Assessment of Circulating lncRNA H19 in Ankylosing Spondylitis Patients and Its Correlation with Disease Activity
Abstract
1. Introduction
2. Subjects and Methods
2.1. Study Design and Participants
2.2. Clinical Examination
2.3. Radiological Evaluation
2.4. Samples
2.5. Methods
2.6. Follow-Up
2.7. Statistical Analysis
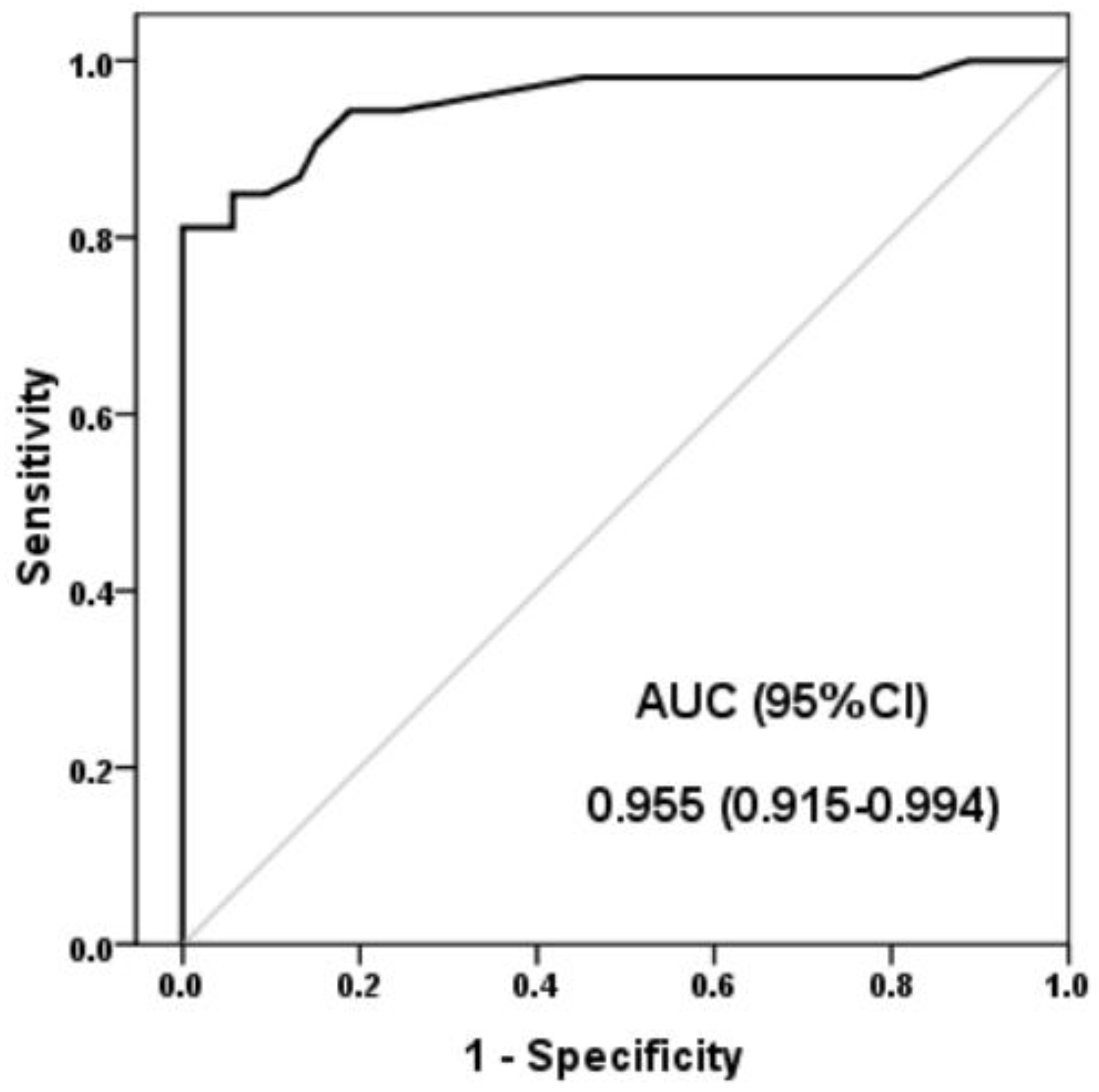
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taurog, J.D.; Chhabra, A.; Colbert, R.A. Ankylosing spondylitis and axial spondyloarthritis. N. Engl. J. Med. 2016, 374, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Montoya, L.; Gul, H.; Emery, P. Recent advances in ankylosing spondylitis: Understanding the disease and management. [version 1; peer review: 2 approved]. F1000Research 2018, 7, 1512. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A. Update on ankylosing spondylitis: Current concepts in pathogenesis. Curr. Allergy Asthma Rep. 2015, 15, 489. [Google Scholar] [CrossRef] [PubMed]
- Hoepken, B.; Serrano, D.; Harris, K.; Hwang, M.C.; Reveille, J. Validation of the Ankylosing Spondylitis Quality of Life Assessment Tool in Patients with Non-Radiographic Axial Spondyloarthritis. Qual. Life Res. 2021, 30, 945–954. [Google Scholar] [CrossRef]
- Zhu, W.; He, X.; Cheng, K.; Zhang, L.; Chen, D.; Wang, X.; Qiu, G.; Cao, X.; Weng, X. Ankylosing Spondylitis: Etiology, Pathogenesis, and Treatments. Bone Res. 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Rudwaleit, M.; van der Heijde, D.; Landewé, R.; Listing, J.; Akkoc, N.; Brandt, J.; Braun, J.; Chou, C.T.; Collantes-Estevez, E.; Dougados, M.; et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): Validation and final selection. Ann. Rheum. Dis. 2019, 68, 777–783. [Google Scholar] [CrossRef]
- Motta, F.; Pederzani, A.; Carena, M.C.; Ceribelli, A.; Wordsworth, P.B.; De Santis, M.; Selmi, C.; Vecellio, M. MicroRNAs in Axial Spondylarthritis: An Overview of the Recent Progresses in the Field with a Focus on Ankylosing Spondylitis and Psoriatic Arthritis. Cur. Rheumatol. Rep. 2021, 23, 59. [Google Scholar] [CrossRef]
- Coates, L.C.; Baraliakos, X.; Blanco, F.J.; Blanco-Morales, E.A.; Braun, J.; Chandran, V.; Fernandez-Sueiro, J.L.; FitzGerald, O.; Gallagher, P.; Gladman, D.D.; et al. The Phenotype of Axial Spondyloarthritis: Is It Dependent on HLA-B27 Status? Arthritis Care Res. 2021, 73, 856–860. [Google Scholar] [CrossRef]
- Doughem, K.; Weisman, M.H.; Ward, M.M.; Gensler, L.S.; Ishimori, M.; Tahanan, A.; Kung, D.C.; Diekman, L.; Lee, M.; Rahbar, M.H.; et al. Chronic back pain in first-degree relatives (FDRs) of patients with ankylosing spondylitis: Predictive value of HLA-B27 and persistence of inflammatory back pain over time. RMD Open 2021, 6, e001418. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Prabhakar, B.; Lee, S.; Bochanis, A.; He, W.; Manautou, J.E.; Rasmussen, T.P. Lnc-RHL, a Novel Long Non-Coding RNA Required for the Differentiation of Hepatocytes from Human Bipotent Progenitor Cells. Cell Prolif. 2021, 54, e12978. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Deng, Y.; Wang, Y.; Sun, X.; Chen, S.; Fu, G. SPAG5-AS1 Inhibited Autophagy and Aggravated Apoptosis of Podocytes via SPAG5/AKT/ mTOR Pathway. Cell Prolif. 2020, 53, e12738. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, B.; Wang, L.; Zhu, Y.; Zhu, X.; Xia, Z.; Zhao, Z.; Xu, L. LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361-3p and HuR to drive cervical cancer progression. Cell Prolif. 2020, 53, e12823. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Shen, J.; Zhang, L.; Chan, M.T.V.; Wu, W.K.K. Emerging Roles of Non-Coding RNAs in Scoliosis. Cell Prolif. 2020, 53, e12736. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Chen, C.; Li, S.; Shen, J.; Tse, G.; Chan, M.T.V.; Wu, W.K.K. Long Non-Coding RNAs in Nucleus Pulposus Cell Function and Intervertebral Disc Degeneration. Cell Prolif. 2018, 51, e12483. [Google Scholar] [CrossRef]
- Zhang, J.; Lei, H.; Li, X. LncRNA SNHG14 Contributes to Proinflammatory Cytokine Production in Rheumatoid Arthritis via the Regulation of the miR 17-5p/MINK1-JNK Pathway. Environ. Toxicol. 2021, 36, 2484–2492. [Google Scholar] [CrossRef]
- Li, Z.; Wong, S.H.; Shen, J.; Chan, M.T.V.; Wu, W.K.K. The Role of MicroRNAS in Ankylosing Spondylitis. Medicine 2016, 95, e3325. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, C.; Jia, Z.; Tong, W.; Liu, D.; He, C.; Huang, X.; Xu, W. Differentially Expressed mRNAs, lncRNAs, and miRNAs With Associated Co-Expression and ceRNA Networks in Ankylosing Spondylitis. Oncotarget 2017, 8, 113543–113557. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, Y.; Yu, Y.; Zhong, C.; Lang, Q.; Liang, Z.; Lv, C.; Xu, F.; Tian, Y. Long Noncoding RNA H19: A Novel Therapeutic Target Emerging in Oncology Via Regulating Oncogenic Signaling Pathways. Front. Cell Dev. Biol. 2021, 9, 796740. [Google Scholar] [CrossRef]
- Zhang, X.U.; Ji, S.; Cai, G.; Pan, Z.; Han, R.; Yuan, Y.; Xu, S.; Yang, J.; Hu, X.; Chen, M.; et al. H19 Increases IL-17a/IL-23 Releases via Regulating VDR by Interacting With Mir675-5p/Mir22-5p in Ankylosing Spondylitis. Mol. Ther. Nucleic Acids 2020, 19, 393–404. [Google Scholar] [CrossRef]
- Salehi-Abari, I. New York Revised Criteria for too Early Diagnosis of Ankylosing Spondylitis (AS). Autoimmune Dis. Ther. Approaches 2016, 3, 126. [Google Scholar] [CrossRef]
- Machado, P.; Landewé, R.; Lie, E.; Kvien, T.K.; Braun, J.; Baker, D.; van der Heijde, D. Ankylosing Spondylitis Disease Activity Score (ASDAS): Defining cut-off values for disease activity states and improvement scores. Ann. Rheum. Dis. 2011, 70, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Machado, P.; Navarro-Compán, V.; Landewé, R.; van Gaalen, F.A.; Roux, C.; van der Heijde, D. Brief Report: Calculating the Ankylosing Spondylitis Disease Activity Score If the Conventional C-Reactive Protein Level Is Below the Limit of Detection or If High-Sensitivity C-Reactive Protein Is Used: An Analysis in the DESIR Cohort. Arthritis Rheumatol. 2015, 67, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Calin, A.; Garrett, S.; Whitelock, H.; Kennedy, L.G.; O′Hea, J.; Mallorie, P.; Jenkinson, T. A new approach to defining functional ability in ankylosing spondylitis: The development of the Bath Ankylosing Spondylitis Functional Index. J. Rheumatol. 1999, 21, 2281–2285. [Google Scholar] [PubMed]
- Garrett, S.; Jenkinson, T.; Kennedy, L.G.; Whitelock, H.; Gaisford, P.; Calin, A. A new approach to defining disease status in ankylosing spondylitis: The Bath Ankylosing Spondylitis Disease Activity Index. J. Rheumatol. 1994, 21, 2286–2291. [Google Scholar] [PubMed]
- Heuft-Dorenbosch, L.; Spoorenberg, A.; van Tubergen, A.; Landewé, R.; van ver Tempel, H.; Mielants, H.; Dougados, M.; van der Heijde, D. Assessment of enthesitis in ankylosing spondylitis. Ann. Rheumatic. Dis. 2003, 62, 127–132. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, G.; Wang, Y.; Yang, J.; Wang, Y.; Zhu, J.; Huang, F. Correlation between magnetic resonance imaging (MRI) findings and the new bone formation factor Dkk-1 in patients with spondyloarthritis. Clin. Rheumatol. 2019, 38, 465–475. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, R.; Wang, X.; Sun, X.; Zhao, B.; Zhang, X.; Gong, X.; Wong, S.H.; Chan, M.T.V.; Wu, W.K.K. Emerging Roles of Long Non-Coding RNAs in Ankylosing Spondylitis. Front. Immunol. 2022, 13, 790924. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Xie, Z.; Li, J.; Wang, P.; Li, Y.; Wu, X.; Wang, S.; Su, H.; Deng, W.; Liu, Z.; Cen, S.; et al. Differential Expression Profiles of Long Noncoding RNA and mRNA of Osteogenically Differentiated Mesenchymal Stem Cells in Ankylosing Spondylitis. J. Rheumatol. 2016, 43, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qu, W.; Yang, X. Comprehensive lncRNA and mRNA profiles in peripheral blood mononuclear cells derived from ankylosing spondylitis patients by RNA-sequencing analysis. Medicine 2022, 101, e27477. [Google Scholar] [CrossRef]
- Abdelrahman, M.H.; Mahdy, S.; Khanjar, I.A.; Siam, A.M.; Malallah, H.A.; Al-Emadi, S.A.; Sarakbi, H.A.; Hammoudeh, M. Prevalence of HLA-B27 in Patients with Ankylosing Spondylitis in Qatar. Int. J. Rheumatol. 2012, 2012, 860213. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.X.; Chung, H.K.; Xiao, L.; Piao, J.J.; Lan, S.; Jaladanki, S.K.; Turner, D.J.; Raufman, J.-P.; Gorospe, M.; Wang, J.-Y. Long Noncoding RNA H19 Impairs the Intestinal Barrier by Suppressing Autophagy and Lowering Paneth and Goblet Cell Function. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-W.; Wang, P.-Y.; Liu, Y.-C.; Sun, L.; Zhu, J.; Zuo, S.; Ma, J.; Li, T.-Y.; Zhang, J.-L.; Chen, G.-W.; et al. Effect of long noncoding RNA H19 overexpression on intestinal barrier function and its potential role in the pathogenesis of ulcerative colitis. Inflamm. Bowel Dis. 2016, 22, 2582–2592. [Google Scholar] [CrossRef]
- Zou, T.; Jaladanki, S.K.; Liu, L.; Xiao, L.; Chung, H.K.; Wang, J.-Y.; Xu, Y.; Gorospe, M. H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Mol. Cell Biol. 2016, 36, 1332–1341. [Google Scholar] [CrossRef]
- Stuhlmüller, B.; Kunisch, E.; Franz, J.; Martinez-Gamboa, L.; Hernandez, M.M.; Pruss, A.; Ulbrich, N.; Erdmann, V.A.; Burmester, G.R.; Kinne, R.W.; et al. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am. J. Pathol. 2003, 163, 901–911. [Google Scholar] [CrossRef]
- Xing, D.; Liang, J.Q.; Li, Y.; Lu, J.; Jia, H.B.; Xu, L.Y.; Ma, X.L. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop. Surg. 2014, 6, 288–293. [Google Scholar] [CrossRef]
- Zhou, L.; Wan, Y.; Cheng, Q.; Shi, B.; Zhang, L.; Chen, S. The Expression and Diagnostic Value of LncRNA H19 in the Blood of Patients with Osteoarthritis. Iran J. Public Health 2020, 49, 1494–1501. [Google Scholar] [CrossRef]
- Chiowchanwisawakit, P.; Lambert, R.G.; Conner-Spady, B.; Maksymowych, W.P. Focal fat lesions at vertebral corners on magnetic resonance imaging predict the development of new syndesmophytes in ankylosing spondylitis. Arthritis Rheum. 2011, 63, 2215–2225. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, Y.; Jia, L.; Li, W. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-b1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells 2015, 33, 3481–3492. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.-C.; Fu, W.-M.; Wang, Y.-B.; Sun, Y.-X.; Xu, L.-L.; Wong, C.-W.; Chan, K.-M.; Li, G.; Waye, M. M-Y.; Zhang, J.-F. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci. Rep. 2016, 6, 20121. [Google Scholar] [CrossRef] [PubMed]
- Uderhardt, S.; Diarra, D.; Katzenbeisser, J.; David, J.P.; Zwerina, J.; Richards, W.; Kronke, G.; Schett, G. Blockade of dickkopf (dkk)-1 induces fusion of sacroiliac joints. Ann. Rheum. Dis. 2010, 69, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Diarra, D.; Stolina, M.; Polzer, K.; Zwerina, J.; Ominsky, M.S.; Dwyer, D.; Korb, A.; Smolen, J.; Hoffmann, M.; Scheinecker, C.; et al. Dickkopf-1 is a master regulator of joint remodeling. Nat. Med. 2007, 13, 156–163. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef]


| Parameters | AS Patients (No.: 53) | Controls (No.: 53) | p | |
|---|---|---|---|---|
| Age | 41 [30–65] | 42 [33–57] | 0.51 | |
| Sex (Male/Female) | 35/18 (66/34) | 33/20 (62.3/37.7) | 0.41 | |
| Smoking | 6 (11.3) | 10 (18.9) | 0.32 | |
| Duration (Years) | 8 [2–20] | |||
| Type: | ||||
| 53 (100) | |||
| 4 (7.5) | |||
| ASDAS-CRP: | 3.3 [1–6.1] | |||
| 8 (15.1) | |||
| 7 (13.2) | |||
| 15 (28.3) 23 (43.4) | |||
| Morning stiffness score | 3 [0–10] | |||
| Patient global assessment score | 5 [0–10] | |||
| MASES | 4 [1–12] | |||
| BASFI | 4.9 [4–6.5] | |||
| Fatigue Score | 3.3 [1–8] | |||
| BASDAI: | 5 [1.5–8] | |||
| 27 (50.9) | |||
| 26 (49.1) | |||
| Associated conditions: | ||||
| 4 (7.5) | |||
| 7 (13.2) | |||
| MRI scores | ||||
| 4.2 [2–8.5] | |||
| 6.5 [2.3–29.5] | |||
| 2.6 [0–9] | |||
| 2.6 [0–10.2] | |||
| Biological Treatment | ||||
| 39 (73.6) | |||
| 14 (26.4) | |||
| Laboratory findings | ||||
| 21 [5–44] | 6.8 [2–16] | <0.001 * | |
| 25.6 [2.1–87.2] | 2.5 [0.9–7] | <0.001 * | |
| 32 (60.4) | 0 (0) | <0.001 * | |
| Parameters of PsA | LncRNA H19 | ||
|---|---|---|---|
| rs | P | ||
| Age | 0.12 | 0.36 | |
| Duration | 0.29 | 0.033 * | |
| ASDAS-CRP | 0.4 | 0.003 * | |
| Morning stiffness score | 0.41 | 0.002 * | |
| Patient global assessment score | 0.33 | 0.017 * | |
| MASES | 0.37 | 0.006 * | |
| BASFI | 0.34 | 0.014 * | |
| BASDAI | 0.57 | <0.001 * | |
| Fatigue Score | 0.33 | 0.015 * | |
| MRI Finding | |||
| 0.34 | 0.012 * | |
| 0.24 | 0.85 | |
| 0.32 | 0.018 * | |
| 0.36 | 0.009 * | |
| ESR | 0.63 | <0.001 * | |
| CRP | 0.59 | <0.001 * | |
| Covariate | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| OR (95%CI) | p-Value | AOR (95%CI) | p-Value | |
| Age | 1.02 (0.97–1.07) | 0.41 | ||
| Sex (Male) | 1.17 (0.53–2.6) | 0.69 | ||
| Smoking | 0.55 (0.18–1.64) | 0.28 | ||
| HLA-B27 | 18.6 (5.8–59.4) | <0.001 * | 0.43 (0–364) | 0.81 |
| CRP | 1.83 (1.36–2.44) | <0.001 * | 1.82 (1.18–2.8) | 0.03 * |
| ESR | 1.32 (1.19–1.47) | 0.001 * | 1.28 (0.95–1.73) | 0.09 |
| LncRNA H19 | 688 (34–1380) | 0.004 * | 211 (4.7–939) | 0.025 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esawy, M.M.; Ebaid, A.M.; Abd-elhameed, A.; Thagfan, F.A.; Mubaraki, M.A.; Alazzouni, A.S.; Dkhil, M.A.; Shabana, M.A. Assessment of Circulating lncRNA H19 in Ankylosing Spondylitis Patients and Its Correlation with Disease Activity. J. Pers. Med. 2023, 13, 914. https://doi.org/10.3390/jpm13060914
Esawy MM, Ebaid AM, Abd-elhameed A, Thagfan FA, Mubaraki MA, Alazzouni AS, Dkhil MA, Shabana MA. Assessment of Circulating lncRNA H19 in Ankylosing Spondylitis Patients and Its Correlation with Disease Activity. Journal of Personalized Medicine. 2023; 13(6):914. https://doi.org/10.3390/jpm13060914
Chicago/Turabian StyleEsawy, Marwa M., Amany M. Ebaid, Amir Abd-elhameed, Felwa A. Thagfan, Murad A. Mubaraki, Ahmed S. Alazzouni, Mohamed A. Dkhil, and Marwa A. Shabana. 2023. "Assessment of Circulating lncRNA H19 in Ankylosing Spondylitis Patients and Its Correlation with Disease Activity" Journal of Personalized Medicine 13, no. 6: 914. https://doi.org/10.3390/jpm13060914
APA StyleEsawy, M. M., Ebaid, A. M., Abd-elhameed, A., Thagfan, F. A., Mubaraki, M. A., Alazzouni, A. S., Dkhil, M. A., & Shabana, M. A. (2023). Assessment of Circulating lncRNA H19 in Ankylosing Spondylitis Patients and Its Correlation with Disease Activity. Journal of Personalized Medicine, 13(6), 914. https://doi.org/10.3390/jpm13060914







