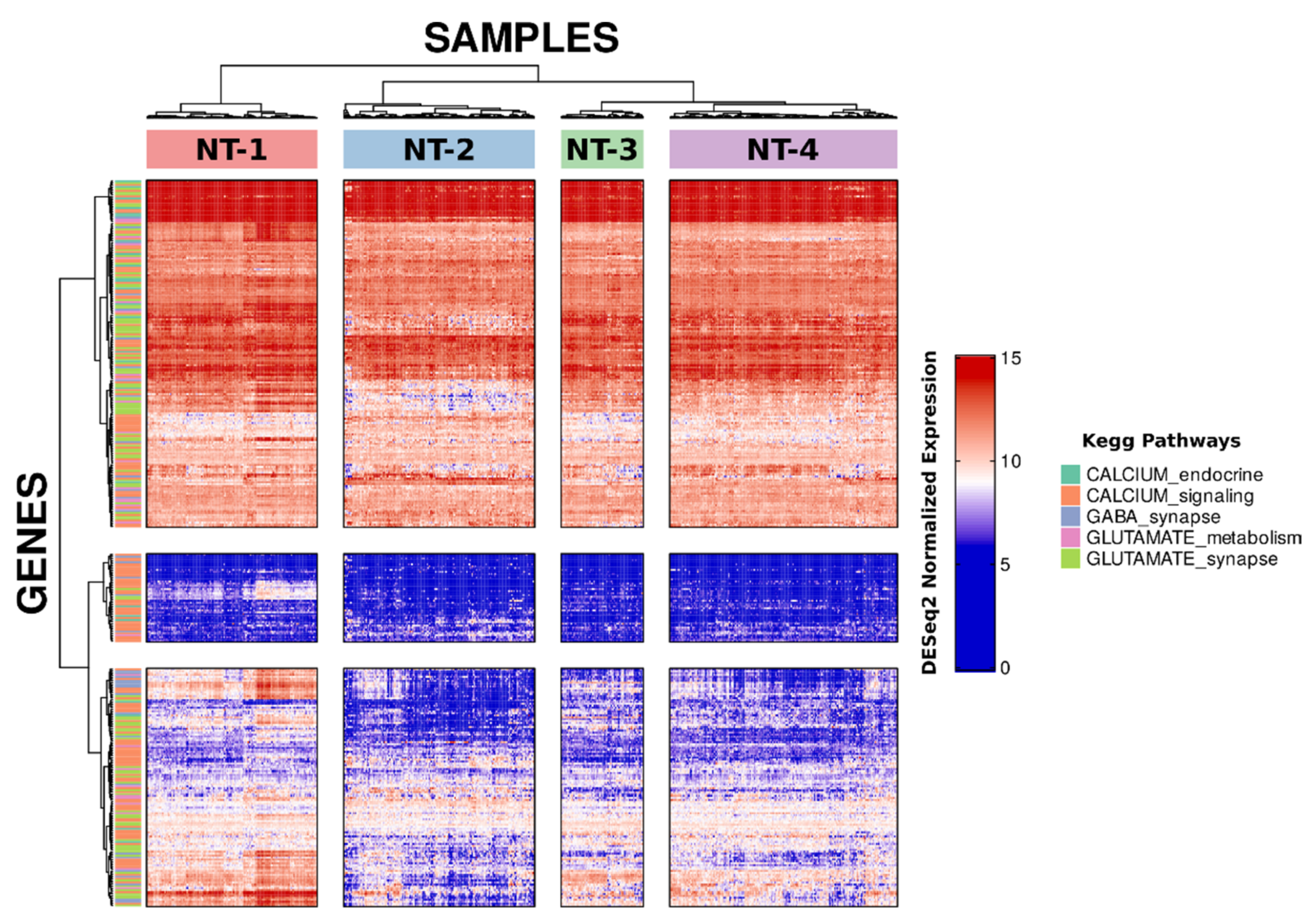
3.1. Unsupervised Clustering Based on GABA, Glutamate and Calcium Gene Expression Distinguishes Four Clusters with Distinct Neurotransmission Profiles
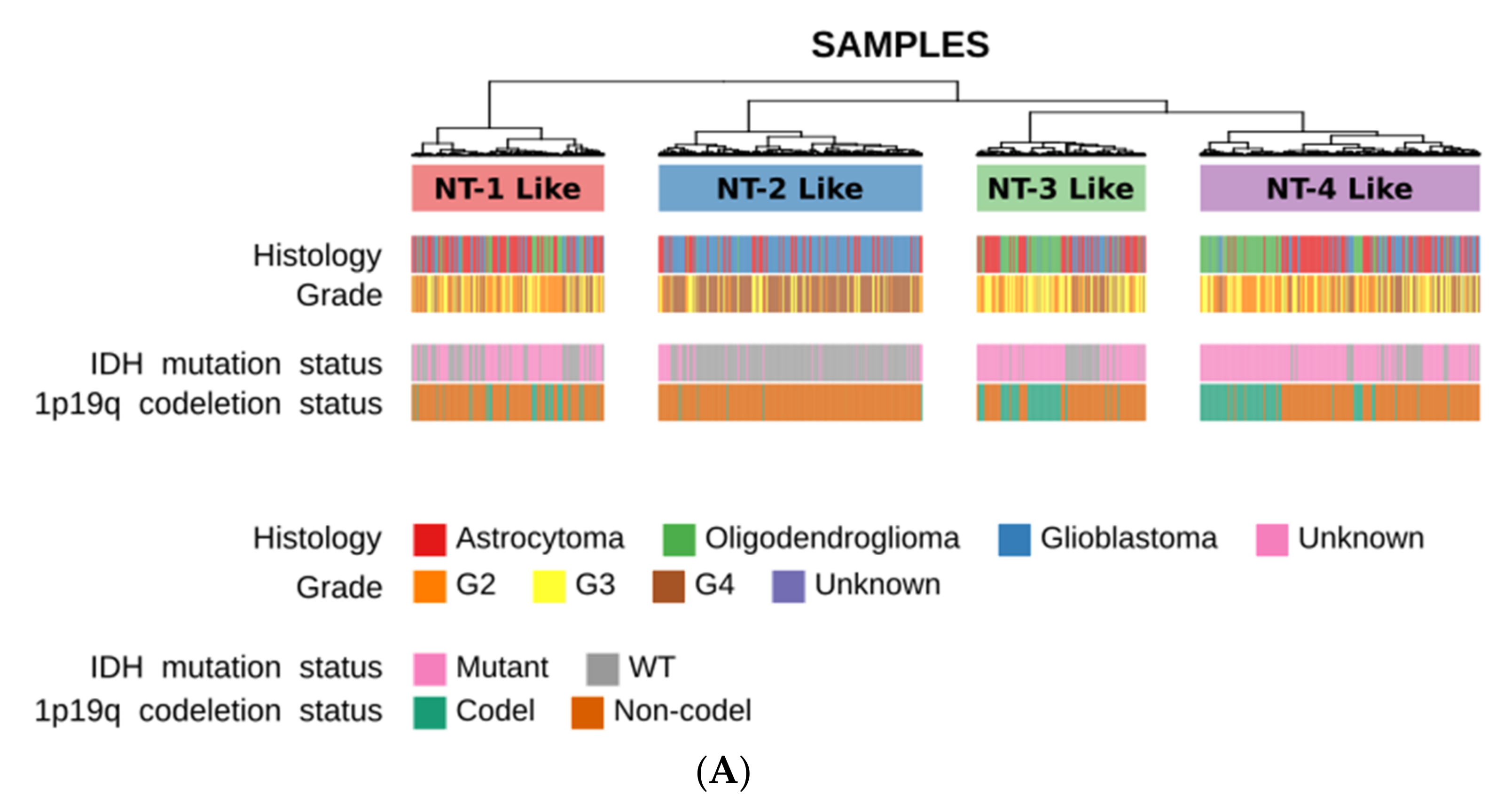
3.2. Neurotransmission-Based Glioma Clustering Recapitulates Current Existing Glioma Molecular Subgroups and Identifies a Novel Subgroup with a Distinct Expression Profile
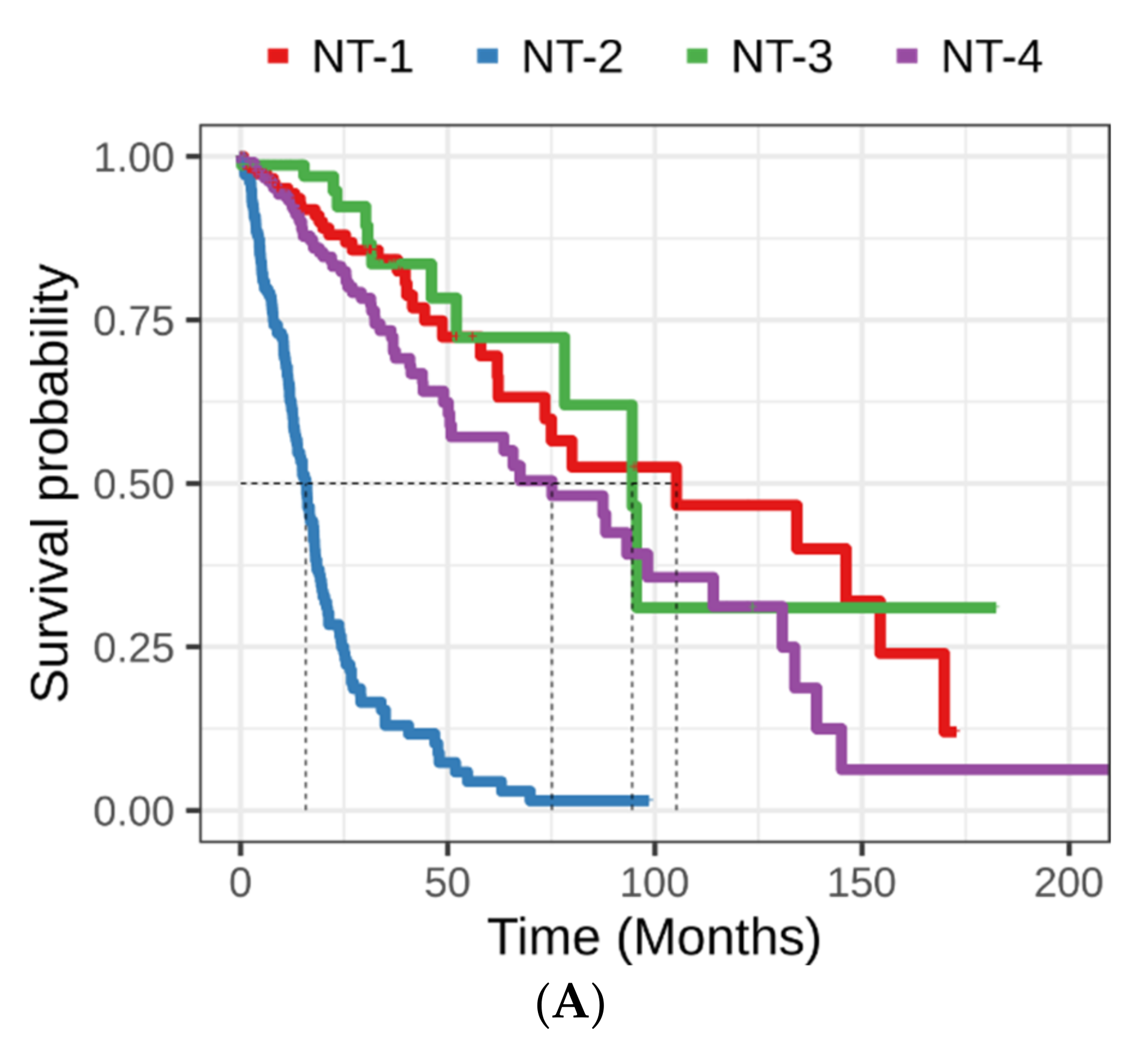
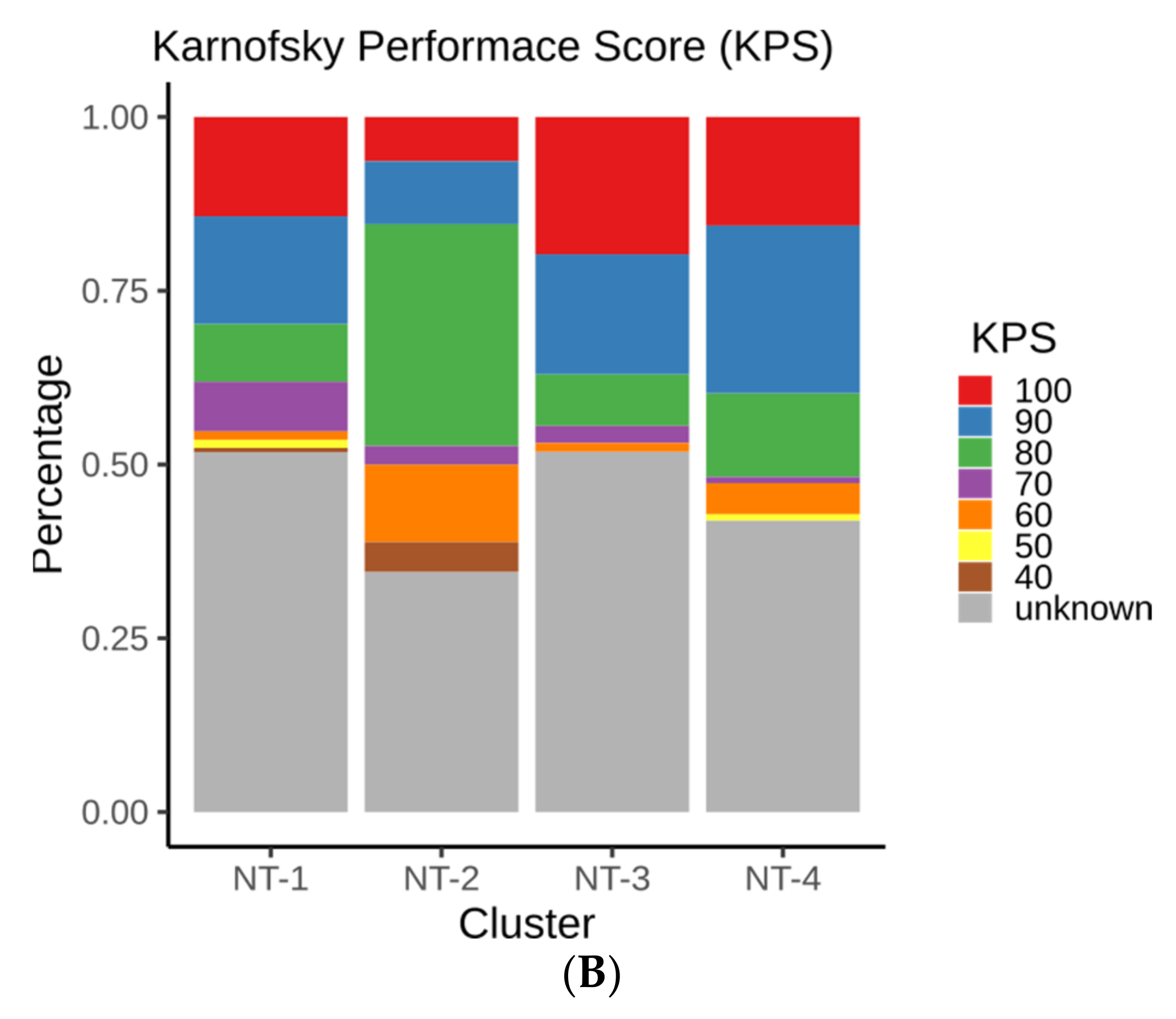
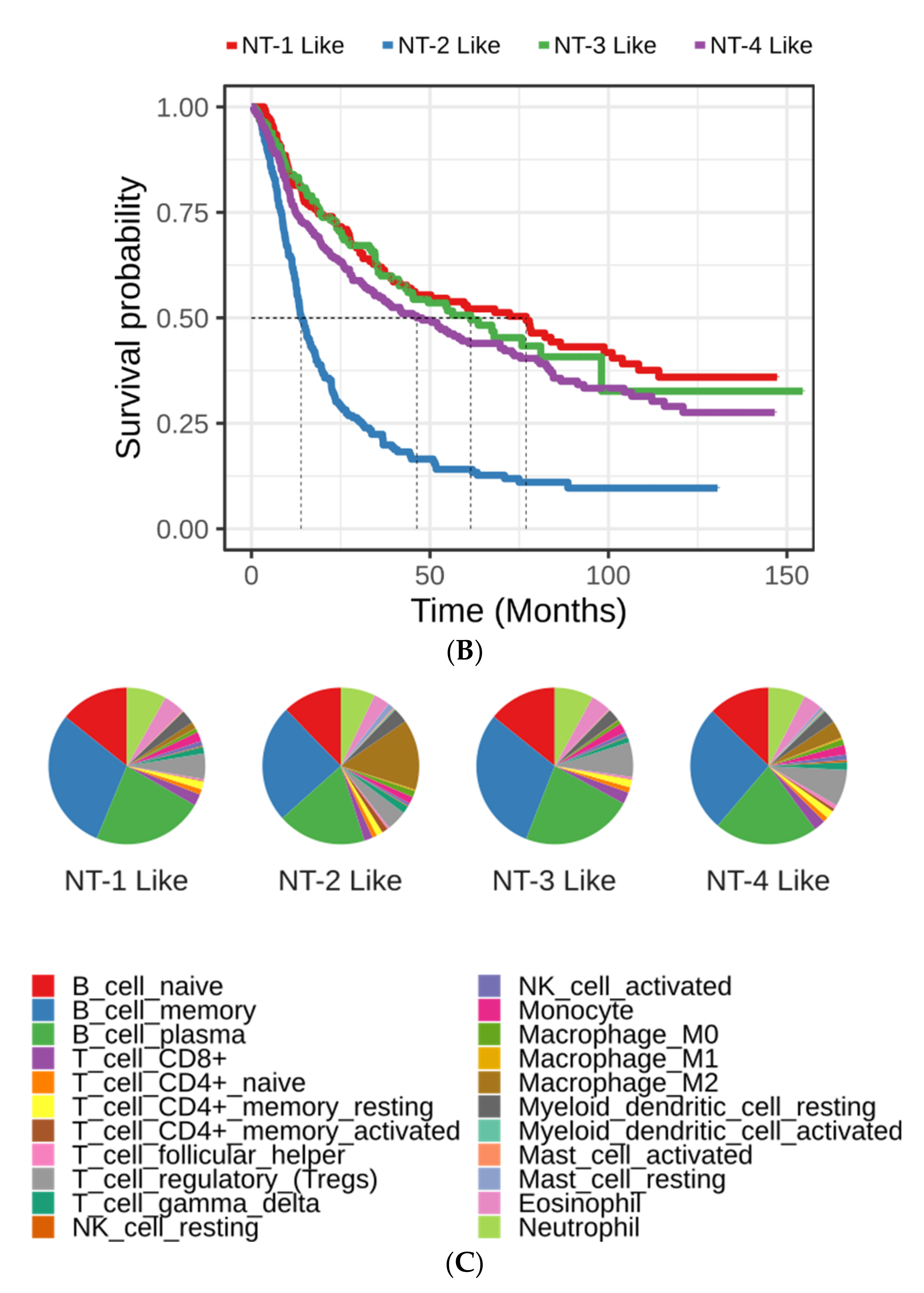
3.3. Clinical and Epidemiological Characterization of Neurotransmission-Based Glioma Clusters
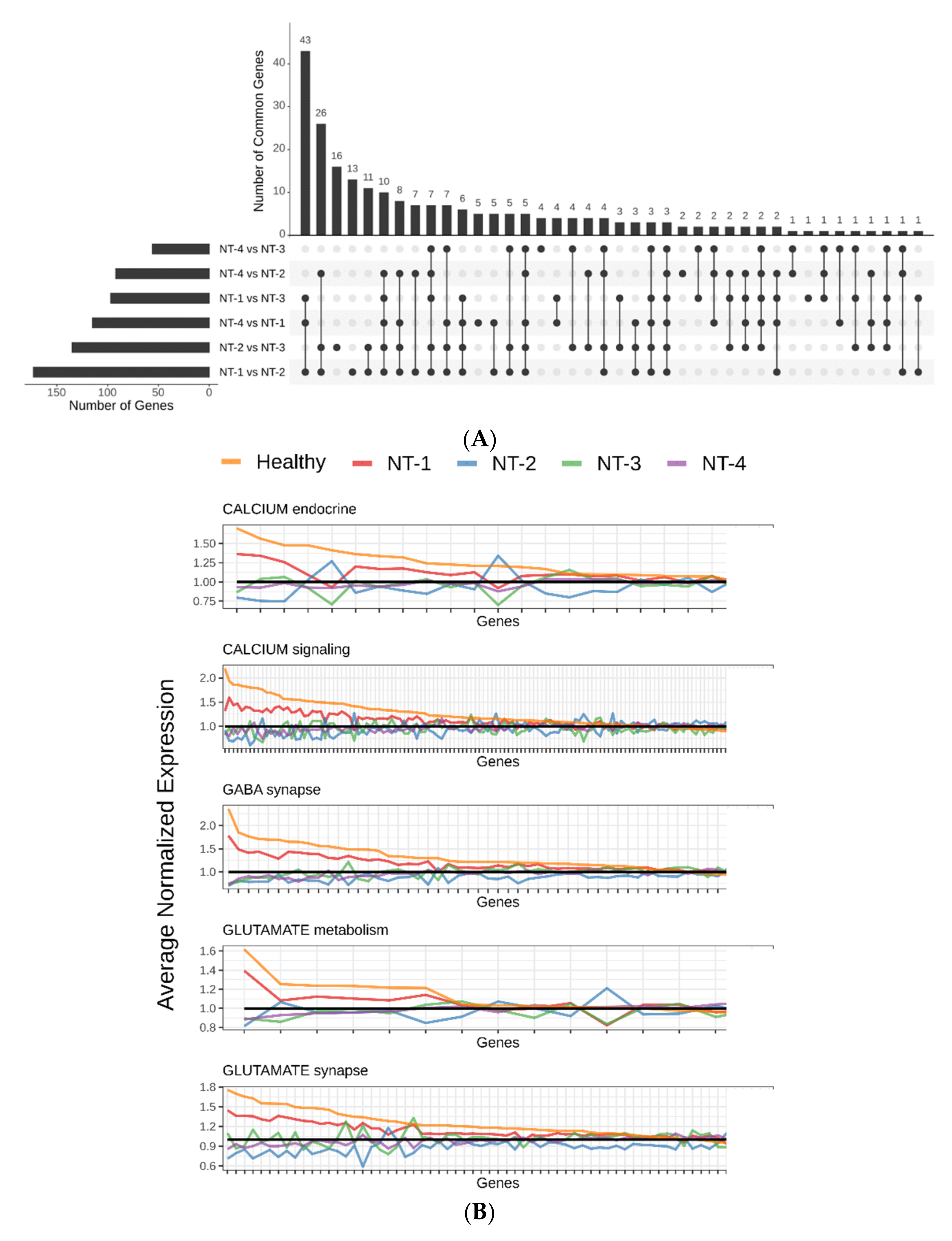
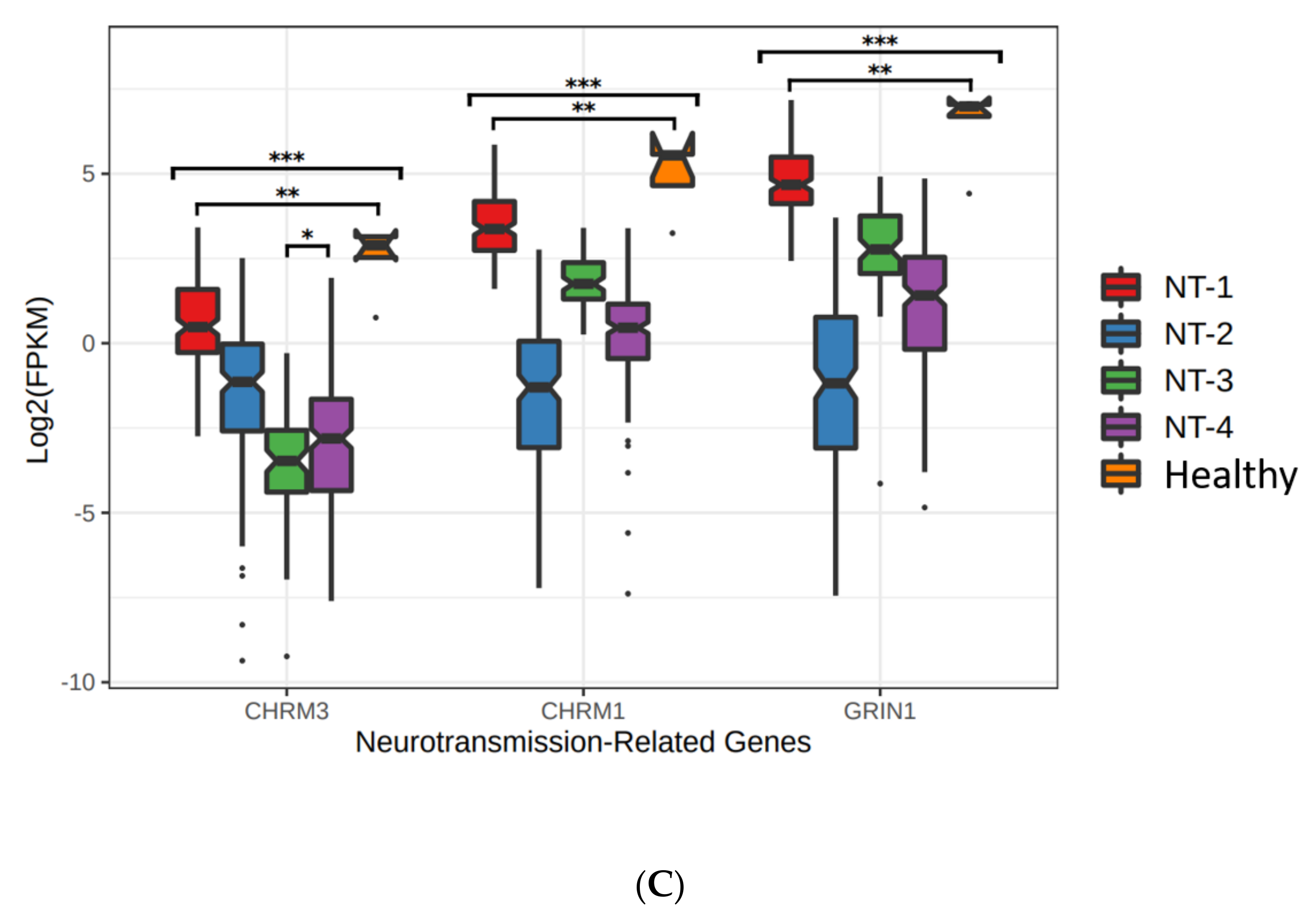
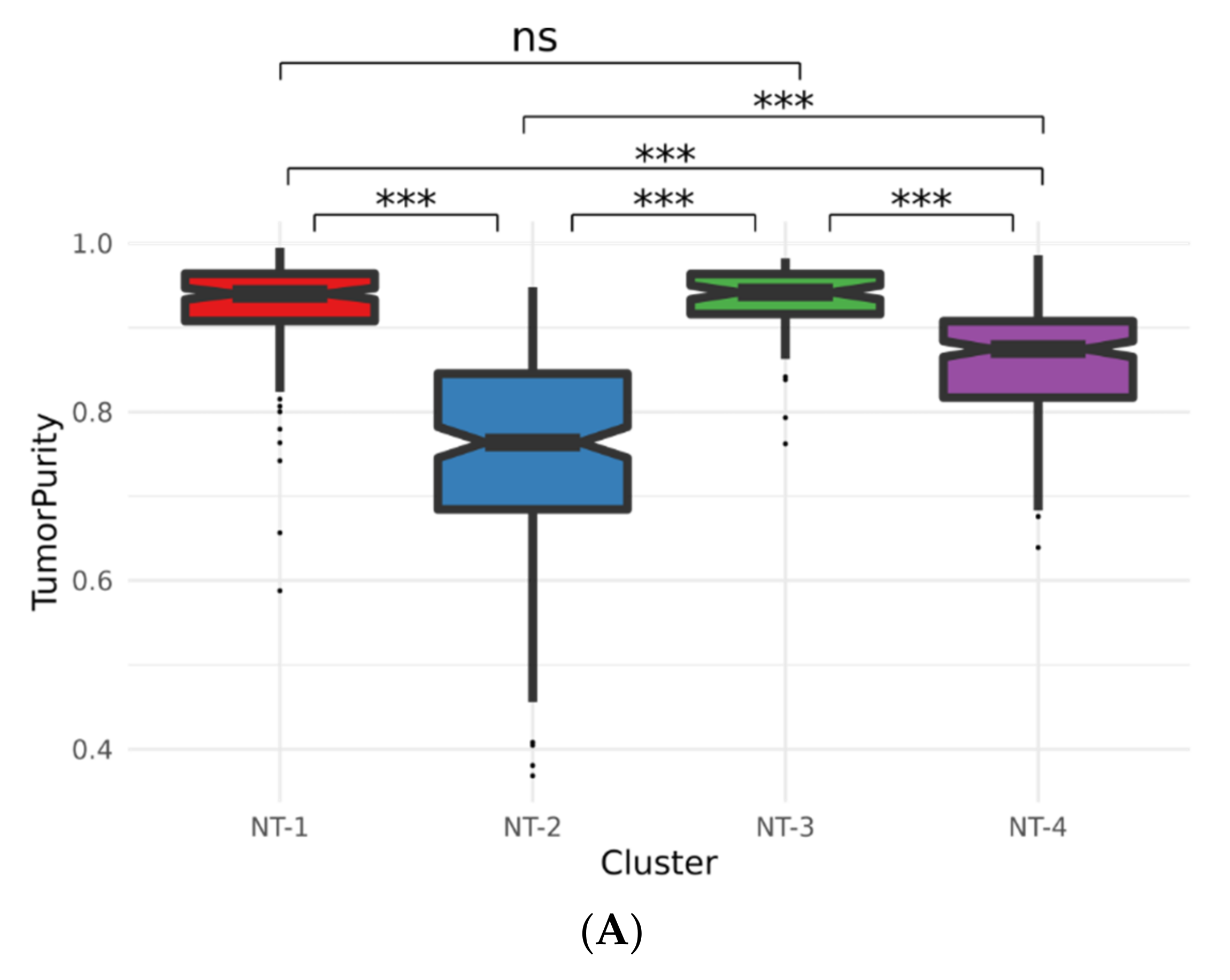
3.4. Lower Expression of Neurotransmission Genes Correlates with Increased Aggressivity in the NT-1, NT-2, NT-3 and NT-4 Gliomas
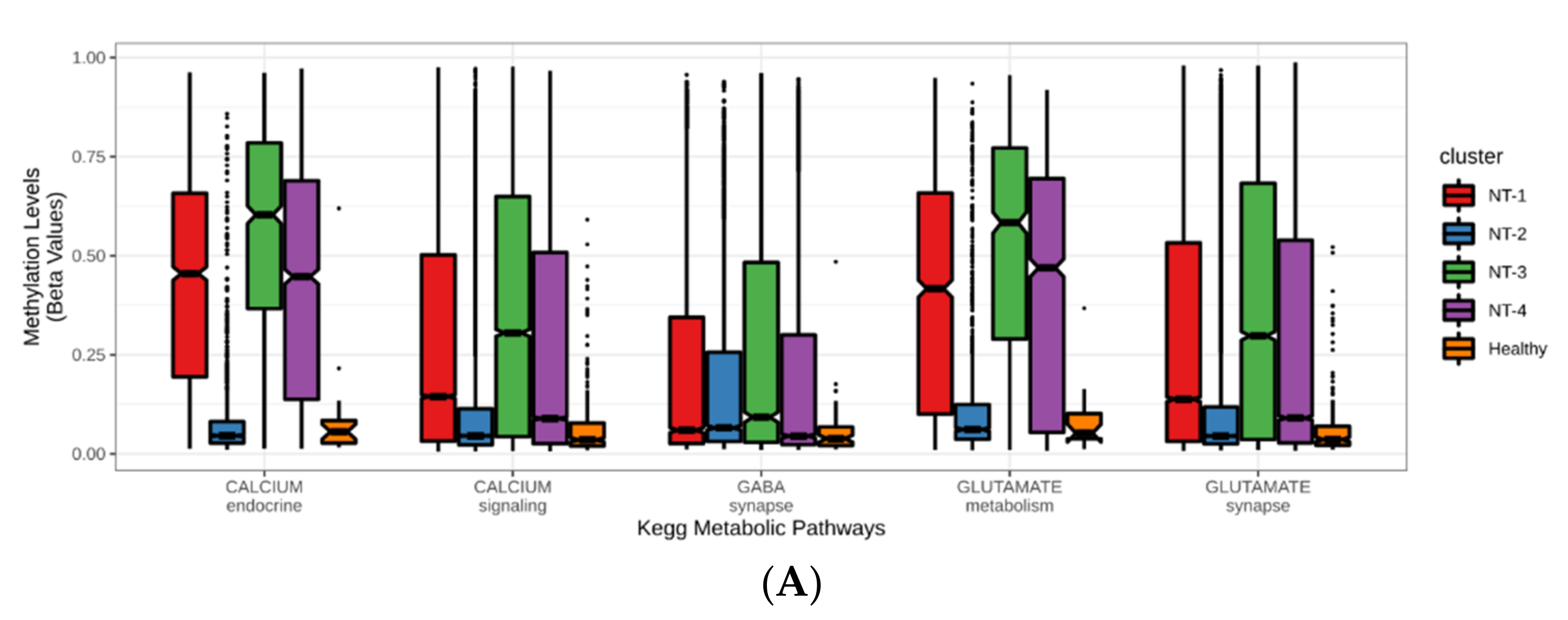
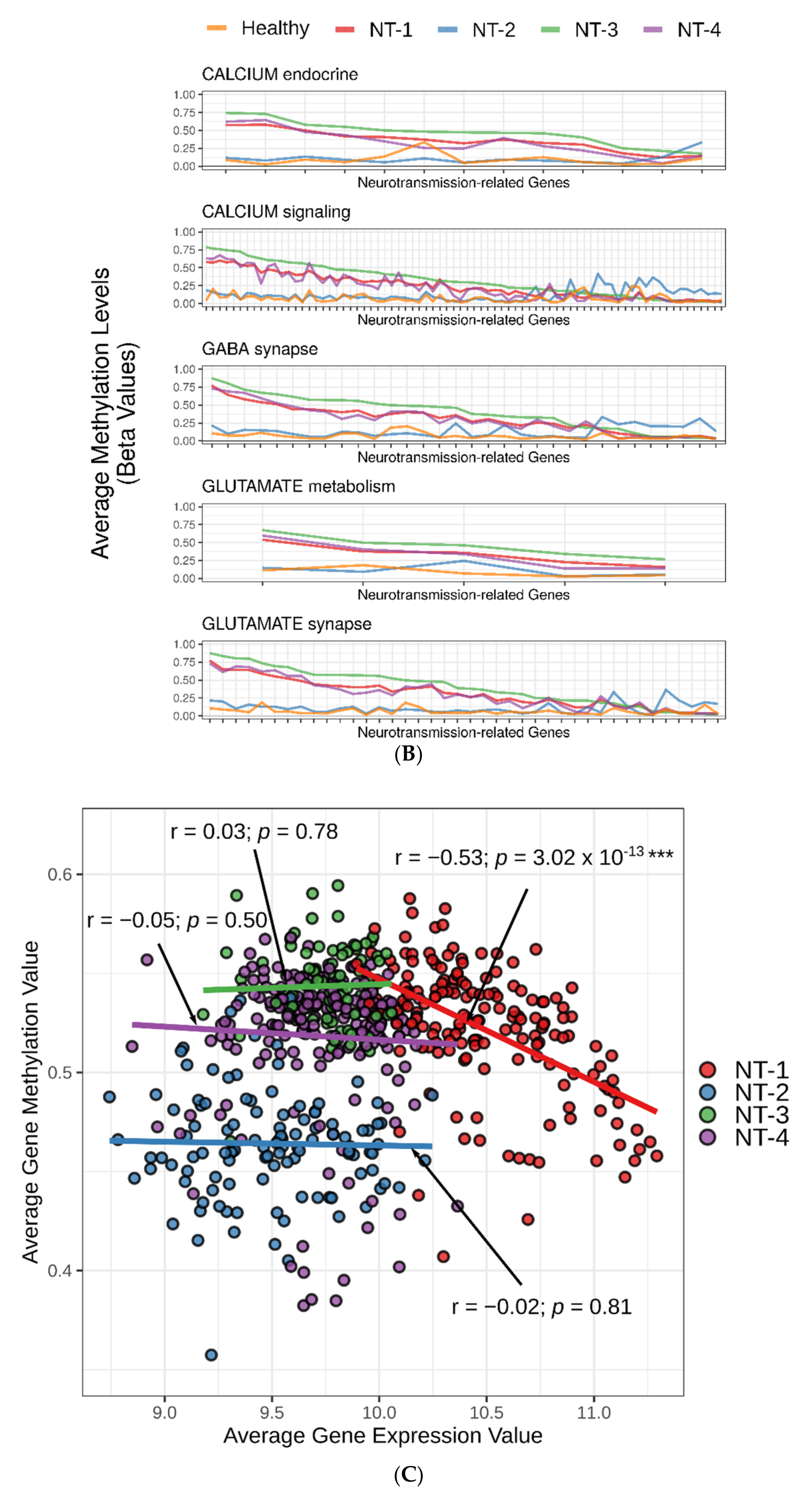
3.5. Correlation between DNA Hypermethylation and Gene Expression Is Preserved in NT-1 Gliomas
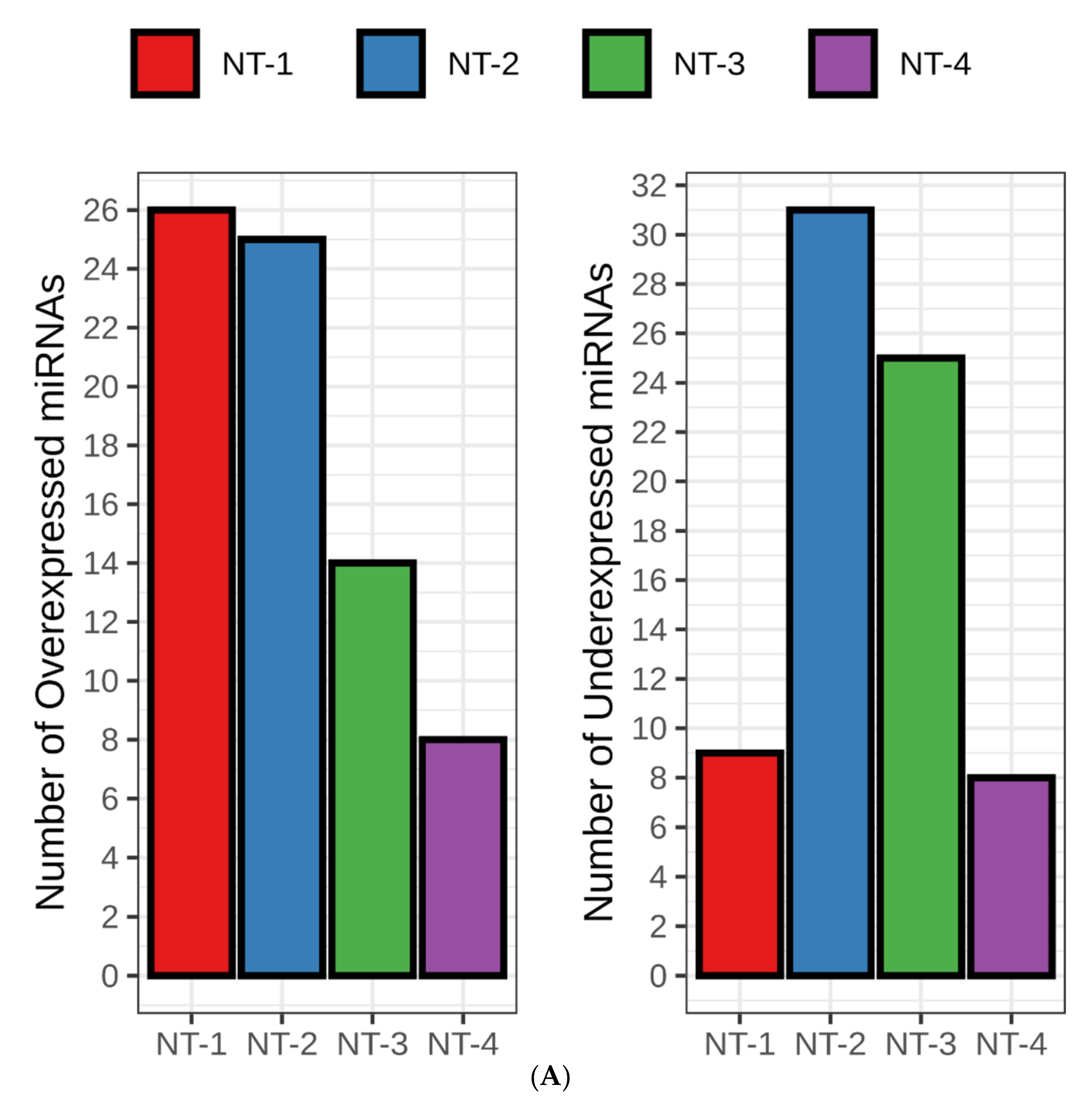
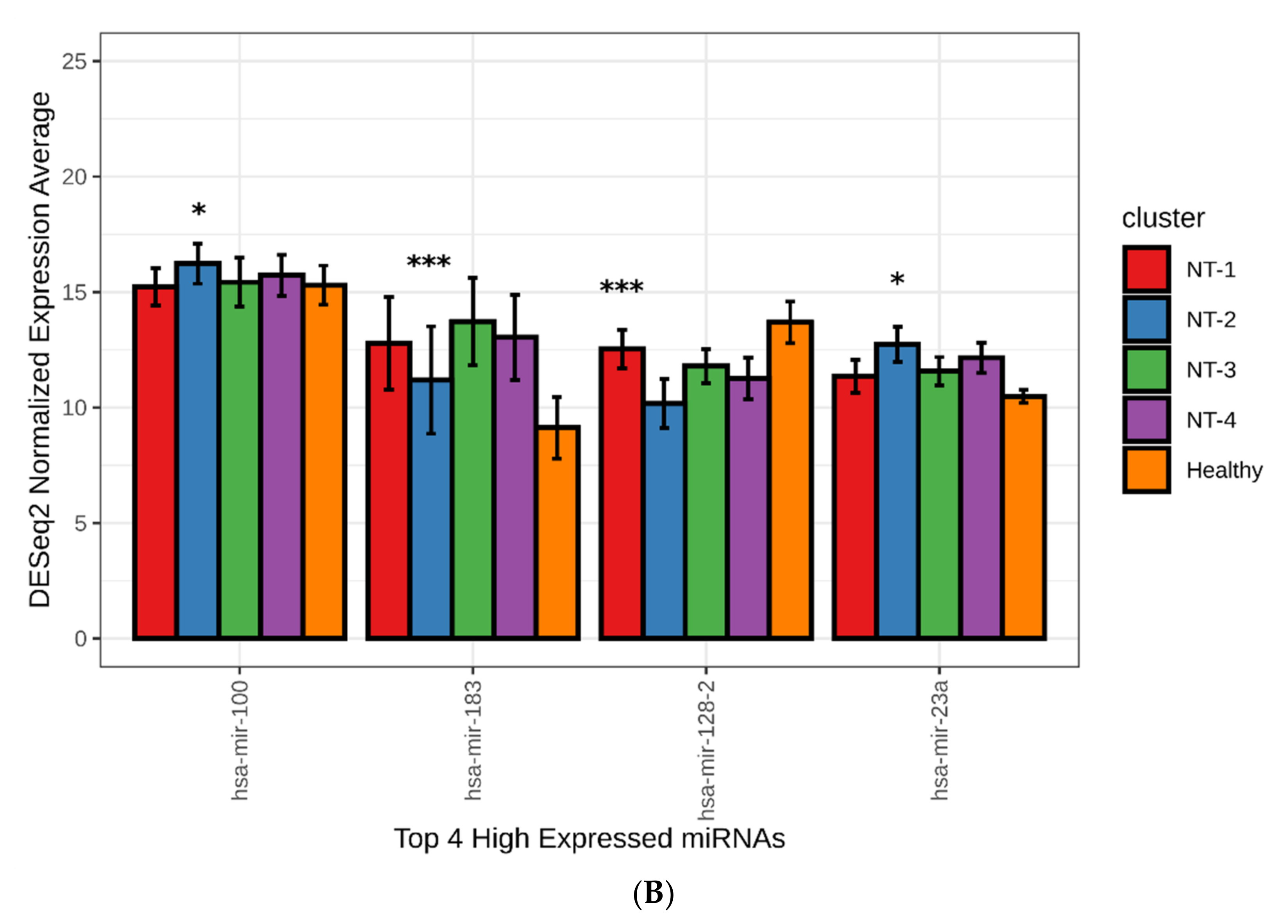
3.6. NT-1 and NT-2 Gliomas Are Regulated by More Complex Epigenetic Mechanisms Involving Differential Expression of microRNA
3.7. Neurotransmission-Related Gene Expression Correlates with the Immune Response Signaling Pathways in NT-1-4 Glioma Clusters
3.8. Immune Cell Characterization Reveals Different Tumor Immune Microenvironment Composition
3.9. Neurotransmission-Related Transcriptomic Profiling on the Chinese Glioma Genome Atlas Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Nuño, M.; Walia, S.; Mukherjee, D.; Black, K.L.; Patil, C.G. Treatment and survival of patients harboring histological variants of glioblastoma. J. Clin. Neurosci. 2014, 21, 1709–1713. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.G.; Fine, H.A. Diffuse Glioma Heterogeneity and Its Therapeutic Implications. Cancer Discov. 2021, 11, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Shirai, K.; Chakravarti, A. Towards personalized therapy for patients with glioblastoma. Expert Rev. Anticancer Ther. 2011, 11, 1935–1944. [Google Scholar] [CrossRef] [Green Version]
- Holland, E.; Ene, C. Personalized Medicine for Gliomas. Surg. Neurol. Int. 2015, 6, 89–95. [Google Scholar] [CrossRef]
- Manini, I.; Caponnetto, F.; Bartolini, A.; Ius, T.; Mariuzzi, L.; Loreto, C.D.; Beltrami, A.P.; Cesselli, D. Role of Microenvironment in Glioma Invasion: What We Learned from In Vitro Models. Int. J. Mol. Sci. 2018, 19, 147. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 Mutations in Gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Berzero, G.; Di Stefano, A.L.; Ronchi, S.; Bielle, F.; Villa, C.; Guillerm, E.; Capelle, L.; Mathon, B.; Laurenge, A.; Giry, M.; et al. IDH-wildtype lower-grade diffuse gliomas: The importance of histological grade and molecular assessment for prognostic stratification. Neuro-Oncology 2021, 23, 955–966. [Google Scholar] [CrossRef]
- Wesseling, P.; Bent, M.V.D.; Perry, A. Oligodendroglioma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 809–827. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Yang, M.; Gaur, U.; Xu, H.; Yao, Y.; Li, D. Alpha-Ketoglutarate: Physiological Functions and Applications. Biomol. Ther. 2016, 24, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Schousboe, A.; Bak, L.K.; Waagepetersen, H.S. Astrocytic Control of Biosynthesis and Turnover of the Neurotransmitters Glutamate and GABA. Front. Endocrinol. 2013, 4, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiter-Brennan, C.; Semmler, L.; Klein, A. The effects of 2-hydroxyglutarate on the tumorigenesis of gliomas. Contemp. Oncol. 2018, 22, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, H.S.; Morishita, W.; Geraghty, A.C.; Silverbush, D.; Gillespie, S.M.; Arzt, M.; Tam, L.T.; Espenel, C.; Ponnuswami, A.; Ni, L.; et al. Electrical and synaptic integration of glioma into neural circuits. Nature 2019, 573, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Crowley, T.; Cryan, J.F.; Downer, E.J.; O’Leary, O.F. Inhibiting neuroinflammation: The role and therapeutic potential of GABA in neuro-immune interactions. Brain. Behav. Immun. 2016, 54, 260–277. [Google Scholar] [CrossRef] [PubMed]
- LoTurco, J.J.; Owens, D.F.; Heath, M.J.S.; Davis, M.B.E.; Kriegstein, A.R. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 1995, 15, 1287–1298. [Google Scholar] [CrossRef] [Green Version]
- Favuzzi, E.; Saldi, G.A.; Binan, L.; Ibrahim, L.A.; Fernández-Otero, M.; Cao, Y.; Zeine, A.; Sefah, A.; Zheng, K.; Xu, Q.; et al. GABA-receptive microglia selectively sculpt developing inhibitory circuits. Cell 2021, 184, 4048–4063.e32. [Google Scholar] [CrossRef] [PubMed]
- Huberfeld, G.; Vecht, C.J. Seizures and gliomas—Towards a single therapeutic approach. Nat. Rev. Neurol. 2016, 12, 204–216. [Google Scholar] [CrossRef]
- Network, T.C.G.A. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Meng, F.; Wang, W.; Wang, Z.; Zhang, C.; Jiang, T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. Data 2017, 4, 170024. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.S.; Chen, H.M.; Yang, M.Y.; Zhang, C.B.; Yu, K.; Ye, W.L.; Hu, B.Q.; Yan, W.; Zhang, W.; Akers, J.; et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014, 24, 1765–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2019, 48, D189–D197. [Google Scholar] [CrossRef]
- Köster, J.; Rahmann, S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.D.; Allaire, A.; Diamandis, P.; Bisaillon, M.; Scott, M.S.; Richer, M. A machine learning analysis of a “normal-like” IDH-WT diffuse glioma transcriptomic subgroup associated with prolonged survival reveals novel immune and neurotransmitter-related actionable targets. BMC Med. 2020, 18, 1–18. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Rusk, N. Expanded CIBERSORTx. Nat. Methods 2019, 16, 577. [Google Scholar] [CrossRef] [PubMed]
- Lakomy, R.; Kazda, T.; Selingerova, I.; Poprach, A.; Pospisil, P.; Belanova, R.; Fadrus, P.; Vybihal, V.; Smrcka, M.; Jancalek, R.; et al. Real-World Evidence in Glioblastoma: Stupp’s Regimen After a Decade. Front. Oncol. 2020, 10, 840. [Google Scholar] [CrossRef]
- Radin, D.P.; Tsirka, S.E. Interactions between Tumor Cells, Neurons, and Microglia in the Glioma Microenvironment. Int. J. Mol. Sci. 2020, 21, 28476. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [Green Version]
- Jalbert, L.E.; Elkhaled, A.; Phillips, J.J.; Neill, E.; Williams, A.; Crane, J.C.; Olson, M.P.; Molinaro, A.M.; Berger, M.S.; Kurhanewicz, J.; et al. Metabolic Profiling of IDH Mutation and Malignant Progression in Infiltrating Glioma. Sci. Rep. 2017, 7, 44792. [Google Scholar] [CrossRef] [Green Version]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Raineri, S.; Mellor, J. IDH1: Linking Metabolism and Epigenetics. Front. Genet. 2018, 9, 493. [Google Scholar] [CrossRef] [Green Version]
- Marquez, V.E.; Kelley, J.A.; Agbaria, R.; Ben-Kasus, T.; Cheng, J.C.; Yoo, C.B.; Jones, P.A. Zebularine: A Unique Molecule for an Epigenetically Based Strategy in Cancer Chemotherapy. Ann. N. Y. Acad. Sci. 2005, 1058, 246–254. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, Z.; Wu, L.; Liu, C.; Chen, Q.; Liu, J.; Wang, X.; Zhuang, Z.; Li, W.; Xu, S.; et al. Upregulation of miR-183 expression and its clinical significance in human brain glioma. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2016, 37, 1341–1347. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Sasayama, T.; Tanaka, K.; Nakamizo, S.; Nishihara, M.; Mizukawa, K.; Kohta, M.; Koyama, J.; Miyake, S.; Taniguchi, M.; et al. MicroRNA-183 upregulates HIF-1α by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J. Neuro-Oncol. 2013, 111, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Yuan, R.; Cheng, S.; Xiong, K.; Zhu, X.; Zhang, Y. Overexpressed miR-183 promoted glioblastoma radioresistance via down-regulating LRIG1. Biomed. Pharmacother. 2018, 97, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Ichiyama, K.; Dong, C. The role of miR-183 cluster in immunity. Cancer Lett. 2018, 443, 108–114. [Google Scholar] [CrossRef]
- Gong, F.-H.; Long, L.; Yang, Y.-S.; Shen, D.-H.; Zhang, Y.-S.; Wang, X.-S.; Zhang, X.-P.; Xiao, X.-Q. Attenuated macrophage activation mediated by microRNA-183 knockdown through targeting NR4A2. Exp. Ther. Med. 2021, 21, 1. [Google Scholar] [CrossRef]
- Muraleedharan, C.K.; McClellan, S.A.; Barrett, R.P.; Li, C.; Montenegro, D.; Carion, T.; Berger, E.; Hazlett, L.D.; Xu, S. Inactivation of the miR-183/96/182 Cluster Decreases the Severity of Pseudomonas aeruginosa-Induced Keratitis. Investig. Ophthalmol. Vis. Sci. 2016, 57, 1506–1517. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Wang, J.; Yan, Z.; You, Y.; Li, C.; Qian, X.; Yin, Y.; Zhao, P.; Wang, Y.; Wang, X.; et al. MiR-128 Inhibits Tumor Growth and Angiogenesis by Targeting p70S6K1. PLoS ONE 2012, 7, e32709. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, F.; Jiao, Y.; Feng, J.; Tang, W.; Yao, H.; Gong, C.; Chen, J.; Su, F.; Zhang, Y.; et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 7105–7115. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.H.; Cheng, C.H.; Shih, C.M.; Ho, K.H.; Lin, C.W.; Lee, C.C.; Liu, A.J.; Chang, C.K.; Chen, K.C. The Inhibition of microRNA-128 on IGF-1-Activating mTOR Signaling Involves in Temozolomide-Induced Glioma Cell Apoptotic Death. PLoS ONE 2016, 11, e0167096. [Google Scholar] [CrossRef]
- She, X.; Yu, Z.; Cui, Y.; Lei, Q.; Wang, Z.; Xu, G.; Xiang, J.; Wu, M.; Li, G. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol. Rep. 2014, 32, 957–964. [Google Scholar] [CrossRef] [Green Version]
- Lian, S.; Shi, R.; Bai, T.; Liu, Y.; Miao, W.; Wang, H.; Liu, X.; Fan, Y. Anti-miRNA-23a oligonucleotide suppresses glioma cells growth by targeting apoptotic protease activating factor-1. Curr. Pharm. Des. 2013, 19, 6382–6389. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Liu, M.; Peng, X.; Zhou, P.; Zhou, J.; Xu, K.; Xu, H.; Jiang, S. MiR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int. J. Oncol. 2013, 42, 757–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Chen, D.; Cui, Y.; Li, Z.; Huang, J. Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10. Sci. Rep. 2013, 3, 3423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, J.; Yang, F.; Wang, Y.; Wang, Y.; Xue, G.; Mei, Q.; Wang, F.; Sun, S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J. Cell. Biochem. 2014, 115, 772–784. [Google Scholar] [CrossRef]
- Li, X.; Liao, D.; Wang, X.; Wu, Z.; Nie, J.; Bai, M.; Fu, X.; Mei, Q.; Han, W. Elevated microRNA-23a Expression Enhances the Chemoresistance of Colorectal Cancer Cells with Microsatellite Instability to 5-Fluorouracil by Directly Targeting ABCF1. Curr. Protein Pept. Sci. 2015, 16, 301–309. [Google Scholar] [CrossRef]
- Alrfaei, B.M.; Vemuganti, R.; Kuo, J.S. microRNA-100 Targets SMRT/NCOR2, Reduces Proliferation, and Improves Survival in Glioblastoma Animal Models. PLoS ONE 2013, 8, e80865. [Google Scholar] [CrossRef]
- Alrfaei, B.M.; Clark, P.; Vemuganti, R.; Kuo, J.S. MicroRNA miR-100 Decreases Glioblastoma Growth by Targeting SMARCA5 and ErbB3 in Tumor-Initiating Cells. Technol. Cancer Res. Treat. 2020, 19. [Google Scholar] [CrossRef]
- Luan, Y.; Zhang, S.; Zuo, L.; Zhou, L. Overexpression of miR-100 inhibits cell proliferation, migration, and chemosensitivity in human glioblastoma through FGFR3. OncoTargets Ther. 2015, 8, 3391–3400. [Google Scholar]
- Bunse, L.; Pusch, S.; Bunse, T.; Sahm, F.; Sanghvi, K.; Friedrich, M.; Alansary, D.; Sonner, J.K.; Green, E.; Deumelandt, K.; et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 2018, 24, 1192–1203. [Google Scholar] [CrossRef]
- Friedrich, M.; Sankowski, R.; Bunse, L.; Kilian, M.; Green, E.; Ramallo Guevara, C.; Pusch, S.; Poschet, G.; Sanghvi, K.; Hahn, M.; et al. Tryptophan metabolism drives dynamic immunosuppressive myeloid states in IDH-mutant gliomas. Nat. Cancer 2021, 2, 723–740. [Google Scholar] [CrossRef]
- Kennedy, B.C.; Showers, C.R.; Anderson, D.E.; Anderson, L.; Canoll, P.; Bruce, J.N.; Anderson, R.C.E. Tumor-associated macrophages in glioma: Friend or foe? J. Oncol. 2013, 2013, 486912. [Google Scholar] [CrossRef] [PubMed]


| Variable | Category | NT-1 (n = 168) | NT-2 (n = 188) | NT-3 (n = 81) | NT-4 (n = 224) |
|---|---|---|---|---|---|
| Gender | Female | 77 (45.8%) | 75 (39.9%) | 39 (48.1%) | 88 (39.3%) |
| Male | 90 (53.6%) | 113 (60.1%) | 42 (51.9%) | 135 (60.3%) | |
| Unknown | 1 (0.6%) | 0 (0.0%) | 0 (0.0%) | 1 (0.4%) | |
| Age at Diagnosis | Min. | 14 | 24 | 22 | 18 |
| 1st Qu. | 32.5 | 51 | 36 | 31 | |
| Median | 40 | 59 | 46 | 38 | |
| Mean | 42.69 | 58.12 | 45.59 | 41.03 | |
| 3rd Qu. | 53 | 66.25 | 53 | 49 | |
| Max. | 87 | 85 | 75 | 89 |
| Univariate Cox Regression | Multivariate Cox Regression | ||||||
|---|---|---|---|---|---|---|---|
| Covariate | Category | Beta | HR (95% CI for HR) | p-value | Beta | HR (95% CI for HR) | p-value |
| Cluster | NT-1 | Reference | Reference | ||||
| NT-2 | 2.216 | 9.169 (6.167–13.633) | 6.62 × 10−28 | 0.901 | 2.461 (1.533–3.952) | 1.93 × 10−4 | |
| NT-3 | −0.262 | 0.769 (0.398–1.485) | 4.35 × 10−1 | −0.456 | 0.634 (0.327–1.229) | 1.77 × 10−1 | |
| NT-4 | 0.433 | 1.543 (1.020–2.333) | 4.00 × 10−2 | 0.31 | 1.363 (0.894–2.079) | 1.51 × 10−1 | |
| Gender | Female | Reference | Reference | ||||
| Male | 0.202 | 1.224 (0.947–1.582) | 1.22 × 10−1 | 0.029 | 1.029 (0.794–1.334) | 8.29 × 10−1 | |
| Age at Diagnosis | 0.066 | 1.068 (1.058–1.079) | 9.31 × 10−42 | 0.039 | 1.040 (1.028–1.052) | 4.32 × 10−11 | |
| Grade | G2 | Reference | Reference | ||||
| G3 | 1.148 | 3.153 (2.054–4.841) | 1.53 × 10−7 | 0.972 | 2.644 (1.717–4.074) | 1.03 × 10−5 | |
| G4 | 2.976 | 19.605 (12.821–29.978) | 6.38 × 10−43 | 1.772 | 5.883 (3.534–9.791) | 9.30 × 10−12 | |
| Unknown | 1.092 | 2.979 (1.652–5.370) | 2.83 × 10−4 | 0.92 | 2.510 (1.381–4.565) | 2.55 × 10−3 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, H.D.; Diamandis, P.; Scott, M.S.; Richer, M. Deciphering of Adult Glioma Vulnerabilities through Expression Pattern Analysis of GABA, Glutamate and Calcium Neurotransmitter Genes. J. Pers. Med. 2022, 12, 633. https://doi.org/10.3390/jpm12040633
Nguyen HD, Diamandis P, Scott MS, Richer M. Deciphering of Adult Glioma Vulnerabilities through Expression Pattern Analysis of GABA, Glutamate and Calcium Neurotransmitter Genes. Journal of Personalized Medicine. 2022; 12(4):633. https://doi.org/10.3390/jpm12040633
Chicago/Turabian StyleNguyen, Hoang Dong, Phedias Diamandis, Michelle S. Scott, and Maxime Richer. 2022. "Deciphering of Adult Glioma Vulnerabilities through Expression Pattern Analysis of GABA, Glutamate and Calcium Neurotransmitter Genes" Journal of Personalized Medicine 12, no. 4: 633. https://doi.org/10.3390/jpm12040633
APA StyleNguyen, H. D., Diamandis, P., Scott, M. S., & Richer, M. (2022). Deciphering of Adult Glioma Vulnerabilities through Expression Pattern Analysis of GABA, Glutamate and Calcium Neurotransmitter Genes. Journal of Personalized Medicine, 12(4), 633. https://doi.org/10.3390/jpm12040633






